PYED-1 Inhibits Biofilm Formation and Disrupts the Preformed Biofilm of Staphylococcus aureus
Abstract
1. Introduction
2. Results and Discussion
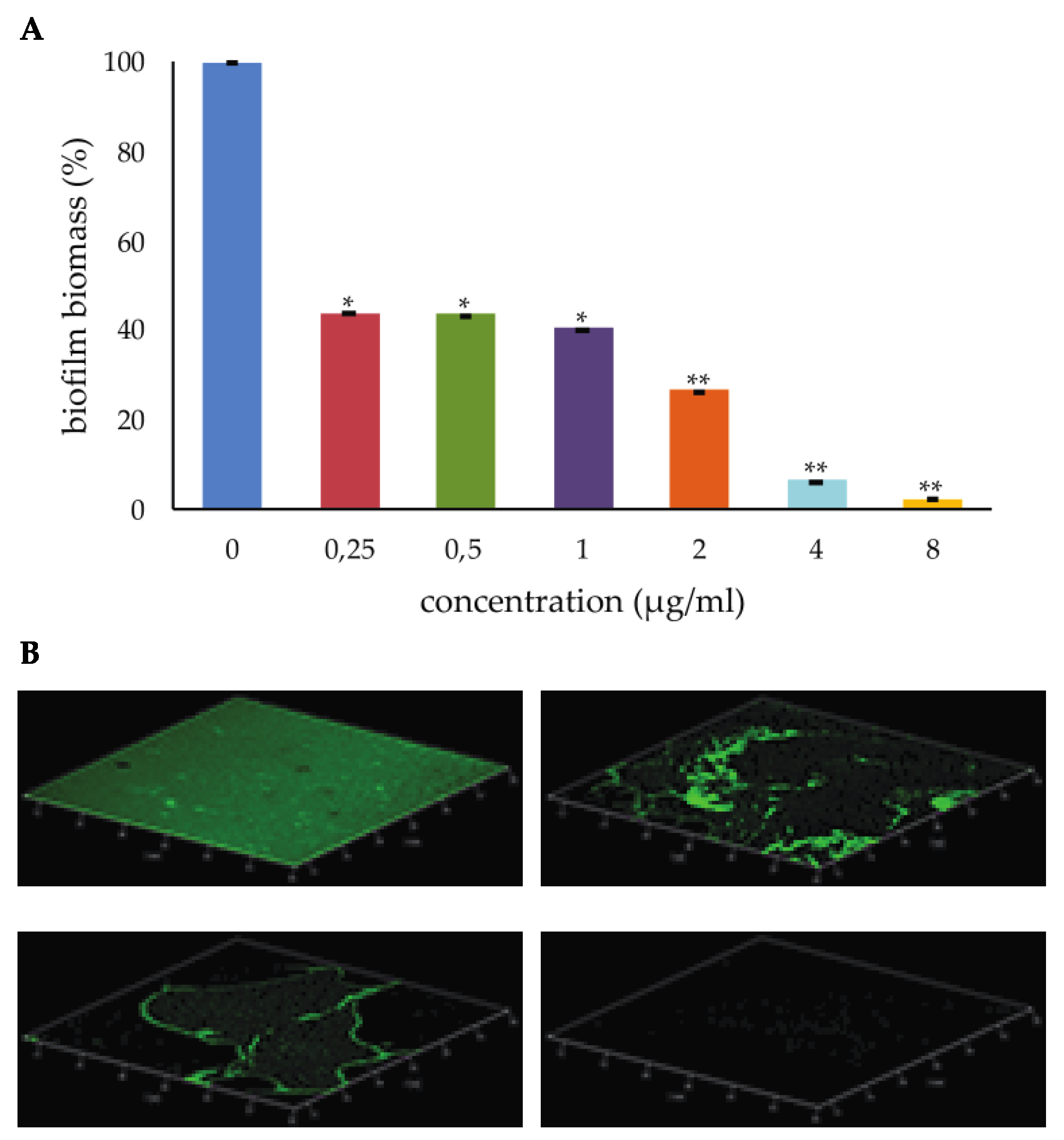
2.1. Effect of PYED-1 on S. aureus Biofilm Formation
2.2. Effect of PYED-1 against S. aureus Preformed Biofilm
2.3. Biofilm Gene Expression
3. Materials and Methods
3.1. Effect of PYED-1 on S. aureus Biofilm Formation
3.2. Effect of PYED-1 against Preformed Biofilm Biomasses
3.3. Effect of PYED-1 against Preformed Biofilm Viability
3.4. Confocal Laser Scanning Microscopy (CLSM)
3.5. Biofilm Gene Expression
3.6. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Ann. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Simoes, M.; Pereira, M.O.; Vieira, M.J. Effect of mechanical stress on biofilms challenged by different chemicals. Water Res. 2005, 39, 5142–5152. [Google Scholar] [CrossRef] [PubMed]
- Parsek, M.R.; Singh, P.K. Bacterial biofilms: An emerging link to disease pathogenesis. Annu. Rev. Microbiol. 2003, 57, 677–701. [Google Scholar] [CrossRef] [PubMed]
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial Biofilm Eradication Agents: A Current Review. Front. Chem. 2019, 28, 824. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013, 136, 1–51. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G.J. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal infections: Mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu. Rev. Med. 2013, 64, 175–188. [Google Scholar] [CrossRef]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef]
- Esposito, A.; De Gregorio, E.; De Fenza, M.; D’Alonzo, D.; Satawani, A.; Guaragna, A. Expeditious synthesis and preliminary antimicrobial activity of deflazacort and its precursors. RSC Adv. 2019, 9, 21519–21524. [Google Scholar] [CrossRef]
- Vollaro, A.; Esposito, A.; Antonaki, E.; Iula, V.D.; D’Alonzo, D.; Guaragna, A.; De Gregorio, E. Steroid Derivatives as potential antimicrobial agents against Staphylococcus aureus planktonic cells. Microorganisms 2020, 8, 468. [Google Scholar] [CrossRef]
- Esposito, A.; Vollaro, A.; Esposito, E.P.; D’Alonzo, D.; Guaragna, A.; Zarrilli, R.; De Gregorio, E. Antibacterial and Antivirulence Activity of Glucocorticoid PYED-1 against Stenotrophomonas maltophilia. Antibiotics (Basel) 2020, 9, 105. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef] [PubMed]
- Goggin, R.; Jardeleza, C.; Wormald, P.J.; Vreugde, S. Corticosteroids directly reduce Staphylococcus aureus biofilm growth: An in vitro study. Laryngoscope 2014, 124, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.R.W.; Gilbert, P. Sensitivity of biofilms to antimicrobial agents. J. Appl. Bacteriol. 1993, 74, 87S–97S. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.; Marques, C. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009, 191, 1393–1403. [Google Scholar] [CrossRef]
- Costerton, J.W. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Rogers, S.; Huigens, R.W., III; Melander, C. A 2-aminobenzimidazole that inhibits and disperses gram-positive biofilms through a zinc-dependent mechanism. J. Am. Chem. Soc. 2009, 131, 9868–9869. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Stoodley, P. Evolving concepts in biofilm infections. Cell Microbiol. 2009, 11, 1034–1043. [Google Scholar] [CrossRef]
- Schwartz, K.; Ganesan, M.; Payne, D.E.; Solomon, M.J.; Boles, B.R. Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Mol. Microbiol. 2016, 99, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Kavanaugh, J.S.; Flack, C.E.; Lister, J.; Ricker, E.B.; Ibberson, C.B.; Jenul, C.; Moormeier, D.E.; Delmain, E.A.; Bayles, K.W.; Horswill, A.R. Identification of extracellular DNA-binding proteins in the biofilm matrix. mBio 2019, 10, e01137-19. [Google Scholar] [CrossRef] [PubMed]
- Okshevsky, M.; Meyer, R.L. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 2015, 41, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Rice, K.C.; Bayles, K.W. Molecular control of bacterial death and lysis. Microbiol. Mol. Biol. Rev. 2008, 72, 85–109. [Google Scholar] [CrossRef] [PubMed]
- Ranjit, D.K.; Endres, J.L.; Bayles, K.W. Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J. Bacteriol. 2011, 193, 2468–2476. [Google Scholar] [CrossRef] [PubMed]
- Sadykov, M.R.; Bayles, K.W. The control of death and lysis in staphylococcal biofilms: A coordination of physiological signals. Curr. Opin. Microbiol. 2012, 15, 211–215. [Google Scholar] [CrossRef]
- Shen, F.; Tang, X.; Wang, Y.; Yang, Z.; Shi, X.; Wang, C.; Zhang, Q.; An, Y.; Cheng, W.; Jin, K.; et al. Phenotype and expression profile analysis of Staphylococcus aureus biofilms and planktonic cells in response to licochalcone A. Appl. Microbiol. Biotechnol. 2015, 99, 359–373. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, Z.; Liu, Z.; Meng, R.; Shi, C.; Chen, X.; Bu, X.; Guo, N. Phenotype and RNA-seq-Based transcriptome profiling of Staphylococcus aureus biofilms in response to tea tree oil. Microb. Pathog. 2018, 123, 304–313. [Google Scholar] [CrossRef]
- Wang, D.; Jin, Q.; Xiang, H.; Wang, W.; Guo, N.; Zhang, K.; Tang, X.; Meng, R.; Feng, H.; Liu, L.; et al. Transcriptional and functional analysis of the effects of magnolol: Inhibition of autolysis and biofilms in Staphylococcus aureus. PLoS ONE 2011, 6, e26833. [Google Scholar] [CrossRef]
- Fernandez, L.; Gonzalez, S.; Campelo, A.B.; Martinez, B.; Rodriguez, A.; Garcia, P. Downregulation of autolysin-encoding genes by phage-derived lytic proteins inhibits biofilm formation in Staphylococcus aureus. Antimicrob. Agents Chemother. 2017, 61, e02724-16. [Google Scholar] [CrossRef]
- Wang, J.; Nong, X.H.; Zhang, X.Y.; Xu, X.Y.; Amin, M.; Qi, S.H. Screening of anti-biofilm compounds from marine-derived fungi and the effects of secalonic acid D on Staphylococcus aureus biofilm. J. Microbiol. Biotechnol. 2017, 27, 1078–1089. [Google Scholar] [CrossRef] [PubMed]
- Paharik, A.E.; Horswill, A.R. The staphylococcal biofilm: Adhesins, regulation, and host response. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Mainiero, M.; Goerke, C.; Geiger, T.; Gonser, C.; Herbert, S.; Wolz, C. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J. Bacteriol. 2010, 192, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Voyich, J.M.; Vuong, C.; DeWald, M.; Nygaard, T.K.; Kocianova, S.; Griffith, S.; Jones, J.; Iverson, C.; Sturdevant, D.E.; Braughton, K.R.; et al. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J. Infect. Dis. 2009, 199, 1698–1706. [Google Scholar] [CrossRef]
- Geiger, T.; Goerke, C.; Mainiero, M.; Kraus, D.; Wolz, C. The virulence regulator Sae of Staphylococcus aureus: Promoter activities and response to phagocytosis-related signals. J. Bacteriol. 2008, 190, 3419–3428. [Google Scholar] [CrossRef] [PubMed]
- Tsang, L.H.; Cassat, J.E.; Shaw, L.N.; Beenken, K.E.; Smeltzer, M.S. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS ONE 2008, 3, e3361. [Google Scholar] [CrossRef] [PubMed]
- Zielinska, A.K.; Beenken, K.E.; Joo, H.S.; Mrak, L.N.; Griffin, L.M.; Luong, T.T.; Lee, C.Y.; Otto, M.; Shaw, L.N.; Smeltze, M.S. Defining the strain-dependent impact of the staphylococcal accessory regulator (sarA) on the alpha-toxin phenotype of Staphylococcus aureus. J. Bacteriol. 2011, 193, 2948–2958. [Google Scholar] [CrossRef]
- Huseby, M.J.; Kruse, A.C.; Digre, J.; Kohler, P.L.; Vocke, J.A.; Mann, E.E.; Bayles, K.W.; Bohach, G.A.; Schlievert, P.M.; Ohlendorf, D.H.; et al. Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms. Proc. Natl. Acad. Sci. USA 2010, 107, 14407–14412. [Google Scholar] [CrossRef]
- Mrak, L.N.; Zielinska, A.K.; Beenken, K.E.; Mrak, I.N.; Atwood, D.N.; Griffin, L.M.; Lee, C.Y.; Smeltzer, M.S. saeRS and sarA act synergistically to repress protease production and promote biofilm formation in Staphylococcus aureus. PLoS ONE 2012, 7, e38453. [Google Scholar] [CrossRef]
- Arya, R.; Ravikumar, R.; Santhosh, R.S.; Princy, S.A. SarA based novel therapeutic candidate against Staphylococcus aureus associated with vascular graft infections. Front. Microbiol. 2015, 6, 416. [Google Scholar] [CrossRef]
- Balamurugan, P.; Praveen Krishna, V.; Bharath, D.; Lavanya, R.; Vairaprakash, P.; Adline Princy, S. Staphylococcus aureus quorum regulator SarA targeted compound, 2-[(Methylamino) methyl] phenol inhibits biofilm and down-regulates virulence genes. Front. Microbiol. 2017, 8, 1290–1300. [Google Scholar] [CrossRef] [PubMed]
- Cheung, G.Y.; Wang, R.; Khan, B.A.; Sturdevant, D.E.; Otto, M. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 2011, 79, 1927–1935. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, M.; Entenza, J.M.; Giachino, P. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 2011, 83, 5171–5179. [Google Scholar] [CrossRef]
- Lauderdale, K.J.; Boles, B.R.; Cheung, A.L.; Horswill, A.R. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 2009, 77, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- O’Riordan, K.; Lee, J.C. Staphylococcus aureus capsular polysaccharides. Clin. Microbiol. Rev. 2004, 17, 218–234. [Google Scholar] [CrossRef] [PubMed]
- Pane, K.; Cafaro, V.; Avitabile, A.; Torres, M.T.; Vollaro, A.; De Gregorio, E.; Catania, M.R.; Di Maro, A.; Bosso, A.; Gallo, G.; et al. Identification of novel cryptic multifunctional antimicrobial peptides from the human stomach enabled by a computational-experimental platform. ACS Synth. Biol. 2018, 7, 2105–2115. [Google Scholar] [CrossRef]
- De Gregorio, E.; Esposito, E.P.; Zarrilli, R.; Di Nocera, P.P. Contact-dependent growth inhibition proteins in Acinetobacter baylyi ADP1. Curr. Microbiol. 2018, 75, 1434–1440. [Google Scholar] [CrossRef]
- Martinucci, M.; Roscetto, E.; Iula, V.D.; Votsi, A.; Catania, M.R.; De Gregorio, E. Accurate identification of members of the Burkholderia cepacia complex in cystic fibrosis sputum. Lett. Appl. Microbiol. 2016, 62, 221–229. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

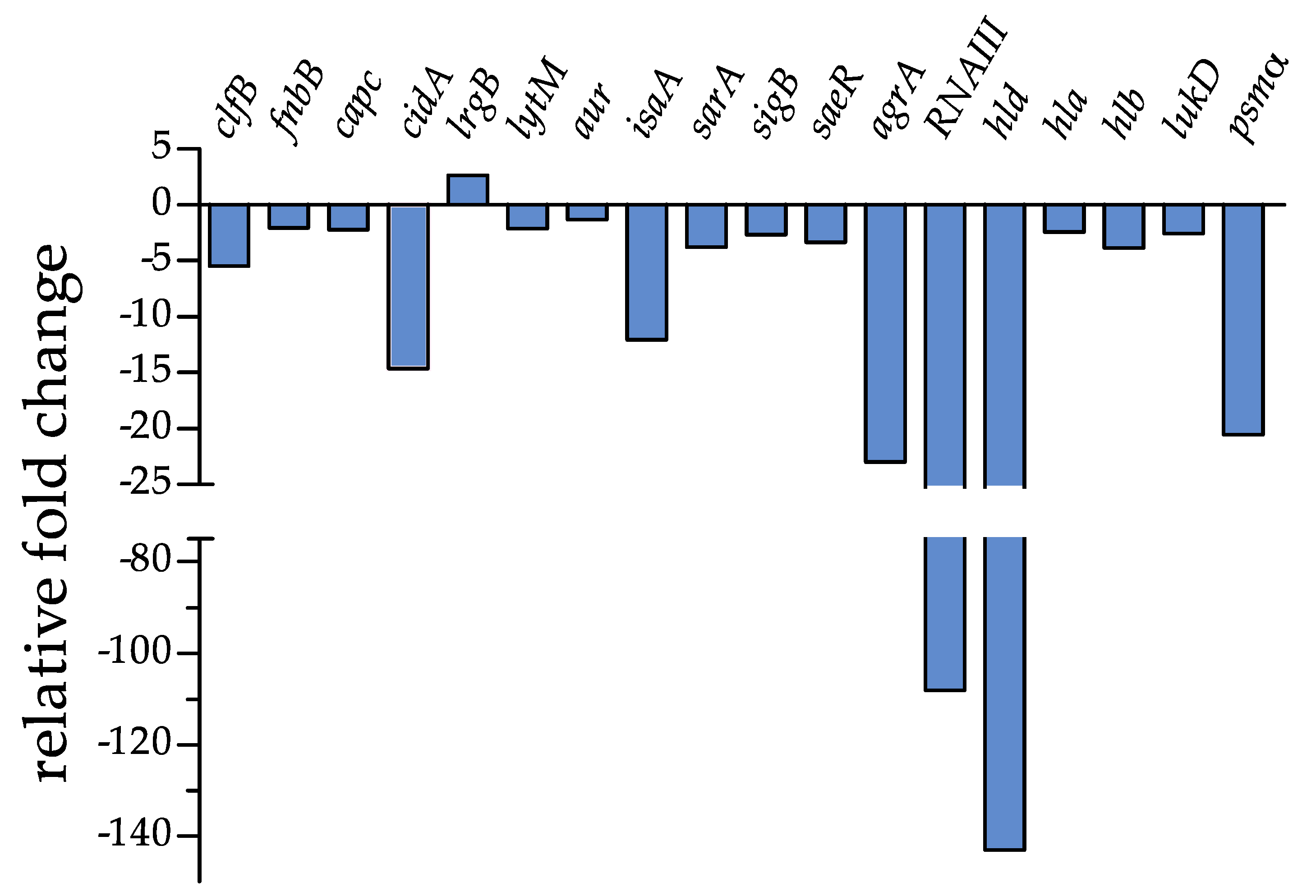
| Gene | Description | Fold Change ± SD | p-Value |
|---|---|---|---|
| clfB | clumping factor B | −5.44 ± 0.011 | 0.0004 |
| fnbB | fibronectin-binding protein B | −2.03 ± 0.029 | 0.0025 |
| capC | capsule biosynthesis protein C | −2.22 ± 0.027 | 0.0019 |
| cidA | holin-like murein hydrolase modulator | −14.67 ± 0.004 | <0.0001 |
| lrgB | antiholin-like protein B | +2.65 ± 0.160 | 0.0013 |
| lytM | peptidoglycan hydrolase | −2.09 ± 0.028 | 0.0022 |
| aur | aureolysin, zinc metalloproteinase | −1.32 ± 0.045 | 0.0155 |
| isaA | immunodominant staphylococcal antigen | −12.06 ± 0.005 | <0.0001 |
| sarA | Transcriptional regulator | −3.77 ± 0.016 | 0.0007 |
| sigB | RNA polymerase sigma factor B | −2.67 ± 0.022 | 0.0013 |
| saeR | response regulator SaeR | −3.34 ± 0.018 | 0.0008 |
| agrA | accessory gene regulator protein A | −22.99 ± 0.002 | 0.0001 |
| RNAIII | small regulatory RNA | −108.14 ± 0.0005 | <0.0001 |
| hld | delta-haemolysin gene | −143.04 ± 0.0004 | <0.0001 |
| hla | alpha-haemolysin | −2.41 ± 0.025 | 0.0016 |
| hlb | beta-haemolysin | −3.83 ± 0.015 | 0.0007 |
| lukD | pore-forming leukocidin | −2.56 ± 0.023 | 0.0014 |
| psmα | Phenol-soluble modulin | −20.54 ± 0.002 | 0.0001 |
| sarA | Transcriptional regulator | −3.77 ± 0.016 | 0.0007 |
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| agrA | TGCGAAGACGATCCAAAAC | TTTAGCTTGCTCAAGCACCTC |
| aur | GATGGTCGCACATTCACAAG | CGCCTGACTGGTCCTTATATTC |
| capC | CATCCAGAGCGGAATAAAGC | CGGAAATACCCGCTAATGAC |
| cidA | CTTAGCCGGCAGTATTGTTG | GTTTGCACCGTCTTCTACCC |
| clfB | TTATGGTGGTGGAAGTGCTG | TGGACTTGGTTCTGGATCTG |
| fnbB | GAACATGGTCAAGCACAAGG | ACGCCATAATTACCGTGACC |
| hla | TCTTGGAACCCGGTATATGG | AGCGAAGTCTGGTGAAAACC |
| hlb | GTGCCAAAGCCGAATCTAAG | ATCAGCGCGTTTATATTGTCC |
| hld | AAGGAAGGAGTGATTTCAATGG | TTTGTTCACTGTGTCGATAATCC |
| isaA | TCCGACAAACACTGTTGACC | AATCCCCAAGCACCTAAACC |
| lrgB | TATTGCCCGAGGATTAGCAC | CAAAGACAGGCACAACTGCTAC |
| lytM | ACGGTGTCGACTATGCAATG | ATTGCCGCCACCATAGTTAC |
| lukD | GTACTTAAGGCAGCCGGAAAC | CGCCCCAATAAAACTGTGAG |
| psmα | TCAAAAGCTTAATCGAACAATTCAC | AATGGCCCCCTTCAAATAAG |
| RNAIII | AAGCCATCCCAACTTAATAACC | GCACTGAGTCCAAGGAAACTAAC |
| rpoB | ACAACCACTTGGCGGTAAAG | ATGCTTCAAGTGCCCATACC |
| sarA | TTGCTTTGAGTTGTTATCAATGG | CAATACAGCGAATTCTTCAAAGC |
| saeR | CCAAGGGAACTCGTTTTACG | ACGCATAGGGACTTCGTGAC |
| sigB | TGATCGCGAACGAGAAATC | ATTGCCGTTCTCTGAAGTCG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vollaro, A.; Esposito, A.; Esposito, E.P.; Zarrilli, R.; Guaragna, A.; De Gregorio, E. PYED-1 Inhibits Biofilm Formation and Disrupts the Preformed Biofilm of Staphylococcus aureus. Antibiotics 2020, 9, 240. https://doi.org/10.3390/antibiotics9050240
Vollaro A, Esposito A, Esposito EP, Zarrilli R, Guaragna A, De Gregorio E. PYED-1 Inhibits Biofilm Formation and Disrupts the Preformed Biofilm of Staphylococcus aureus. Antibiotics. 2020; 9(5):240. https://doi.org/10.3390/antibiotics9050240
Chicago/Turabian StyleVollaro, Adriana, Anna Esposito, Eliana Pia Esposito, Raffaele Zarrilli, Annalisa Guaragna, and Eliana De Gregorio. 2020. "PYED-1 Inhibits Biofilm Formation and Disrupts the Preformed Biofilm of Staphylococcus aureus" Antibiotics 9, no. 5: 240. https://doi.org/10.3390/antibiotics9050240
APA StyleVollaro, A., Esposito, A., Esposito, E. P., Zarrilli, R., Guaragna, A., & De Gregorio, E. (2020). PYED-1 Inhibits Biofilm Formation and Disrupts the Preformed Biofilm of Staphylococcus aureus. Antibiotics, 9(5), 240. https://doi.org/10.3390/antibiotics9050240









