Influence of Single Nucleotide Polymorphisms on Rifampin Pharmacokinetics in Tuberculosis Patients
Abstract
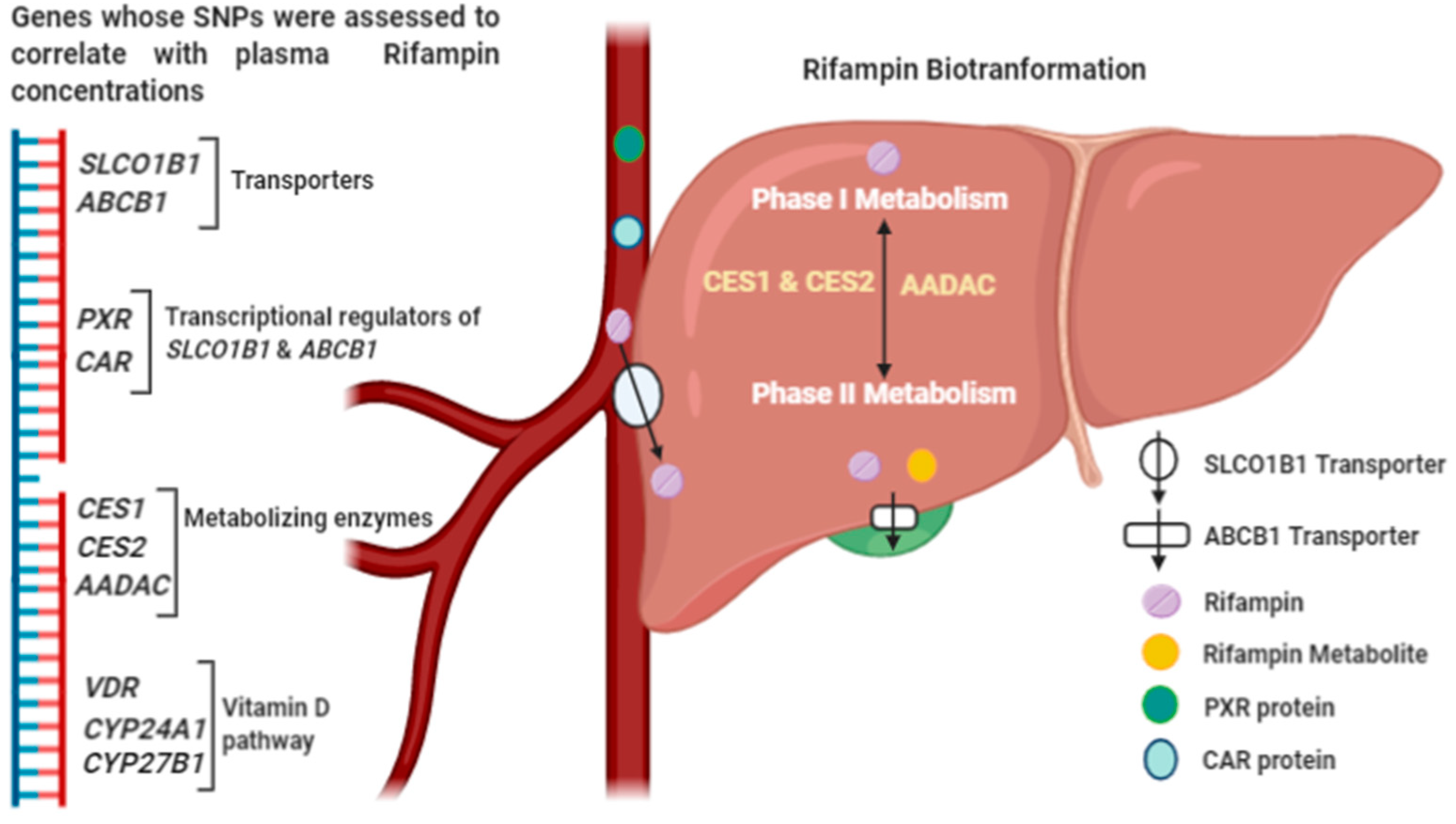
1. Introduction
2. SLCO1B1
3. ABCB1
4. PXR and CAR
5. CES1 and CES2
6. AADAC
7. Vitamin D Pathway Gene Polymorphisms
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Murray, J.F.; Schraufnagel, D.E.; Hopewell, P.C. Treatment of tuberculosis. A historical perspective. Ann. Am. Thorac. Soc. 2015, 12, 1749–1759. [Google Scholar] [CrossRef] [PubMed]
- Maggi, N.; Pasqualucci, C.; Ballotta, R.; Sensi, P. Rifampicin: A new orally active rifamycin. Chemotherapy 1966, 11, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Gumbo, T.; Louie, A.; DeZiel, M.R.; Liu, W.; Parsons, L.M.; Salfinger, M.; Drusano, G.L. Concentration-dependent mycobacterium tuberculosis killing and prevention of resistance by rifampin. Antimicrob. Agents Chemother. 2007, 51, 3781–3788. [Google Scholar] [CrossRef] [PubMed]
- Alsultan, A.; Peloquin, C.A. Therapeutic drug monitoring in the treatment of tuberculosis: An update. Drugs 2014, 74, 839–854. [Google Scholar] [CrossRef] [PubMed]
- McClure, W.R.; Cech, C.L. On the mechanism of rifampicin inhibition of RNA synthesis. J. Biol. Chem. 1978, 253, 8949–8956. [Google Scholar]
- Rastogi, N.; David, H. Mode of action of antituberculous drugs and mechanisms of drug resistance in Mycobacterium tuberculosis. Res. Microbiol. 1993, 144, 133–143. [Google Scholar] [CrossRef]
- Telenti, A.; Imboden, P.; Marchesi, F.; Matter, L.; Schöpfer, K.; Bodmer, T.; Lowrie, D.; Colston, M.; Cole, S. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 1993, 341, 647–651. [Google Scholar] [CrossRef]
- Zaw, M.T.; Emran, N.A.; Lin, Z. Mutations inside rifampicin-resistance determining region of rpoB gene associated with rifampicin-resistance in Mycobacterium tuberculosis. J. Infect. Public Health 2018, 11, 605–610. [Google Scholar] [CrossRef]
- Seijger, C.; Hoefsloot, W.; Guchteneire, I.B.-D.; Brake, L.T.; Van Ingen, J.; Kuipers, S.; Van Crevel, R.; Aarnoutse, R.; Boeree, M.; Magis-Escurra, C.; et al. High-dose rifampicin in tuberculosis: Experiences from a Dutch tuberculosis centre. PLoS ONE 2019, 14, e0213718. [Google Scholar] [CrossRef]
- Svensson, E.M.; Svensson, R.J.; Brake, L.H.M.T.; Boeree, M.J.; Heinrich, N.; Konsten, S.; Churchyard, G.; Dawson, R.; Diacon, A.H.; Kibiki, G.S.; et al. The potential for treatment shortening with higher rifampicin doses: Relating drug exposure to treatment response in patients with pulmonary tuberculosis. Clin. Infect. Dis. 2018, 67, 34–41. [Google Scholar] [CrossRef]
- Stott, K.; Pertinez, H.; Sturkenboom, M.; Boeree, M.J.; Aarnoutse, R.; Ramachandran, G.; Requena-Méndez, A.; Peloquin, C.; Koegelenberg, C.F.N.; Alffenaar, J.W.C.; et al. Pharmacokinetics of rifampicin in adult TB patients and healthy volunteers: A systematic review and meta-analysis. J. Antimicrob. Chemother. 2018, 73, 2305–2313. [Google Scholar] [CrossRef]
- Wang, D.G.; Fan, J.B.; Siao, C.J.; Berno, A.; Young, P.; Sapolsky, R.; Ghandour, G.; Perkins, N.; Winchester, E.; Spencer, J.; et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 1998, 280, 1077–1082. [Google Scholar] [CrossRef]
- Acocella, G. Clinical pharmacokinetics of rifampicin. Clin. Pharmacokinet. 1978, 3, 108–127. [Google Scholar] [CrossRef]
- Liederer, B.M.; Borchardt, R.T. Enzymes involved in the bioconversion of ester-based prodrugs. J. Pharm. Sci. 2006, 95, 1177–1195. [Google Scholar] [CrossRef]
- Nakajima, A.; Fukami, T.; Kobayashi, Y.; Watanabe, A.; Nakajima, M.; Yokoi, T. Human arylacetamide deacetylase is responsible for deacetylation of rifamycins: Rifampicin, rifabutin, and rifapentine. Biochem. Pharmacol. 2011, 82, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Giacomini, K.M.; Huang, S.M.; Tweedie, D.J.; Benet, L.Z.; Brouwer, K.L.; Chu, X.; Dahlin, A.; Evers, R.; Fischer, V.; Hillgren, K.M.; et al. Membrane transporters in drug development. Nat. Rev. Drug Discov. 2010, 9, 215–236. [Google Scholar] [CrossRef] [PubMed]
- Hillgren, K.M.; Keppler, D.; A Zur, A.; Giacomini, K.M.; Stieger, B.; Cass, C.E.; Zhang, L. Emerging transporters of clinical importance: An update from the international transporter consortium. Clin. Pharmacol. Ther. 2013, 94, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xie, W. Pregnane X receptor and constitutive androstane receptor at the crossroads of drug metabolism and energy metabolism. Drug Metab. Dispos. 2010, 38, 2091–2095. [Google Scholar] [CrossRef]
- Hagenbuch, B.; Meier, P.J. Organic anion transporting polypeptides of the OATP/ SLC21 family: Phylogenetic classification as OATP/ SLCO superfamily, new nomenclature and molecular/functional properties. Pflüger Arch. 2004, 447, 653–665. [Google Scholar] [CrossRef]
- Mills, J.B.; Rose, K.A.; Sadagopan, N.; Sahi, J.; De Morais, S.M.F. Induction of drug metabolism enzymes and MDR1 using a novel human hepatocyte cell line. J. Pharmacol. Exp. Ther. 2004, 309, 303–309. [Google Scholar] [CrossRef]
- Wang, X.; Sykes, D.B.; Miller, D.S. Constitutive androstane receptor-mediated up-regulation of ATP-driven xenobiotic efflux transporters at the blood-brain barrier. Mol. Pharmacol. 2010, 78, 376–383. [Google Scholar] [CrossRef]
- Guo, Y.X.; Xu, X.F.; Zhang, Q.Z.; Li, C.; Deng, Y.; Jiang, P.; He, L.Y.; Peng, W.X. The inhibition of hepatic bile acids transporters Ntcp and Bsep is involved in the pathogenesis of isoniazid/rifampicin-induced hepatotoxicity. Toxicol. Mech. Methods 2015, 25, 1–6. [Google Scholar] [CrossRef]
- Brake, L.H.T.; Russel, F.G.; Heuvel, J.J.V.D.; De Knegt, G.J.; De Steenwinkel, J.; Burger, D.M.; Aarnoutse, R.E.; Koenderink, J.B. Inhibitory potential of tuberculosis drugs on ATP-binding cassette drug transporters. Tuberculosis 2016, 96, 150–157. [Google Scholar] [CrossRef]
- Shugarts, S.; Benet, L.Z. The role of transporters in the pharmacokinetics of orally administered drugs. Pharm. Res. 2009, 26, 2039–2054. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Kesterson, R.A.; Yamamoto, H.; Taketani, Y.; Nishiwaki, E.; Tatsumi, S.; Inoue, Y.; Morita, K.; Takeda, E.; Pike, J.W.; et al. Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Mol. Endocrinol. 1997, 11, 1165–1179. [Google Scholar] [CrossRef] [PubMed]
- Koivisto, O.; Hanel, A.; Carlberg, C. Key Vitamin D target genes with functions in the immune system. Nutrients 2020, 12, 1140. [Google Scholar] [CrossRef] [PubMed]
- Caraba, A.; Crişan, V.; Romoşan, I.; Mozos, I.; Murariu, M.-S. Vitamin D status, disease activity, and endothelial dysfunction in early rheumatoid arthritis patients. Dis. Markers 2017, 2017, 1–7. [Google Scholar] [CrossRef]
- Gao, L.; Tao, Y.; Zhang, L.; Jin, Q. Vitamin D receptor genetic polymorphisms and tuberculosis: Updated systematic review and meta-analysis. Int. J. Tuberc. Lung Dis. 2010, 14, 15–23. [Google Scholar]
- Saeki, M.; Kurose, K.; Tohkin, M.; Hasegawa, R. Identification of the functional vitamin D response elements in the human MDR1 gene. Biochem. Pharmacol. 2008, 76, 531–542. [Google Scholar] [CrossRef]
- Wang, Z.; Lin, Y.S.; Zheng, X.E.; Senn, T.; Hashizume, T.; Scian, M.; Dickmann, L.J.; Nelson, S.D.; Baillie, T.A.; Hebert, M.F.; et al. An inducible cytochrome P450 3A4-dependent vitamin D catabolic pathway. Mol. Pharmacol. 2011, 81, 498–509. [Google Scholar] [CrossRef]
- Xu, Y.; Hashizume, T.; Shuhart, M.C.; Davis, C.L.; Nelson, W.L.; Sakaki, T.; Kalhorn, T.F.; Watkins, P.B.; Schuetz, E.G.; Thummel, K.E.; et al. Intestinal and hepatic CYP3A4 catalyze hydroxylation of 1α,25-dihydroxyvitamin D3: Implications for drug-induced osteomalacia. Mol. Pharmacol. 2005, 69, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Niemi, M. Role of OATP transporters in the disposition of drugs. Pharmacogenomics 2007, 8, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Vavricka, S.; Van Montfoort, J.; Ha, H.R.; Meier, P.J.; Fattinger, K. Interactions of rifamycin SV and rifampicin with organic anion uptake systems of human liver. Hepatology 2002, 36, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Treiber, A.; Schneiter, R.; Häusler, S.; Stieger, B. Bosentan is a substrate of human OATP1B1 and OATP1B3: Inhibition of hepatic uptake as the common mechanism of its interactions with Cyclosporin A, rifampicin, and sildenafil. Drug Metab. Dispos. 2007, 35, 1400–1407. [Google Scholar] [CrossRef] [PubMed]
- Oshiro, C.; Mangravite, L.M.; Klein, T.; Altman, R. PharmGKB very important pharmacogene: SLCO1B1. Pharm. Genom. 2010, 20, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Niemi, M.; Pasanen, M.K.; Neuvonen, P.J. Organic anion transporting polypeptide 1B1: A genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol. Rev. 2011, 63, 157–181. [Google Scholar] [CrossRef]
- Pasanen, M.K.; Backman, J.T.; Neuvonen, P.J.; Niemi, M. Frequencies of single nucleotide polymorphisms and haplotypes of organic anion transporting polypeptide 1B1 SLCO1B1 gene in a Finnish population. Eur. J. Clin. Pharmacol. 2006, 62, 409–415. [Google Scholar] [CrossRef]
- Allegra, S.; Fatiguso, G.; Calcagno, A.; Baietto, L.; Motta, I.; Favata, F.; Cusato, J.; Bonora, S.; Di Perri, G.; D’Avolio, A.; et al. Role of vitamin D pathway gene polymorphisms on rifampicin plasma and intracellular pharmacokinetics. Pharmacogenomics 2017, 18, 865–880. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.C.; Soares, R.A.; Nascimento, R.M.; Machado-Coelho, G.L.; Mill, J.G.; Krieger, J.E.; Pereira, A.D.C. SLCO1B1 rs4149056 polymorphism associated with statin-induced myopathy is differently distributed according to ethnicity in the Brazilian general population: Amerindians as a high risk ethnic group. BMC Med. Genet. 2011, 12, 136. [Google Scholar] [CrossRef] [PubMed]
- Mwinyi, J.; Köpke, K.; Schaefer, M.; Roots, I.; Gerloff, T. Comparison of SLCO1B1 sequence variability among German, Turkish, and African populations. Eur. J. Clin. Pharmacol. 2008, 64, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Link, E.; Parish, S.; Armitage, J.; Bowman, L.; Heath, S.; Matsuda, F.; Gut, I.; Lathrop, M.; Collins, R. SLCO1B1Variants and statin-induced myopathy—A genomewide study. N. Engl. J. Med. 2008, 359, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Ho, R.H. Interindividual and interethnic variability in drug disposition: Polymorphisms in organic anion transporting polypeptide 1B1 (OATP1B1;SLCO1B1). Br. J. Clin. Pharmacol. 2017, 83, 1176–1184. [Google Scholar] [CrossRef]
- Dompreh, A.; Tang, X.; Zhou, J.; Yang, H.; Topletz, A.; Ahwireng, E.A.; Antwi, S.; Enimil, A.; Langaee, T.; Peloquin, C.A.; et al. Effect of genetic variation of NAT2 on isoniazid and SLCO1B1 and CES2 on rifampin pharmacokinetics in ghanaian children with tuberculosis. Antimicrob. Agents Chemother. 2017, 62. [Google Scholar] [CrossRef]
- Prado, Y.; Saavedra, N.; Zambrano, T.; Lagos, J.; Rosales, A.; Salazar, L. SLCO1B1 c.388A>G Polymorphism is associated with HDL-C levels in response to atorvastatin in chilean individuals. Int. J. Mol. Sci. 2015, 16, 20609–20619. [Google Scholar] [CrossRef]
- Grapci, A.D.; Dimovski, A.J.; Kapedanovska, A.; Vavlukis, M.; Eftimov, A.; Matevska-Geshkovska, N.; Labachevski, N.; Jakjovski, K.; Gorani, D.; Kedev, S.; et al. Frequencies of single-nucleotide polymorphisms and haplotypes of the SLCO1B1 gene in selected populations of the western balkans. Balk. J. Med. Genet. 2015, 18, 5–21. [Google Scholar] [CrossRef]
- Chigutsa, E.; Visser, M.E.; Swart, E.C.; Denti, P.; Pushpakom, S.; Egan, D.; Holford, N.H.; Smith, P.J.; Maartens, G.; Owen, A.; et al. TheSLCO1B1rs4149032 polymorphism is highly prevalent in South Africans and is associated with reduced rifampin concentrations: Dosing implications. Antimicrob. Agents Chemother. 2011, 55, 4122–4127. [Google Scholar] [CrossRef] [PubMed]
- Gengiah, T.; Botha, J.H.; Soowamber, D.; Naidoo, K.; Karim, S.S.A. Low rifampicin concentrations in tuberculosis patients with HIV infection. J. Infect. Dev. Ctries. 2014, 8, 987–993. [Google Scholar] [CrossRef]
- Mukonzo, J.K.; Kengo, A.; Kutesa, B.; Nanzigu, S.; Pohanka, A.; McHugh, T.D.; Zumla, A.; Aklillu, E. Role of pharmacogenetics in rifampicin pharmacokinetics and the potential effect on TB–rifampicin sensitivity among Ugandan patients. Trans. R. Soc. Trop. Med. Hyg. 2019, 114, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Weiner, M.; Peloquin, C.; Burman, W.; Luo, C.-C.; Engle, M.; Prihoda, T.J.; Mac Kenzie, W.R.; Bliven-Sizemore, E.; Johnson, J.L.; Vernon, A. Effects of tuberculosis, race, and human gene SLCO1B1 polymorphisms on rifampin concentrations. Antimicrob. Agents Chemother. 2010, 54, 4192–4200. [Google Scholar] [CrossRef] [PubMed]
- Dudenkov, T.M.; Ingle, J.N.; Buzdar, A.U.; Robson, M.E.; Kubo, M.; Ibrahim-Zada, I.; Batzler, A.; Jenkins, G.D.; Pietrzak, T.L.; Carlson, E.E.; et al. SLCO1B1 polymorphisms and plasma estrone conjugates in postmenopausal women with ER+ breast cancer: Genome-wide association studies of the estrone pathway. Breast Cancer Res. Treat. 2017, 164, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, A.; Chirehwa, M.T.; Ramsuran, V.; McIlleron, H.; Naidoo, K.; Yende-Zuma, N.; Singh, R.; Ngcapu, S.; Adamson, J.; Govender, K.; et al. Effects of genetic variability on rifampicin and isoniazid pharmacokinetics in South African patients with recurrent tuberculosis. Pharmacogenomics 2019, 20, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Calcagno, A.; Cusato, J.; Sekaggya-Wiltshire, C.; Von Braun, A.; Motta, I.; Turyasingura, G.; Castelnuovo, B.; Fehr, J.; Di Perri, G.; Lamorde, M.; et al. The influence of pharmacogenetic variants in HIV/Tuberculosis coinfected patients in uganda in the SOUTH study. Clin. Pharmacol. Ther. 2019, 106, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, K.; Kumar, A.K.H.; Kannan, T.; Vijayalakshmi, R.; Sudha, V.; Nesakumar, S.M.; Bharathiraja, T.; Lavanya, J.; Swaminathan, S.; Ramachandran, G.; et al. SLCO1B1 gene polymorphisms do not influence plasma rifampicin concentrations in a South Indian population. Int. J. Tuberc. Lung Dis. 2016, 20, 1231–1235. [Google Scholar] [CrossRef] [PubMed]
- Jeremiah, K.; Denti, P.; Chigutsa, E.; Faurholt-Jepsen, D.; PrayGod, G.; Range, N.; Castel, S.; Wiesner, L.; Hagen, C.M.; Christiansen, M.; et al. Nutritional supplementation increases rifampin exposure among tuberculosis patients coinfected with HIV. Antimicrob. Agents Chemother. 2014, 58, 3468–3474. [Google Scholar] [CrossRef]
- Sloan, D.; McCallum, A.D.; Schipani, A.; Egan, D.; Mwandumba, H.C.; Ward, S.; Waterhouse, D.; Banda, G.; Allain, T.J.; Owen, A.; et al. Genetic determinants of the pharmacokinetic variability of rifampin in malawian adults with pulmonary tuberculosis. Antimicrob. Agents Chemother. 2017, 61, e00210-17. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.M.; George, A.M. The ABC transporter structure and mechanism: Perspectives on recent research. Cell. Mol. Life Sci. 2004, 61, 682–699. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Hrycyna, C.; Schoenlein, P.V.; Germann, U.; Pastan, I. Genetic analysis of the multidrug transporter. Annu. Rev. Genet. 1995, 29, 607–649. [Google Scholar] [CrossRef]
- Wolking, S.; Schaeffeler, E.; Lerche, H.; Schwab, M.; Nies, A.T. Impact of genetic polymorphisms of ABCB1 (MDR1, P-Glycoprotein) on drug disposition and potential clinical implications: Update of the literature. Clin. Pharmacokinet. 2015, 54, 709–735. [Google Scholar] [CrossRef]
- Schuetz, E.G.; Schinkel, A.H.; Relling, M.V.; Schuetz, J.D. P-glycoprotein: A major determinant of rifampicin-inducible expression of cytochrome P4503A in mice and humans. Proc. Natl. Acad. Sci. USA 1996, 93, 4001–4005. [Google Scholar] [CrossRef]
- Hodges, L.M.; Markova, S.M.; Chinn, L.W.; Gow, J.M.; Kroetz, D.L.; Klein, T.E.; Altman, R.B. Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein). Pharm. Genom. 2011, 21, 152–161. [Google Scholar] [CrossRef]
- Fung, K.L.; Gottesman, M.M. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim. Biophys. Acta 2009, 1794, 860–871. [Google Scholar] [CrossRef]
- Moore, D.D.; Kato, S.; Xie, W.; Mangelsdorf, D.J.; Schmidt, D.R.; Xiao, R.; Kliewer, S.A. International union of pharmacology. LXII. The NR1H and NR1I receptors: Constitutive androstane receptor, pregnene X receptor, farnesoid X receptor α, farnesoid X receptor β, liver X receptor α, liver X receptor β, and vitamin D receptor. Pharmacol. Rev. 2006, 58, 742–759. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tang, Y.; Guo, C.; Wang, J.; Boral, D.; Nie, D. Nuclear receptors in the multidrug resistance through the regulation of drug-metabolizing enzymes and drug transporters. Biochem. Pharmacol. 2012, 83, 1112–1126. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kuehl, P.; Green, E.D.; Touchman, J.W.; Watkins, P.B.; Daly, A.K.; Hall, S.D.; Maurel, P.; Relling, M.; Brimer, C.; et al. The human pregnane X receptor: Genomic structure and identification and functional characterization of natural allelic variants. Pharmacogenetics 2001, 11, 555–572. [Google Scholar] [CrossRef] [PubMed]
- Lamba, J.; Lamba, V.; Strom, S.; Venkataramanan, R.; Schuetz, E. Novel single nucleotide polymorphisms in the promoter and intron 1 of human pregnane X receptor/NR1I2 and their association with CYP3A4 expression. Drug Metab. Dispos. 2007, 36, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Moreira, R.; Jorge, A.A.L.; Mendonca, B.B.; Bachega, T.A. Frequency of genetic polymorphisms of PXR gene in the Brazilian population. Clinics (Sao Paulo) 2011, 66, 1041–1044. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, S.S. Alternatively spliced isoforms of the human constitutive androstane receptor. Nucleic Acids Res. 2003, 31, 3194–3207. [Google Scholar] [CrossRef]
- Swart, M.; Whitehorn, H.; Ren, Y.; Smith, P.J.; Ramesar, R.; Dandara, C. PXR and CAR single nucleotide polymorphisms influence plasma efavirenz levels in South African HIV/AIDS patients. BMC Med. Genet. 2012, 13, 112. [Google Scholar] [CrossRef]
- Jinno, H.; Tanaka-Kagawa, T.; Hanioka, N.; Ishida, S.; Saeki, M.; Soyama, A.; Itoda, M.; Nishimura, T.; Saito, Y.; Ozawa, S.; et al. Identification of novel alternative splice variants of human constitutive androstane receptor and characterization of their expression in the liver. Mol. Pharmacol. 2004, 65, 496–502. [Google Scholar] [CrossRef]
- Jamis-Dow, C.A.; Katki, A.G.; Collins, J.M.; Klecker, R.W. Rifampin and rifabutin and their metabolism by human liver esterases. Xenobiotica 1997, 27, 1015–1024. [Google Scholar] [CrossRef]
- Ross, M.K.; Crow, J.A. Human carboxylesterases and their role in xenobiotic and endobiotic metabolism. J. Biochem. Mol. Toxicol. 2007, 21, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zou, L.; Jin, Q.; Hou, J.; Ge, G.-B.; Yang, L. Human carboxylesterases: A comprehensive review. Acta Pharm. Sin. B 2018, 8, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Merali, Z.; Ross, S.; Paré, G. The pharmacogenetics of carboxylesterases: CES1 and CES2 genetic variants and their clinical effect. Drug Metab. Drug Interact. 2014, 29, 143–151. [Google Scholar] [CrossRef]
- Song, S.H.; Chang, H.E.; Jun, S.H.; Park, K.U.; Lee, J.H. Relationship between CES2 genetic variations and rifampicin metabolism. J. Antimicrob. Chemother. 2013, 68, 1281–1284. [Google Scholar] [CrossRef]
- Fukami, T.; Yokoi, T. The emerging role of human esterases. Drug Metab. Pharmacokinet. 2012, 27, 466–477. [Google Scholar] [CrossRef]
- Andraos, C.; Koorsen, G.; Knight, J.C.; Bornman, L. Vitamin D receptor gene methylation is associated with ethnicity, tuberculosis, and TaqI polymorphism. Hum. Immunol. 2010, 72, 262–268. [Google Scholar] [CrossRef]
- Zmuda, J.M.; Cauley, J.A.; E Ferrell, R. Molecular epidemiology of vitamin D receptor gene variants. Epidemiol. Rev. 2000, 22, 203–217. [Google Scholar] [CrossRef]
- Jurutka, P.W.; Remus, L.S.; Whitfield, G.K.; Thompson, P.D.; Hsieh, J.C.; Zitzer, H.; Tavakkoli, P.; Galligan, M.A.; Dang, H.T.; Haussler, C.A.; et al. The polymorphic N terminus in human vitamin D receptor isoforms influences transcriptional activity by modulating interaction with transcription factor IIB. Mol. Endocrinol. 2000, 14, 401–420. [Google Scholar] [CrossRef]
- Shaik, A.P.; Alsaeed, A.H.; Alsaeed, M.A.; Alyousef, A.; Bammidi, V.K.; Sultana, A. Vitamin D Receptor FokI, ApaI and TaqI polymorphisms in lead exposed subjects from Saudi Arabia. Front. Genet. 2019, 10, 388. [Google Scholar] [CrossRef]
- Vieira, L.A.; De Marchi, P.L.; Dos Santos, A.A.; Christofolini, D.M.; Barbosa, C.P.; Fonseca, F.L.A.; Bianco, B.; Rodrigues, L.M.R. Analysis of FokI polymorphism of vitamin D Receptor gene in intervertebral disc degeneration. Genet. Test. Mol. Biomark. 2014, 18, 625–629. [Google Scholar] [CrossRef]
- DeLuca, H.F. Evolution of our understanding of vitamin D. Nutr. Rev. 2008, 66, S73–S87. [Google Scholar] [CrossRef]
- Sakaki, T.; Sawada, N.; Komai, K.; Shiozawa, S.; Yamada, S.; Yamamoto, K.; Ohyama, Y.; Inouye, K. Dual metabolic pathway of 25-hydroxyvitamin D3 catalyzed by human CYP24. J. Biol. Inorg. Chem. 2000, 267, 6158–6165. [Google Scholar] [CrossRef] [PubMed]
- Penna-Martinez, M.; Ramos-Lopez, E.; Stern, J.; Kahles, H.; Hinsch, N.; Hansmann, M.-L.; Selkinski, I.; Grünwald, F.; Vorländer, C.; Bechstein, W.O.; et al. Impaired vitamin D activation and association with CYP24A1 haplotypes in differentiated thyroid carcinoma. Thyroid 2012, 22, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Bailey, R.; Cooper, J.D.; Zeitels, L.; Smyth, D.; Yang, J.H.; Walker, N.; Hyppönen, E.; Dunger, D.; Ramos-Lopez, E.; Badenhoop, K.; et al. Association of the vitamin D metabolism gene CYP27B1 with type 1 diabetes. Diabetes 2007, 56, 2616–2621. [Google Scholar] [CrossRef] [PubMed]
- Rinkwitz, S.; Geng, F.; Manning, E.; Suster, M.; Kawakami, K.; Becker, T.S.T.S. BAC transgenic zebrafish reveal hypothalamic enhancer activity around obesity associated SNP rs9939609 within the humanFTOgene. Genesis 2015, 53, 640–651. [Google Scholar] [CrossRef]
- Seo, S.; Takayama, K.; Uno, K.; Ohi, K.; Hashimoto, R.; Nishizawa, D.; Ikeda, K.; Ozaki, N.; Nabeshima, T.; Miyamoto, Y.; et al. Functional analysis of deep intronic SNP rs13438494 in intron 24 of PCLO gene. PLoS ONE 2013, 8, e76960. [Google Scholar] [CrossRef]
- Rose, K.; Penna-Martinez, M.; Klahold, E.; Karger, D.; Shoghi, F.; Kahles, H.; Bayer, M.; Hintermann, E.; Pfeilschifter, J.M.; Badenhoop, K.; et al. Influence of the vitamin D plasma level and vitamin D-related genetic polymorphisms on the immune status of patients with type 1 diabetes: A pilot study. Clin. Exp. Immunol. 2013, 171, 171–185. [Google Scholar] [CrossRef]
- Jin, C.H.; Pike, J.W. Human vitamin D receptor-dependent transactivation in Saccharomyces cerevisiae requires retinoid X receptor. Mol. Endocrinol. 1996, 10, 196–205. [Google Scholar]
- Väisänen, S.; Dunlop, T.W.; Sinkkonen, L.; Frank, C.; Carlberg, C. Spatio-temporal activation of chromatin on the human CYP24 gene promoter in the presence of 1α,25-dihydroxyvitamin D3. J. Mol. Biol. 2005, 350, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Roff, A.; Wilson, R.T. A novel SNP in a vitamin D response element of the CYP24A1 promoter reduces protein binding, transactivation, and gene expression. J. Steroid Biochem. Mol. Biol. 2008, 112, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Hibler, E.; Klimentidis, Y.C.; Jurutka, P.; Kohler, L.N.; Lance, P.; Roe, D.J.; Thompson, P.A.; Jacobs, E.T. CYP24A1 and CYP27B1 polymorphisms, concentrations of vitamin D metabolites, and odds of colorectal adenoma recurrence. Nutr. Cancer 2015, 67, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
| Sl No. | Author, Year | Population | SNP ID | Criteria of Sample Collection | Time of Sample Collection | RF Concentration in Plasma |
|---|---|---|---|---|---|---|
| 1 | Mukonzo et al., 2020 [48] | 50 TB patients from Uganda | rs4149056 | After 21 days of ATT initiation | Predose, 1, 2, 4, 6 and 12 h postdose | No change |
| rs2306283 | No change | |||||
| rs4149032 | No change | |||||
| 2 | Naidoo et al., 2019 [51] | 172 recurrent TB patients in South Africa | rs2306283 | 1 and/or 2 months and at 6 months during ATT | Predose, 2.5, 6 and 24 h postdose | No change |
| rs4149032 | No change | |||||
| rs4149056 | No change | |||||
| rs4149015 | No change | |||||
| 3 | Calcagno et al., 2019 [52] | 221 PTB with HIV patients in Uganda | rs4149032 | At 2nd, 4th and 8th week of ATT | 1, 2 and 4 h postdose | No change |
| 4 | Dompreh et al., 2018 [43] | 113 pediatric TB patients in Ghana | rs2306283 | After 4 weeks of ATT | Predose, 1, 2, 4 and 8 h postdose | Decreased |
| rs11045819 | No change | |||||
| rs4149056 | No change | |||||
| rs4149032 | No change | |||||
| 5 | Allegra et al., 2017 [38] | 24 TB patients in Italy | rs4149056 | At 2nd week and 4th week of ATT | Plasma Cmax (end of 3 infusions for IV route and 2 h postdose for oral) and Ctrough | Increased |
| 6 | Sloan et al., 2017 [55] | 174 adult PTB patients in Malawi | rs11045819 | Day 14 or 21 of ATT | Predose, 2 and 6 h postdose | No change |
| rs4149032 | No change | |||||
| 7 | Ramesh et al., 2016 [53] | 256 South Indian adult PTB/EPTB patients | rs11045819 | After a minimum of 2 weeks of ATT | 2 h postdose | No change |
| rs4149032 | No change | |||||
| rs4149033 | No change | |||||
| 8 | Jeremiah et al., 2014 [54] | PTB patients in Tanzania | rs4149032 | 1st occasion: 7 ± 2 days after ATT 2nd occasion: Around 56 days after ATT | 2, 4 and 6 h postdose | No change |
| 9 | Gengiah et al., 2014 [47] | 57 TB with HIV patients in South Africa | rs4149032 | At 4th, 8th and 12th week of TB treatment | 2.5 h postdose | Decreased |
| 10 | Chigutsa et al., 2011 [46] | 60 PTB patients in South Africa | rs4149032 | At least 1 month after the start of ATT | 4 to 8 samples per patient, randomly collected over a 7 h period | Decreased |
| rs4149056 | No change | |||||
| rs11045819 | No change | |||||
| 11 | Weiner et al., 2010 [49] | 72 TB Patients (37 from Africa and 35 from the United States and Spain) | rs4149015 | Between the 9th and 40th doses in TB patients | Just before dose and 1, 2, 6, 8 to 10, 11 to 13 and 23 to 25 h after dose | No change |
| rs2306283 | No change | |||||
| rs11045819 | Decreased | |||||
| rs4149056 | No change | |||||
| rs59502379 | No change |
| Sl No. | Author, Year | Population | SNP ID | Criteria of Sample Collection | Time of Sample Collection | RF Concentration in Plasma |
|---|---|---|---|---|---|---|
| 1 | Naidoo et al., 2019 [51] | 172 recurrent TB patients in South Africa | rs10276036 | 1 and/or 2 months and at 6 months during ATT | Predose, 2.5, 6 and 24 h postdose | No change |
| rs1128503 | No change | |||||
| rs2032582 | No change | |||||
| rs1045642 | No change | |||||
| rs2235033 | No change | |||||
| rs2235013 | No change | |||||
| 2 | Calcagno et al., 2019 [52] | 221 PTB with HIV patients in Uganda | rs1045642 | At 2nd, 4th and 8th week of ATT | 1, 2 and 4 h postdose | No change |
| 3 | Allegra et al., 2017 [38] | 24 TB patients in Italy | rs1045642 | At 2nd week and 4th week of ATT | Plasma Cmax (end of 3 infusions for IV route and 2 h postdose for oral) and Ctrough | No change |
| 4 | Chigutsa et al., 2011 [46] | 60 PTB patients in South Africa | rs1045642 | At least 1 month after the start of ATT | 4 to 8 samples per patient, randomly collected over a 7 h period | No change |
| rs2032582 | ||||||
| rs1128503 | ||||||
| rs3842 |
| Sl No. | Author, Year | Population | SNP ID | Criteria of Sample Collection | Time of Sample Collection | RF Concentration in Plasma |
|---|---|---|---|---|---|---|
| 1 | Naidoo et al., 2019 [51] | 172 recurrent TB patients in South Africa | rs2472677 | 1 and/or 2 months and at 6 months during ATT | Predose, 2.5, 6 and 24 h postdose | No change |
| rs1523130 | No change | |||||
| 2 | Calcagno et al., 2019 [52] | 221 PTB with HIV patients in Uganda | rs2472677 | At 2nd, 4th and 8th week of ATT | 1, 2 and 4 h postdose | No change |
| 3 | Allegra et al., 2017 [38] | 24 TB patients in Italy | rs2472677 | At 2nd week and 4th week of ATT | Plasma Cmax (end of 3 infusions for IV route and 2 h postdose for oral) and Ctrough | No change |
| 4 | Chigutsa et al., 2011 [46] | 60 PTB patients in South Africa | rs2472677 | At least 1 month after the start of ATT | 4 to 8 samples per patient, randomly collected over a 7 h period | No change |
| rs1523130 | No change |
| Sl No. | Author, Year | Population | SNP ID | Criteria of Sample Collection | Time of Sample Collection | RF Concentration in Plasma |
|---|---|---|---|---|---|---|
| 1 | Chigutsa et al., 2011 [46] | 60 PTB patients in South Africa | rs2307424 | At least 1 month after the start of ATT | 4 to 8 samples per patient, randomly collected over a 7 h period | No change |
| CES1 | ||||||
|---|---|---|---|---|---|---|
| Sl No. | Author, Year | Population | SNP ID/ Nucleotide Change | Criteria of Sample Collection | Time of Sample Collection | RF Concentration in Plasma |
| 1 | Sloan et al., 2017 [55] | 174 Adult PTB patients in Malawi | rs12149368 | Day 14 or 21 of ATT | Predose, 2 and 6 h postdose | No change |
| CES2 | ||||||
| 1 | Dompreh et al., 2018 [43] | 113 Pediatric TB patients in Ghana | rs3759994 | After 4 weeks of ATT | Predose, 1, 2, 4 and 8 h postdose | No change |
| 2 | Song et al., 2013 [74] | 35 TB patients in South Korea | c.-2548C>T | - | 2 h postdose | No change |
| c.-2263A>G | Increased | |||||
| c.269-965A>G | No change | |||||
| c.474-152T>C | No change | |||||
| c.615+120G>A | No change | |||||
| c.1612+136G>A | No change | |||||
| c.1613-87G>A | No change | |||||
| c.1872*69A>G | No change | |||||
| c.1872*302_304delGAA | No change | |||||
| c.1872*445C>T | No change | |||||
| Sl No. | Author, Year | Population | SNPs Investigated | Criteria of Sample Collection | Time of Sample Collection | RF Concentration in Plasma |
|---|---|---|---|---|---|---|
| 1 | Sloan et al., 2017 [55] | 174 Adult PTB patients in Malawi | rs1803155 | Day 14 or 21 of ATT | Predose, 2 and 6 h postdose | No change |
| rs61733693 |
| Sl No. | Author, Year | Population | Gene | SNP ID | Pharmacokinetic Sampling | Sample Timing | RF Concentration in Plasma |
|---|---|---|---|---|---|---|---|
| 1 | Calcagno et al., 2019 [52] | 221 PTB with HIV patients in Uganda | VDR | rs11568820 (Cdx2) | At 2nd, 4th and 8th week of ATT | 1, 2 and 4 h postdose | No change |
| 2 | Allegra et al., 2017 [38] | 24 TB patients in Italy | VDR | rs731236 (TaqI) | At 2nd week and 4th week of ATT | Plasma Cmax (end of 3 infusions for IV route and 2 h postdose for oral) and Ctrough | No change |
| rs10735810 (FokI) | Decreased | ||||||
| rs1544410 (BsmI) | No change | ||||||
| rs11568820 (Cdx2) | No change | ||||||
| rs7975232 (ApaI) | No change | ||||||
| CYP24A1 | rs927650 | Increased | |||||
| rs2248359 | No change | ||||||
| rs2585428 | No change | ||||||
| CYP27B11 | rs4646536 | Increased | |||||
| rs10877012 | No change |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, L.; Sekhar Miraj, S.; Surulivelrajan, M.; Varma, M.; Sanju, C.S.V.; Rao, M. Influence of Single Nucleotide Polymorphisms on Rifampin Pharmacokinetics in Tuberculosis Patients. Antibiotics 2020, 9, 307. https://doi.org/10.3390/antibiotics9060307
Thomas L, Sekhar Miraj S, Surulivelrajan M, Varma M, Sanju CSV, Rao M. Influence of Single Nucleotide Polymorphisms on Rifampin Pharmacokinetics in Tuberculosis Patients. Antibiotics. 2020; 9(6):307. https://doi.org/10.3390/antibiotics9060307
Chicago/Turabian StyleThomas, Levin, Sonal Sekhar Miraj, Mallayasamy Surulivelrajan, Muralidhar Varma, Chidananda S. V. Sanju, and Mahadev Rao. 2020. "Influence of Single Nucleotide Polymorphisms on Rifampin Pharmacokinetics in Tuberculosis Patients" Antibiotics 9, no. 6: 307. https://doi.org/10.3390/antibiotics9060307
APA StyleThomas, L., Sekhar Miraj, S., Surulivelrajan, M., Varma, M., Sanju, C. S. V., & Rao, M. (2020). Influence of Single Nucleotide Polymorphisms on Rifampin Pharmacokinetics in Tuberculosis Patients. Antibiotics, 9(6), 307. https://doi.org/10.3390/antibiotics9060307





