Omics for Bioprospecting and Drug Discovery from Bacteria and Microalgae
Abstract
1. Introduction
- (1)
- What factors determine the organism’s ability to express target molecules under natural or laboratory conditions?
- (2)
- What factors could promote de novo biosynthesis of a metabolite of interest?
- (3)
- How accurate are the methods applied in screening the microorganism bearing potential for compounds of interest?
- (4)
- What approaches can enhance the likelihood of encountering a compound of interest from the strain under research?
2. Metabarcoding and Metagenomics in Discovery of Strains of Interest
2.1. Metabarcoding
2.2. Metagenomics
3. Genomics and Metagenomics as Quick Guides to Discover Compounds of Interest
4. Transcriptomics
4.1. Transcriptomics in the Discovery of Noncoding RNAs with a Metabolic Regulatory Role
4.2. CRISPR-Cas Systems and Their Relevance to Transcriptomics and Bioprospecting
5. Proteomics
6. Glycomics
7. Lipidomics
8. Metabolomics
9. The Potential of Mass Spectrometry in Omics
10. Biosynthetic Pathways of Drug Leads and Heterologous Expression
11. One Strain Many Compounds (OSMAC) Approach in Omics
12. Bioinformatics and Chemoinformatics Crosstalk in Drug Discovery from Bacteria and Microalgae
13. Future Prospects
14. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Buenz, E.J.; Verpoorte, R.; Bauer, B.A. The ethnopharmacologic contribution to bioprospecting natural products. Annu. Rev. Pharmacol. Toxicol. 2018, 5, 509–530. [Google Scholar] [CrossRef] [PubMed]
- Soejarto, D.D.; Fong, H.H.S.; Tan, G.T.; Zhang, H.J.; Ma, C.Y.; Franzblau, S.G.; Gyllenhaal, C.; Riley, M.C.; Kadushin, M.R.; Pezzuto, J.M.; et al. Ethnobotany/ethnopharmacology and mass bioprospecting: Issues on intellectual property and benefit-sharing. J. Ethnopharmacol. 2005, 100, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Mate, N.; Nader, W.; Tamayo, G. Encyclopeida of Biodiversity; Levin, S.E., Ed.; Academic Press: Cambridge, MA, USA, 2013; Volume 1, pp. 571–579. [Google Scholar]
- Maldonado, L.A.; Stach, J.; Pathom-aree, W.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Diversity of cultivable actinobacteria in geographically widespread marine sediments. Antonie Van Leeuwenhoek 2005, 87, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Wu, Z.; Bian, L.; Feng, D.; Leung, D.Y.C. Cultivation of Spirulina platensis for biomass production and nutrient removal from synthetic human urine. Appl. Energy 2013, 102, 427–431. [Google Scholar] [CrossRef]
- Aoi, Y.; Kinoshita, T.; Hata, T.; Ohta, H.; Obokata, H.; Tsuneda, S. Hollow-fiber membrane chamber as a device for in situ environmental cultivation. Appl. Environ. Microbiol. 2009, 75, 3826–3833. [Google Scholar] [CrossRef]
- Stengel, D.B.; Connan, S.; Popper, Z.A. Algal chemodiversity and bioactivity: Sources of natural variability and implications for commercial application. Biotechnol. Adv. 2011, 29, 483–501. [Google Scholar] [CrossRef]
- Aziz, F.H. Fikrat; M.H.; Rasul, B.H. An ecological observation on inland water ecosystem in Erbil–Iraq Kurdistan with particular reference to blue green algae Glaucospira. J. Baghdad Sci. 2014, 11, 1385–1387. [Google Scholar]
- Szubert, K.; Wiglusz, M.; Mazur-Marzec, H. Bioactive metabolites produced by Spirulina subsalsa from the Baltic Sea. Oceanologia 2018, 60, 245–255. [Google Scholar] [CrossRef]
- Huang, X.; Pinto, D.; Fritz, G.; Mascher, T. Environmental sensing in actinobacteria: A comprehensive survey on the signaling capacity of this phylum. J. Bacteriol. 2015, 197, 2517–2535. [Google Scholar] [CrossRef]
- Vester, J.K.; Glaring, M.A.; Stougaard, P. Improved cultivation and metagenomics as new tools for bioprospecting in cold environments. Extremophiles 2015, 19, 17–29. [Google Scholar] [CrossRef]
- Hemaiswarya, S.; Raja, R.; Ravikumar, R.; Kumar, A.; Carvalho, I. Omics: Applications in Biomedical, Agricultural, and Environmental Sciences; Barh, D., Zambare, V., Azevedo, V., Eds.; CRC Press: Boca Raton, FL, USA; Taylor and Francis: London, UK, 2013; Chapter 18; p. 439. [Google Scholar] [CrossRef]
- Marco, D.E.; Abram, F. Using Genomics, Metagenomics and Other “Omics” to Assess Valuable Microbial Ecosystem Services and Novel Biotechnological Applications. Front. Microbiol. 2019, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Munang’andu, H.M.; Mugimba, K.K.; Byarugaba, D.K.; Mutoloki, S.; Evensen, Ø. Current advances on virus discovery and diagnostic role of viral metagenomics in aquatic organisms. Front. Microbiol. 2017, 8, e406. [Google Scholar] [CrossRef] [PubMed]
- Carruthers, M.; Yurchenko, A.A.; Augley, J.J.; Adams, C.E.; Herzyk, P.; Elmer, K.R. De novo transcriptome assembly, annotation and comparison of four ecological and evolutionary model salmonid fish species. BMC Genom. 2018, 19, 1–39. [Google Scholar] [CrossRef]
- Mills, J.D.; Kawahara, Y.; Janitz, M. Strand-specific RNA-Seq provides greater resolution of transcriptome profiling. Curr. Genom. 2013, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Kodzius, R.; Gojobori, T. Marine genomics marine metagenomics as a source for bioprospecting. Mar. Genom. 2015, 24, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, D.J.; Andreote, F.D.; Chaves, D.; Montan, S.; Jime, D.J.; Zambrano, M.; Baena, S.; Osorio-forero, C.; Junca, H. Structural and functional insights from the metagenome of an acidic hot spring microbial planktonic community in the Colombian Andes. PLoS ONE 2012, 7, e52069. [Google Scholar] [CrossRef]
- Chaudhary, A.K.; Dhakal, D.; Sohng, J.K. An insight into the “-omics” based engineering of streptomycetes for secondary metabolite overproduction. BioMed Res. Int. 2013, 968518. [Google Scholar] [CrossRef]
- Rai, V.; Karthikaichamy, V.; Das, D.; Noronha, S.; Wangikar, P.P.; Srivastava, S. Multi-omics Frontiers in Algal Research: Techniques and Progress to Explore Biofuels in the Postgenomics World. OMICS 2016, 20, 387–399. [Google Scholar] [CrossRef]
- Ramos, P.I.; Porto, D.F.; Lanzatotti, E.; Sosa, E.J.; Burguener, G.; Pardo, A.M.; Klein, C.C.; Sagot, M.-F.; De Vasconcelos, A.T.R.; Gales, A.C.; et al. An integrative, multi-omics approach towards the prioritization of Klebsiella pneumoniae drug targets. Sci. Rep. 2018, 8, 10755. [Google Scholar] [CrossRef]
- Kuppusamy, P.; Soundharrajan, I.; Srigopalram, S.; Yusoff, M.M. Potential pharmaceutical and biomedical applications of Diatoms microalgae—An overview. Ind. J. Geo Mar. Sci. 2017, 46, 663–667. [Google Scholar]
- Wang, M.; Zhang, J.; He, S.; Yan, X. A review study on macrolides isolated from cyanobacteria. Mar. Drugs 2017, 15, 126. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, J. Amphidinolides and its related macrolides from marine dinoflagellates. J. Antibiot. (Tokyo) 2008, 61, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Challis, G.L.; Hopwood, D.A. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. USA 2003, 100, 14555–14561. [Google Scholar] [CrossRef] [PubMed]
- Hielscher-Michael, S.; Griehl, C.; Buchholz, M.; Demuth, H.U.; Arnold, N.; Wessjohann, L.A. Natural products from microalgae with potential against Alzheimer’s disease: Sulfolipids are potent glutaminyl cyclase inhibitors. Mar. Drugs 2016, 14, 203. [Google Scholar] [CrossRef]
- Lin, P.-Y.; Tsai, C.-T.; Chuang, W.-L.; Chao, Y.-H.; Pan, I.-H.; Chen, Y.-K.; Lin, C.-C.; Wang, B.-Y. Chlorella sorokiniana induces mitochondrial-mediated apoptosis in human non-small cell lung cancer cells and inhibits xenograft tumor growth in vivo. BMC Complement. Altern. Med. 2017, 17, 1–88. [Google Scholar] [CrossRef]
- Shannon, E.; Abu-Ghannam, N. Antibacterial derivatives of marine algae: An overview of pharmacological mechanisms and applications. Mar. Drugs 2016, 14, 81. [Google Scholar] [CrossRef]
- Yamada, Y.; Kuzuyama, T.; Komatsu, M.; Shin-ya, K.; Omura, S.; Cane, D.E.; Ikeda, H. Terpene synthases are widely distributed in bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 857–862. [Google Scholar] [CrossRef]
- López, Y.; Soto, S.M. The preventing usefulness biofilm microalgae infections compounds for preventing biofilm infections. Antibiotics 2020, 9, 9. [Google Scholar] [CrossRef]
- Sambanthamoorthy, K.; Gokhale, A.A.; Lao, W.; Parashar, V.; Neiditch, M.B.; Semmelhack, M.F.; Lee, I.; Waters, C.M. Identification of a novel benzimidazole that inhibits bacterial biofilm formation in a broad-spectrum manner. Antimicrob. Agents Chemother. 2011, 55, 4369–4378. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Berben, T. Microbial diversity and biogeochemical cycling in soda lakes. Extremophiles 2014, 791–809. [Google Scholar] [CrossRef]
- Nolla-ardèvol, V.; Strous, M.; Tegetmeyer, H.E.; Oren, A. Anaerobic digestion of the microalga Spirulina at extreme alkaline conditions: Biogas production, metagenome, and metatranscriptome. Front. Microbiol. 2015, 6, 597. [Google Scholar] [CrossRef] [PubMed]
- Alkhalili, R.N.; Hatti-kaul, R.; Canbäck, B. Genome Sequence of Geobacillus sp. Strain ZGt-1, an Antibacterial Peptide-Producing Bacterium from Hot Springs in Jordan. Genome Announc. 2015, 3, e00799-15. [Google Scholar] [CrossRef] [PubMed]
- Alkhalili, R.N.; Bernfur, K.; Dishisha, T.; Mamo, G.; Schelin, J.; Canbäck, B.; Emanuelsson, C.; Hatti-Kaul, R. Antimicrobial protein candidates from the thermophilic Geobacillus sp. Strain ZGt-1: Production, proteomics, and bioinformatics analysis. Int. J. Mol. Sci. 2016, 17, 1363. [Google Scholar] [CrossRef] [PubMed]
- Kwei, C.K.; Lewis, D.; King, K.; Donohue, W.; Neilan, B.A. Molecular classification of commercial Spirulina strains and identification of their sulfolipid biosynthesis genes. J. Microbiol. Biotechnol. 2011, 21, 359–365. [Google Scholar] [PubMed]
- Groendahl, S.; Kahlert, M.; Fink, P. The best of both worlds: A combined approach for analyzing microalgal diversity via metabarcoding and morphology-based methods. PLoS ONE 2017, 12, e0172808. [Google Scholar] [CrossRef]
- Franzén, O.; Hu, J.; Bao, X.; Itzkowitz, S.H.; Peter, I.; Bashir, A. Improved OTU-picking using long-read 16S rRNA gene amplicon sequencing and generic hierarchical clustering. Microbiome 2015, 43. [Google Scholar] [CrossRef]
- Preetha, K.; John, L.; Subin, C.S.; Vijayan, K.K. Phenotypic and genetic characterization of Dunaliella (Chlorophyta) from Indian salinas and their diversity. Aquat. Biosyst. 2012, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Jovel, J.; Patterson, J.; Wang, W.; Hotte, N.; O’Keefe, S.; Mitchel, T.; Perry, T.; Kao, D.; Mason, A.L.; Madsen, K.L.; et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 2016, 7, 459. [Google Scholar] [CrossRef] [PubMed]
- Godoy-Vitorino, F.; Romaguera, J.; Zhao, C.; Vargas-Robles, D.; Ortiz-Morales, G.; Vázquez-Sánchez, F.; Sanchez-Vázquez, M.; De La Garza-Casillas, M.; Martinez-Ferrer, M.; White, J.R.; et al. Cervicovaginal fungi and bacteria associated with cervical intraepithelial neoplasia and high-risk human papillomavirus infections in a hispanic population. Front. Microbiol. 2018, 9, 2533. [Google Scholar] [CrossRef]
- Perkerson, R.B., III; Perkerson, E.A.; Casamatta, D.A. Phylogenetic examination of the cyanobacterial genera Geitlerinema and Limnothrix (Pseudanabaenaceae) using 16S rDNA gene sequence data. Arch. Hydrobiol. Suppl. Algol. Stud. 2010, 134, 1–16. [Google Scholar] [CrossRef]
- Scheldeman, P.; Baurain, D.; Bouhy, R.; Scott, M.; Mühling, M.; Whitton, B.A.; Belay, A.; Wilmotte, A. Arthrospira ('Spirulina’) strains from four continents are resolved into only two clusters, based on amplified ribosomal DNA restriction analysis of the internally transcribed spacer. FEMS Microbiol. Lett. 1999, 172, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.A.; Flint, S.H.; Lindsay, D.; Cox, M.P.; Biggs, P.J. Insights into the Geobacillus stearothermophilus species based on phylogenomic principles. BMC Microbiol. 2017, 17, 140. [Google Scholar] [CrossRef] [PubMed]
- Chèneby, D.; Philippot, L.; Hartmann, A.; Hénault, C.; Germon, J.C. 16S rDNA analysis for characterization of denitrifying bacteria isolated from three agricultural soils. FEMS Microbiol. Ecol. 2000, 34, 121–128. [Google Scholar] [CrossRef]
- Alanagreh, L.; Pegg, C.; Harikumar, A.; Buchheim, M. Assessing intragenomic variation of the internal transcribed spacer two: Adapting the Illumina metagenomics protocol. PLoS ONE 2017, 12, e0181491. [Google Scholar] [CrossRef] [PubMed]
- Lafontaine, D.L.J.; Tollervey, D. The function and synthesis of ribosomes. Nat. Rev. Mol. Cell Biol. 2001, 2, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Tesson, S.V.M.; Šantl-temkiv, T.; Dillon, J.G. Ice nucleation activity and aeolian dispersal success in airborne and aquatic microalgae. Front Microbiol. 2018, 9, e2681. [Google Scholar] [CrossRef]
- Hadi, S.I.I.A.; Santana, H.; Brunale, P.P.M.; Gomes, T.G. DNA barcoding green microalgae isolated from neotropical inland waters. PLoS ONE 2016, 11, e0149284. [Google Scholar] [CrossRef]
- Patel, A.; Chaudhary, S.; Syed, B.A.; Gami, B.; Patel, P. Rbcl marker based approach for molecular identification of Arthrospira and Dunaliella isolates from non-axenic cultures. J. Genet. Genet. Eng. 2018, 2, 24–34. [Google Scholar]
- Duong, V.T.; Ahmed, F.; Thomas-hall, S.R.; Quigley, S.; Nowak, E.; Schenk, P.M. High protein- and high lipid-producing microalgae from northern Australia as potential feedstock for animal feed and biodiesel. Front. Bioeng. Biotechnol. 2015, 3, 53. [Google Scholar] [CrossRef]
- Gantner, S.; Stenlid, J. Utilizing ITS1 and ITS2 to study environmental fungal diversity using pyrosequencing. FEMS Microbiol. Ecol. 2012, 84, 165–175. [Google Scholar] [CrossRef]
- Walter, J.M.; Coutinho, F.H.; Dutilh, B.E.; Thompson, C.C.; Thompson, F.L. Proposal of a new genome-based taxonomy for Cyanobacteria. Front. Microbiol. 2016, 8, e02132. [Google Scholar] [CrossRef]
- Nolla-ardèvol, V.; Peces, M.; Strous, M.; Tegetmeyer, H.E. Metagenome from a Spirulina digesting biogas reactor: Analysis via binning of contigs and classification of short reads. BMC Microbiol. 2015, 15, e277. [Google Scholar] [CrossRef] [PubMed]
- Krohn-molt, I.; Wemheuer, B.; Alawi, M.; Poehlein, A.; Schmeisser, C.; Grundhoff, A.; Daniel, R.; Hanelt, D.; Streit, W.R. Metagenome Survey of a multispecies and alga-associated biofilm revealed key elements of bacterial-algal interactions in photobioreactors. Appl. Environ. Microbiol. 2013, 79, 6196–6206. [Google Scholar] [CrossRef] [PubMed]
- Pryszcz, L.P.; Santos, F. Interactions between closely related bacterial strains are revealed by deep transcriptome sequencing. Appl. Environ. Microbiol. 2015, 81, 8445–8456. [Google Scholar] [CrossRef]
- Sharma, G.; Dua, P.; Agarwal, S. A comprehensive review of dysregulated miRNAs involved in cervical cancer. Curr. Genom. 2014, 15, 310–323. [Google Scholar] [CrossRef]
- Peter, A.P.; Lakshmanan, K.; Mohandass, S. Cyanobacterial KnowledgeBase ( CKB ), a compendium of cyanobacterial genomes and proteomes. PLoS ONE 2015, 10, e0136262. [Google Scholar] [CrossRef]
- Xu, T.; Qin, S.; Hu, Y.; Song, Z.; Ying, J.; Li, P.; Dong, W.; Zhao, F.; Yang, H.; Bao, Q. Whole genomic DNA sequencing and comparative genomic analysis of Arthrospira platensis: High genome plasticity and genetic diversity. DNA Res. 2016, 23, 325–338. [Google Scholar] [CrossRef]
- Parks, D.; Chuvochina, M.; Waite, D.; Rinke, C.; Skarshewisk, A.; Chaumeil, P.-A.; Hungeholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Rückert, C.; Albersmeier, A.; Busche, T.; Jaenicke, S.; Winkler, A.; Fri, Ó.H.; Lambert, C.; Badcock, D.; Bernaerts, K.; Anne, J.; et al. Complete genome sequence of Streptomyces lividans TK24. J. Biotechnol. 2015, 199, 21–22. [Google Scholar] [CrossRef]
- Guarnieri, M.T.; Levering, J.; Henard, C.A.; Boore, J.L.; Betenbaugh, M.J.; Zengler, K.; Knoshaug, E.P. Genome sequence of the oleaginous green slga, Chlorella vulgaris UTEX 395. Front Bioeng Biotechnol. 2018, 6, 7. [Google Scholar] [CrossRef]
- Xu, L.; Sun, C.; Huang, M.; Wu, Y.; Yuan, C.; Dai, W. Marine Genomics Complete genome sequence of Euzebya sp. DY32-46, a marine Actinobacteria isolated from the Pacific Ocean. Mar. Genom. 2018, 44, 65–69. [Google Scholar] [CrossRef]
- Abd, H.H.; Baky, E.; El Baroty, G.S.; Mostafa, E.M. Optimization growth of Spirulina (Arthrospira) platensis in photobioreac- tor under varied nitrogen concentration for maximized biomass, carotenoids and lipid contents. Recent Pat. Food Nutr. Agric. 2018, 10, 70–81. [Google Scholar] [CrossRef]
- Cuperlovic-culf, M.; Culf, A.S.; Morin, P.J.; Touaibia, M. Application of metabolomics in drug discovery, development and theranostics. Curr. Metab. 2013, 1, 41–57. [Google Scholar] [CrossRef]
- Guljamow, A.; Kreische, M.; Ishida, K.; Liaimer, A. crossm high-density cultivation of terrestrial Nostoc strains leads to reprogramming of secondary metabolome. Appl. Environ. Microbiol. 2017, 83, e01510-17. [Google Scholar] [CrossRef]
- Cho, D.H.; Choi, J.W.; Kang, Z.; Kim, B.H.; Oh, H.M.; Kim, H.S.; Ramanan, R. Microalgal diversity fosters stable biomass productivity in open ponds treating wastewater. Sci. Rep. 2017, 7, 1979. [Google Scholar] [CrossRef]
- Hagihara, R.; Katsuyama, Y.; Sugai, Y.; Onaka, H.; Ohnishi, Y. Novel desferrioxamine derivatives synthesized using the secondary metabolism-specific nitrous acid biosynthetic pathway in Streptomyces davawensis. J. Antibiot. 2018, 71, 911–919. [Google Scholar] [CrossRef]
- Jones, M.B.; Nierman, W.C.; Shan, Y.; Frank, B.C.; Spoering, A.; Ling, L.; Peoples, A.; Zullo, A.; Lewis, K.; Nelson, K.E. Reducing the bottleneck in discovery of novel antibiotics. Microb. Ecol. 2016, 73, 658–667. [Google Scholar] [CrossRef]
- Setoain, J.; Mart, M.; Tabas-madrid, D.; Sorzano, C.O.S.; Bakker, A.; Gonzalez-couto, E.; Elvira, J.; Pascual-montano, A.; Elmer, P. NFFinder: An online bioinformatics tool for searching similar transcriptomics experiments in the context of drug repositioning. Nucleic Acids Res. 2015, 43, W193–W199. [Google Scholar] [CrossRef]
- Heueis, N.; Vockenhuber, M.; Suess, B. Small non-coding RNAs in Streptomycetes. RNA Biol. 2014, 11, 464–469. [Google Scholar] [CrossRef][Green Version]
- Zhang, C. Novel functions for small RNA molecules. Curr. Opin. Mol. Ther. 2009, 11, 641–651. [Google Scholar]
- Liu, W.; Shi, Y.; Yao, L.; Zhou, Y.; Ye, B. Prediction and characterization of small non-coding RNAs related to secondary metabolites in Saccharopolyspora erythraea. PLoS ONE 2013, 8, e8067611. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Xue, Q.; Tang, L.; Carney, J.R.; Betlach, M.; Mcdaniel, R. Cloning, characterization and heterologous expression of a polyketide synthase of oleandomycin and p-450 oxidase involved in the biosynthesis the antibiotic encoding the 6-deoxyerythronolide. J. Antibiot. (Tokyo) 2000, 53, 502–508. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Peano, C.; Talà, A.; Corti, G.; Pasanisi, D.; Durante, M.; Mita, G.; Bicciato, S. Comparative genomics and transcriptional profiles of Saccharopolyspora erythraea NRRL 2338 and a classically improved erythromycin over-producing strain. Microb. Cell Fact. 2012, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Hou, S.; Brenes-álvarez, M.; Reimann, V.; Omer, S.; Backofen, R.; Muro-pastor, A.M.; Hess, W.R.; Backofen, R.; Muro-pastor, A.M.; Crispr-cas, W.R.H. CRISPR-Cas systems in multicellular cyanobacteria. RNA Biol. 2018, 16, 518–529. [Google Scholar] [CrossRef]
- Tong, Y.; Charusanti, P.; Zhang, L.; Weber, T.; Lee, S.Y. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth. Biol. 2015, 4, 1020–1029. [Google Scholar] [CrossRef]
- Keun, S.; Hwan, G.; Seong, W.; Kim, H.; Kim, S.; Lee, D.; Lee, S. CRISPR interference-guided balancing of a biosynthetic mevalonate pathway increases terpenoid production. Metab. Eng. 2016, 38, 228–240. [Google Scholar]
- Crawley, A.B.; Henriksen, J.R. CRISPRdisco: An automated pipeline for the discovery and analysis of CRISPR-Cas systems. CRIAPR J. 2018, 1, 171–181. [Google Scholar] [CrossRef]
- Jhanker, Y.M.; Kadir, M.F.; Khan, R.I.; Hasan, R. Proteomics in drug discovery. J. Appl. Pharm. Sci. 2012, 2, 1–12. [Google Scholar] [CrossRef][Green Version]
- Lindsay, M.A. Target discovery. Nat. Rev. Drug Discov. 2003, 2, 831–838. [Google Scholar] [CrossRef]
- Scheepstra, M.; Hekking, K.F.W.; Van Hijfte, L.; Folmer, R.H.A. Bivalent ligands for protein degradation in drug discovery. Comput. Struct. Biotechnol. J. 2019, 17, 160–176. [Google Scholar] [CrossRef]
- Frantzi, M.; Latosinska, A.; Mischak, H. Proteomics in drug development: The dawn of a new era? Proteom. Clin. Appl. 2019, 13, e1800087. [Google Scholar] [CrossRef] [PubMed]
- Ctortecka, C.; Palve, V.; Kuenzi, B.M.; Fang, B.; Sumi, N.J.; Izumi, V.; Novakova, S.; Kinose, F.; Rix, L.L.R.; Haura, E.B.; et al. Functional Proteomics and deep network interrogation reveal a complex mechanism of action of midostaurin in lung cancer cells. Mol. Cell Proteom. 2018, 17, 2434–2447. [Google Scholar] [CrossRef] [PubMed]
- Svozil, J.; Baerenfaller, K. A Cautionary tale on the inclusion of variable posttranslational modifications in database-dependent searches of mass spectrometry data. Methos Enzymol. 2017, 586, 433–452. [Google Scholar] [CrossRef]
- Adibekian, A.; Stallforth, P.; Hecht, M.; Werz, D.B.; Seeberger, P.H. Comparative bioinformatics analysis of the mammalian and bacterial glycomes. Chem. Sci. 2011, 2, 337–344. [Google Scholar] [CrossRef]
- Buck, C.B.; Thompson, C.D.; Roberts, J.N.; Müller, M.; Lowy, D.R.; Schiller, J.T. Carrageenan is a potent inhibitor of papillomavirus infection. PLoS Pathog. 2006, 2, e69. [Google Scholar] [CrossRef]
- Aoki-Kinoshita, K.F. Using glycome databases for drug discovery. Expert Opin. Drug Discov. 2008, 3, 877–890. [Google Scholar] [CrossRef]
- Furmaniak, M.A.; Misztak, A.E.; Franczuk, M.D.; Wilmotte, A.; Waleron, M.; Waleron, K.F. Edible cyanobacterial genus Arthrospira: Actual state of the art in cultivation methods, genetics, and application in medicine. Front. Microbiol. 2017, 8, 541. [Google Scholar] [CrossRef]
- Santoyo, S.; Jaime, L.; Plaza, M.; Herrero, M.; Rodriguez-Meizoso, I.; Ibañez, E.; Reglero, G. Antiviral compounds obtained from microalgae commonly used as carotenoid sources. J. Appl. Phycol. 2012, 24, 731–741. [Google Scholar] [CrossRef]
- Okuyama, H.; Tominaga, A.; Fukuoka, S.; Taguchi, T.; Kusumoto, Y.; Ono, S. Spirulina lipopolysaccharides inhibit tumor growth in a Toll-like receptor 4-dependent manner by altering the cytokine milieu from interleukin-17/interleukin-23 to interferon-γ. Oncol. Rep. 2017, 37, 684–694. [Google Scholar] [CrossRef]
- Ahmadi, A.; Zorofchian Moghadamtousi, S.; Abubakar, S.; Zandi, K. Antiviral potential of algae polysaccharides isolated from marine sources: A review. BioMed Res. Int. 2015, 2015, e825203. [Google Scholar] [CrossRef]
- Haubrich, B.A. Microbial Sterolomics as a Chemical Biology Tool. Molecules 2018, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.; Silva, J.; Mendonça, S.H.; Abreu, M.H.; Domingues, M.R. Lipidomic approaches towards deciphering glycolipids from microalgae as a reservoir of bioactive lipids. Mar. Drugs 2016, 14, 101. [Google Scholar] [CrossRef]
- Kohli, G.S.; John, U.; Van Dolah, F.M.; Murray, S.A. Evolutionary distinctiveness of fatty acid and polyketide synthesis in eukaryotes. ISME J. 2016, 10, 1877–1890. [Google Scholar] [CrossRef] [PubMed]
- Boukhris, S.; Athmouni, K.; Hamza-mnif, I.; Siala-elleuch, R.; Ayadi, H.; Nasri, M.; Sellami-kamoun, A. The Potential of a brown microalga cultivated in high salt medium for the production of high-value compounds. BioMed Res. Int. 2017, 2017, e4018562. [Google Scholar] [CrossRef] [PubMed]
- Vaz, B.; da, S.; Moreira, J.B.; de Morais, M.G.; Costa, J.A.V. Microalgae as a new source of bioactive compounds in food supplements. Curr. Opin. Food Sci. 2016, 7, 73–77. [Google Scholar] [CrossRef]
- Villarruel-López, A.; Ascencio, F.; Nuño, K. Microalgae, a Potential Natural Functional Food Source—A Review. Pol. J. Food Nutr. Sci. 2017, 67, 251–263. [Google Scholar] [CrossRef]
- Maciel, E.; Leal, M.C.; Lillebø, A.I.; Domingues, P.; Domingues, M.R.; Calado, R. Bioprospecting of marine macrophytes using ms-based lipidomics as a new approach. Mar Drugs 2016, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Deshnium, P.; Paithoonrangsarid, K.; Suphatrakul, A.; Meesapyodsuk, D.; Tanticharoen, M.; Cheevadhanarak, S. Temperature-independent and -dependent expression of desaturase genes in filamentous cyanobacterium Spirulina platensis strain C1 (Arthrospira sp. PCC 9438). FEMS Microbiol. Lett. 2000, 184, 207–213. [Google Scholar] [CrossRef]
- Stonik, V.A.; Stonik, I.V. Sterol and sphingoid glycoconjugates from microalgae. Mar Drugs 2018, 16, 554. [Google Scholar] [CrossRef]
- Lauritano, C.; Ferrante, M.I.; Rogato, A. Marine Natural Products from Microalgae. An Omics Overview. Mar. Drugs 2019, 17, 269. [Google Scholar] [CrossRef]
- Zeng, C.; Wen, B.; Hou, G.; Lei, L.; Mei, Z.; Jia, X.; Chen, X.; Zhu, W.; Li, J.; Kuang, Y.; et al. Lipidomics profiling reveals the role of glycerophospholipid metabolism in psoriasis. Gigascience 2017, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Manzo, E.; Cutignano, A.; Pagano, D.; Gallo, C.; Barra, G.; Nuzzo, G.; Sansone, C.; Ianora, A.; Urbanek, K.; Fenoglio, D.; et al. A new marine-derived sulfoglycolipid triggers dendritic cell activation and immune adjuvant response. Sci. Rep. 2017, 7, e6286. [Google Scholar] [CrossRef] [PubMed]
- Morgese, M.G.; Mhillaj, E.; Francavilla, M.; Bove, M.; Morgano, L.; Tucci, P.; Trabace, L.; Schiavone, S. Chlorella sorokiniana Extract Improves Short-Term Memory in Rats. Molecues 2016, 21, 1311. [Google Scholar] [CrossRef]
- Blondeau, N.; Lipsky, R.H.; Bourourou, M.; Duncan, M.W.; Gorelick, P.B.; Marini, A.M. Alpha-Linolenic Acid: An Omega-3 Fatty Acid with Neuroprotective Properties—Ready for Use in the Stroke Clinic? Biomed Res Int. 2015, 2015, 519830. [Google Scholar] [CrossRef] [PubMed]
- Salvador-reyes, L.A.; Luesch, H. Biological targets and mechanisms of action of natural products from marine cyanobacteria. Nat. Prod. Rep. 2015, 32, 478–503. [Google Scholar] [CrossRef]
- Matthews, H.; Hanison, J.; Nirmalan, N. “Omics”-Informed drug and biomarker discovery: Opportunities, challenges and future perspectives. Proteomics 2016, 4, 28. [Google Scholar] [CrossRef]
- Tuyiringire, N.; Tusubira, D.; Munyampundu, J.P.; Tolo, C.U.; Muvunyi, C.M.; Ogwang, P.E. Application of metabolomics to drug discovery and understanding the mechanisms of action of medicinal plants with anti-tuberculosis activity. Clin. Transl. Med. 2018, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Applications of metabolomics in drug discovery and development. Drugs RD 2008, 9, 307–322. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; Purdy, J.G.; Vastag, L.; Shenk, T.; Koyuncu, E. Metabolomics in drug target discovery. Cold Spring Harb. Symp. Quant. Biol. 2011, 76, 235–246. [Google Scholar] [CrossRef]
- Culf, A.S. Applied metabolomics in drug discovery. Expert Opin Drug Discov. 2016, 11, 759–770. [Google Scholar] [CrossRef]
- Puddick, J.; Prinsep, M.R. MALDI-TOF mass spectrometry of cyanobacteria: A global approach to the discovery of novel secondary metabolites. Chem NZ 2008, 72, 68–71. [Google Scholar]
- Rai, V.; Muthuraj, M.; Gandhi, M.N.; Das, D. Real-time iTRAQ-based proteome profiling revealed the central metabolism involved in nitrogen starvation induced lipid accumulation in microalgae. Nat. Publ. Gr. 2017, 7, e45732. [Google Scholar] [CrossRef] [PubMed]
- Salv, F.; Jae, D.; Karlsson, A.; Id, F.B.; Kristiansson, E.; Moore, E.R.B. Proteotyping bacteria: Characterization, differentiation and identification of pneumococcus and other species within the Mitis Group of the genus Streptococcus by tandem mass spectrometry proteomics. PLoS ONE 2018, 13, e0208804. [Google Scholar] [CrossRef]
- Fenselau, C. Rapid characterizaion of microorganisms by mass spectrometry. What can be learned and how? J. Am. Soc. Mass Spectrom. 2013, 24, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Marvin, L.F.; Roberts, M.A.; Fay, L.B. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in clinical chemistry. Clin. Chim. Acta 2003, 337, 11–21. [Google Scholar] [CrossRef]
- El-Aneed, A.; Cohen, A.; Banoub, J. Mass spectrometry, review of the basics: Electrospray, MALDI, and commonly used mass analyzers. Appl. Spectrosc. Rev. 2009, 44, 210–230. [Google Scholar] [CrossRef]
- Wang, H.; Yang, Y.; Chen, W.; Ding, L.; Li, P.; Zhao, X.; Wang, X.; Li, A.; Bao, Q. Identification of differentially expressed proteins of Arthrospira (Spirulina) plantensis-YZ under salt-stress conditions by proteomics and qRT-PCR analysis. Proteome Sci. 2013, 11, 6. [Google Scholar] [CrossRef]
- Jahoda, E.; Raus, M.; Has, P. Intact cell MALDI-TOF mass spectrometric analysis of Chroococcidiopsis cyanobacteria for classification purposes and identification of possible marker proteins. PLoS ONE 2018, 13, e0208275. [Google Scholar] [CrossRef]
- Sun, L.; Jiang, W.; Sato, H.; Kawachi, M.; Lu, X. Rapid classification and identification of microcystis aeruginosa strains using MALDI—TOF MS and polygenetic analysis. PLoS ONE 2016, 11, e0170637. [Google Scholar] [CrossRef][Green Version]
- Johnston, S.; Rees, E.; Thomas, I.; Jones, C.; Reid, M.; Davies, A.P.; Harris, L.G.; Mack, D.; Johnston, S.; Rees, E.; et al. Comparison of bacterial identification by MALDI- TOF mass spectrometry and conventional diagnostic microbiology methods. Br. J. Biomed. 2018, 4845, 47–55. [Google Scholar]
- Esquenazi, E.; Coates, C.; Simmons, L.; Gonzalez, D.; Gerwick, W.H.; Dorrestein, P.C. Visualizing the spatial distribution of secondary metabolites produced by marine cyanobacteria and sponges via MALDI-TOF imaging. Mol. Biosyst. Mol. Biosyst. 2008, 4, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Dashti, Y.; Grkovic, T.; Abdelmohsen, U.R.; Hentschel, U.; Quinn, R.J. Actinomycete metabolome induction/suppression with N-acetylglucosamine. J. Nat. Prod. 2017, 80, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Gautam, K.; Tripathi, J.K.; Pareek, A.; Sharma, D.K. Growth and secretome analysis of possible synergistic interaction between green algae and cyanobacteria. J. Biosci. Bioeng. 2018, 127, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Neubauer, P. Escherichia coli as a cell factory for heterologous production of nonribosomal peptides and polyketides. New Biotechnol. 2014, 31, 579–585. [Google Scholar] [CrossRef]
- Huo, L.; Hug, J.J.; Fu, C.; Bian, X.; Zhang, Y.; Müller, R. Natural Product Reports Heterologous expression of bacterial natural product biosynthetic pathways. Nat. Prod. Rep. 2019, 36, 1412–1436. [Google Scholar] [CrossRef]
- Said, J.S.; Trybala, E.; Görander, S.; Ekblad, M.; Liljeqvist, J.Å.; Jennische, E.; Lange, S.; Bergström, T. The cholestanol-conjugated sulfated oligosaccharide PG545 disrupts the lipid envelope of herpes simplex virus particles. Antimicrob. Agents Chemother. 2016, 60, 1049–1057. [Google Scholar] [CrossRef]
- Yang, F.; Tang, Q.; Zhong, X.; Bai, Y.; Chen, T.; Zhang, Y.; Li, Y.; Zheng, W. Surface decoration by Spirulina polysaccharide enhances the cellular uptake and anticancer efficacy of selenium nanoparticles. Int. J. Nanomed. 2012, 7, 835–844. [Google Scholar] [CrossRef]
- Blin, K.; Medema, M.H.; Kottmann, R.; Lee, S.Y.; Weber, T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2016, 45, D555–D559. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. AntiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Schorn, M.; Zettler, J.; Noel, J.P.; Dorrestein, P.C.; Moore, B.S.; Kaysser, L. The genetic basis for the biosynthesis of the pharmaceutically important class of epoxyketone proteasome inhibitors. ACS Chem. Biol. 2014, 9, 301–309. [Google Scholar] [CrossRef]
- Sun, J.; Su, Y.; Wang, T. Expression, purification and identification of CtCVNH, a novel anti-HIV ( Human Immunodeficiency Virus ) protein from Ceratopteris thalictroides. Int. J. Mol. Sci. 2013, 14, 7506–7514. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Rather, M.A.; Dar, M.S.; Aga, M.A.; Ahmad, N.; Manzoor, A.; Qayum, A.; Shah, A.; Mushtaq, S.; Ahmad, Z.; et al. Novel bioactive molecules from Lentzea violacea strain AS 08 using one strain-many compounds (OSMAC) approach. Bioorg. Med. Chem. Lett. 2017, 27, 2579–2582. [Google Scholar] [CrossRef] [PubMed]
- Scherlach, K.; Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 2009, 7, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Bode, H.B.; Bethe, B.; Hˆfs, R.; Zeeck, A. Big effects from small changes: Possible ways to explore nature’s chemical diversity. Chembiochem 2002, 3, 619–627. [Google Scholar] [CrossRef]
- Kçnig, C.C.; Scherlach, K.; Schroeckh, V.; Horn, F.; Nietzsche, S.; Brakhage, A.A.; Hertweck, C. Bacterium induces cryptic meroterpenoid pathway in the pathogenic fungus Aspergillus fumigatus. ChemBioChem 2013, 14, 938–942. [Google Scholar] [CrossRef]
- Abdelwahab, M.F.; Kurtán, T.; Mándi, A.; Müller, W.E.G.; Fouad, M.A.; Kamel, M.S.; Liu, Z.; Ebrahim, W.; Daletos, G.; Proksch, P. Induced secondary metabolites from the endophytic fungus Aspergillus versicolor through bacterial co-culture and OSMAC approaches. Tetrahedron Lett. 2018, 59, 2647–2652. [Google Scholar] [CrossRef]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H.; Aon, J.C. Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: A literature review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef]
- Romano, S. Extending the “one strain many compounds” (OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef]
- Eckelmann, D.; Spiteller, M.; Kusari, S. Spatial-temporal profiling of prodiginines and serratamolides produced by endophytic Serratia marcescens harbored in Maytenus serrata. Sci. Rep. 2018, 8, 5283. [Google Scholar] [CrossRef]
- Kusari, S.; Lamshoeft, M.; Kusari, P.; Gottfried, S.; Zuehlke, S.; Louven, K.; Hentschel, U.; Kayser, O.; Spiteller, M. Endophytes are hidden producers of maytansine in Putterlickia roots. J. Nat. Prod. 2014, 77, 2577–2584. [Google Scholar] [CrossRef]
- Wang, W.L.; Xu, S.Y.; Ren, Z.G.; Tao, L.; Jiang, J.W.; Zheng, S. Application of metagenomics in the human gut microbiome. World J. Gastroenterol. 2015, 21, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Gadea, A.; Le Lamer, A.; Le Gall, S.; Jonard, C.; Ferron, S.; Catheline, D.; Ertz, D.; Le Pogam, P.; Boustie, J.; Lohézic, F.; et al. Intrathalline metabolite profiles in the lichen Argopsis friesiana shape gastropod grazing patterns. J. Chem. Ecol. 2018, 44, 471–482. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, L.S.; Gregoracci, G.B.; Gueiros, G.; Silva, Z.; Salgado, L.T.; Filho, G.A.; Alves-ferreira, M.; Pereira, R.C.; Thompson, F.L. Transcriptomic analysis of the red seaweed Laurencia dendroidea (Florideophyceae, Rhodophyta) and its microbiome. BMC Genom. 2012, 13, 487. [Google Scholar] [CrossRef] [PubMed]
- Segev, E.; Wyche, T.P.; Kim, K.H.; Ellebrandt, C.; Vlamakis, H.; Barteneva, N.; Paulson, J.N.; Chai, L.; Clardy, J.; Kolter, R. Dynamic metabolic exchange governs a marine algal-bacterial interaction. eLife 2016, 5, e17473. [Google Scholar] [CrossRef] [PubMed]
- Aiyar, P.; Schaeme, D.; García-Altares, M.; Carrasco Flores, D.; Dathe, H.; Hertweck, C.; Sasso, S.; Mittag, M. Antagonistic bacteria disrupt calcium homeostasis and immobilize algal cells. Nat. Commun. 2017, 8, 1756. [Google Scholar] [CrossRef]
- Salim, A.; Chin, Y.; Kinghorn, A. Bioactive Molecules and Medicinal Plants; Ramawat, K., Merillon, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 1–24. [Google Scholar] [CrossRef]
- Shoemaker, R.H. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef]
- Cong, F.; Cheung, A.K.; Huang, S.A. Chemical Genetics-Based Target Identification in Drug Discovery. Annu. Rev. Pharmacol. 2012, 52, 57–78. [Google Scholar] [CrossRef]
- Sundarrajan, S.; Lulu, S.; Arumugam, M. Computational evaluation of phytocompounds for combating drug resistant tuberculosis by multi-targeted therapy. J. Mol. Model 2015, 21, 247. [Google Scholar] [CrossRef]
- Pereira, F.; Aires-de-Sousa, J. Computational methodologies in the exploration of marine natural product leads. Mar. Drugs 2018, 16, 236. [Google Scholar] [CrossRef]
- Paiva, F.C.R.; Ferreira, G.M.; Trossini, G.H.G.; Pinto, E. Identification, In vitro testing and molecular docking studies of microginins’ mechanism of angiotensin-converting enzyme inhibition. Molecules 2017, 22, 1884. [Google Scholar] [CrossRef]
- Verma, E.; Mishra, A.K.; Singh, A.K.; Singh, V.K. Structural elucidation and molecular docking of a novel antibiotic compound from cyanobacterium Nostoc MGL001. Front. Microbiol. 2016, 7, 1899. [Google Scholar] [CrossRef]
- Gkelis, S.; Panou, M.; Konstantinou, D.; Apostolidis, P.; Kasampali, A.; Papadimitriou, S.; Kati, D.; Di Lorenzo, G.M.; Ioakeim, S.; Zervou, S.K.; et al. Diversity, cyanotoxin production, and bioactivities of cyanobacteria isolated from freshwaters of greece. Toxins (Basel) 2019, 11, 436. [Google Scholar] [CrossRef]
- Mudimu, O.; Rybalka, N.; Bauersachs, T.; Born, J.; Friedl, T.; Schulz, R. Biotechnological screening of microalgal and cyanobacterial strains for biogas production and antibacterial and antifungal effects. Metabolites 2014, 4, 373–393. [Google Scholar] [CrossRef] [PubMed]
- Lauritano, C.; Andersen, J.H.; Hansen, E.; Albrigtsen, M.; Escalera, L.; Esposito, F.; Helland, K.; Hanssen, K.Ø.; Romano, G.; Ianora, A. Bioactivity screening of microalgae for antioxidant, anti-inflammatory, anticancer, anti-diabetes, and antibacterial activities. Front. Mar. Sci. 2016, 3, 68. [Google Scholar] [CrossRef]
- Koleva, M.N.; Fernandez-ballester, G. In silico approaches for TRP channel modulation. Methods Mol. Biol. 2019, 1987, 187–206. [Google Scholar] [CrossRef]
- Rehman, N.U.; Rafiq, K.; Khan, A.; Halim, S.A.; Ali, L.; Al-Saady, N.; Al-Balushi, A.H.; Al-Busaidi, H.K.; Al-Harrasi, A. α-Glucosidase inhibition and molecular docking studies of natural brominated metabolites from marine macro brown alga Dictyopteris hoytii. Mar. Drugs 2019, 17, 666. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.R.; Standl, E.; Tong, N.; Shah, P.; Kalra, S.; Rathod, R. Therapeutic potential of α-glucosidase inhibitors in type 2 diabetes mellitus: An evidence-based review. Expert Opin. Pharm. 2015, 16, 1959–1981. [Google Scholar] [CrossRef] [PubMed]
- Chapel, C.; Garcia, C.; Roingeard, P.; Zitzmann, N.; Dubuisson, J.; Dwek, R.A.; Trépo, C.; Zoulim, F.; Durantel, D. Antiviral effect of α-glucosidase inhibitors on viral morphogenesis and binding properties of hepatitis C virus-like particles. J. Gen. Virol. 2006, 87, 861–871. [Google Scholar] [CrossRef]
- Davis, G.D.J.; Vasanthi, A.H.R. QSAR based docking studies of marine algal anticancer compounds as inhibitors of protein kinase B (PKBβ). Eur. J. Pharm. Sci. 2015, 76, 110–118. [Google Scholar] [CrossRef]
- Montone, C.M.; Capriotti, A.L.; Cavaliere, C.; La Barbera, G.; Piovesana, S.; Chiozzi, R.Z.; Laganà, A. Peptidomic strategy for purification and identification of potential ACE-inhibitory and antioxidant peptides in Tetradesmus obliquus microalgae. Anal. Bioanal. Chem. 2018, 410, 3573–3586. [Google Scholar] [CrossRef]
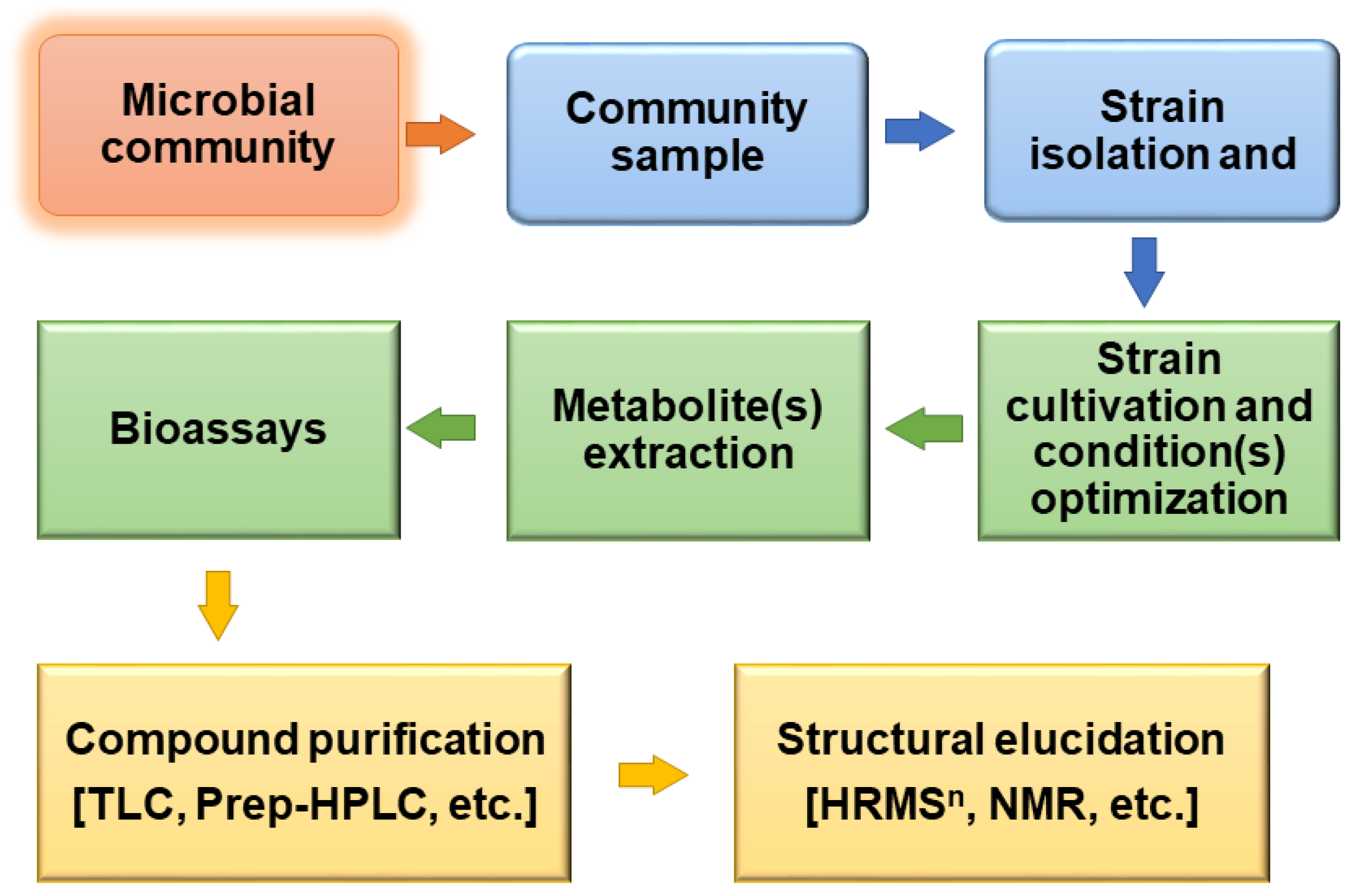


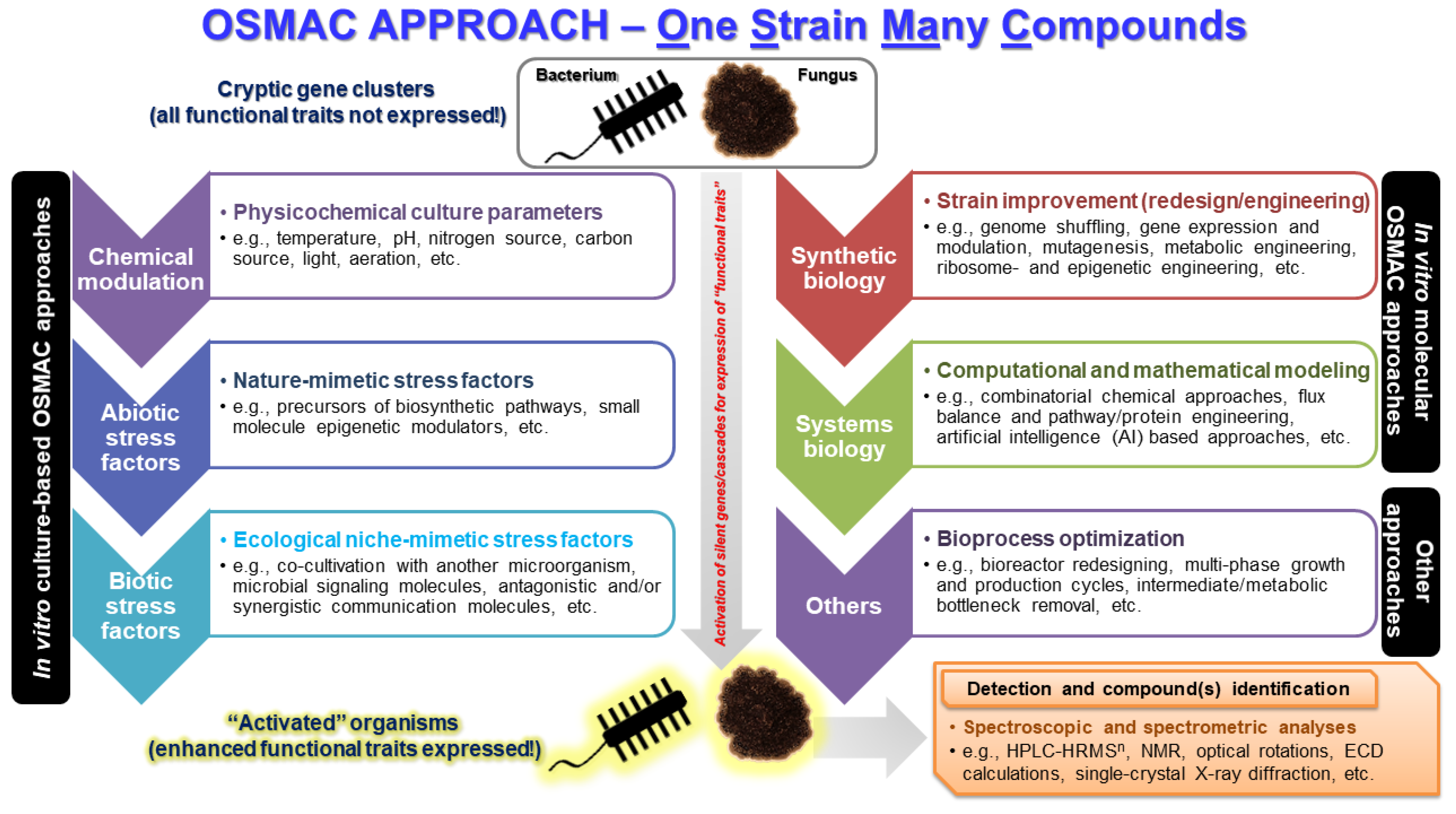
| Strain | Domain | Phylum | Genome Size | Reference |
|---|---|---|---|---|
| Arthrospira platensis | Prokaryota | Cyanobacteria | 6.0 Mb | [53] |
| Arthrospira platensis | Prokaryota | Cyanobacteria | 6.62 Mb | [59] |
| Arthrospira maxima | Prokaryota | Cyanobacteria | 6.0 Mb | NCBI |
| Chlorella sp. A99 | Eukaryota | Chlorophyta | 40.934 | NCBI |
| Chlorella vulgaris UTEX 395 | Eukaryota | Chlorophyta | 37.34 Mb | [62] |
| Oscillatoria nigro-viridis PCC 7112 | Prokaryota | Cyanobacteria | 7.97 Mb | [58] |
| Streptomyces lividans TK24 | Prokaryota | Actinobacteria | 8.345 Mbp | [61] |
| Euzebya sp. DY32-46 | Prokaryota | Actinobacteria | 5.715 Mb | [63] |
| Geobacillus sp. ZGt-1 | Prokaryota | Firmicutes | 3.7 Mb | [37] |
| Lipid | Source Microorganisms | Bioactivity | Reference |
|---|---|---|---|
| Sulfoquinovosyldiacyl glycerol (SQDG) | Spirulina spp., Chlorella spp., Pavlova lutheri | Antiviral and immunomodulatory | [38,94,103] |
| Sulfoquinovosylmonoacyl glycerol (SQMG) | Spirulina spp., Chlorella spp., Pavlova lutheri | Antiviral and immunomodulatory | [94,104] |
| Gamma-linoleic acid | Arthrospira spp., Chlorella spp. | Immunomodulatory and neurological | [105] |
| Alpha-linoleic acid | Arthrospira spp., Chlorella spp. | Neuroprotective | [106] |
| Kalkitoxin | Lyngbya majuscula | Neurotoxin | [107] |
| Antillatoxin | Lyngbya majuscula | Neurotoxin | [107] |
| Metabolite | Metabolite Classes | Gene | Source Microorganism | Bioactivities | Factory | Reference |
|---|---|---|---|---|---|---|
| Lyngbyatoxin | NRP | NRPS | Lyngbya majuscula | Anticancer | E. coli | [18] |
| Epoxomicin | NRP/PK | NRPS/PKS complex | S. hygroscopicus ATCC 53904 | Anti-inflammatory, Anticancer, Antiplasmodium | S. albus J1046 | [132] |
| Eponemycin | NRP | NRPS/PKS complex | S. hygroscopicus ATCC 53709 | Anti-inflammatory, Anticancer, Antiplasmodium | S. albus J1046 | [132] |
| Cyanovirin N | RP | RiPPs | Nostoc ellipsosporum | Antiviral | E. coli | [133] |
| Oleandomycin | PKS | OlePKS | Streptomyces antibioticus | Antibacterial | Saccharopolyspora erythraea | [74] |
| Cinnamycin | RP | RiPPs | Antibiotic | S. albus | [130] |
| Tool | Database/Software | Application | URL |
|---|---|---|---|
| DrugBank | Drug Database | Pharmacological assessment of compounds through search | https://www.drugbank.ca |
| BinBase | Metabolomic database | Similarity search for metabolites | http://fiehnlab.ucdavis.edu/projects/binbase_setupx#binbase |
| MetaboLights database | Metabolomic database | Search for metabolites | https://www.ebi.ac.uk/metabolights/index |
| HMDB | Metabolomic database | Clinical chemistry, biomarker discovery and general education | http://www.hmdb.ca/ |
| Click2Drug | Browser/Database | Search for integrated tools for CADD | https://www.click2drug.org/ |
| PubChem | Database | Chemical molecule search | https://pubchem.ncbi.nlm.nih.gov/ |
| SciFinder | Database | Chemical molecule search | https://sso.cas.org/pf/metadata.ping |
| LiSiCA | Software | Searches for 2D and 3D similarities between a reference compound and a database of target compounds | http://insilab.org/lisica/ |
| MedChem Studio | Software | Data visualization, compound clustering, high throughput screening analysis, lead identification and prioritization, de novo design, scaffold hopping, lead optimization | https://www.simulations-plus.com/software/admetpredictor/medchem-studio/ |
| PyRx | Software | Virtual Screening for Computational Drug Discovery, target screening | https://pyrx.sourceforge.io/ |
| CRISPRdisco | Software | Identification of CRISPR repeat-spacer arrays and cas genes in genome data sets | https://github.com/crisprlab/CRISPRdisco |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maghembe, R.; Damian, D.; Makaranga, A.; Nyandoro, S.S.; Lyantagaye, S.L.; Kusari, S.; Hatti-Kaul, R. Omics for Bioprospecting and Drug Discovery from Bacteria and Microalgae. Antibiotics 2020, 9, 229. https://doi.org/10.3390/antibiotics9050229
Maghembe R, Damian D, Makaranga A, Nyandoro SS, Lyantagaye SL, Kusari S, Hatti-Kaul R. Omics for Bioprospecting and Drug Discovery from Bacteria and Microalgae. Antibiotics. 2020; 9(5):229. https://doi.org/10.3390/antibiotics9050229
Chicago/Turabian StyleMaghembe, Reuben, Donath Damian, Abdalah Makaranga, Stephen Samwel Nyandoro, Sylvester Leonard Lyantagaye, Souvik Kusari, and Rajni Hatti-Kaul. 2020. "Omics for Bioprospecting and Drug Discovery from Bacteria and Microalgae" Antibiotics 9, no. 5: 229. https://doi.org/10.3390/antibiotics9050229
APA StyleMaghembe, R., Damian, D., Makaranga, A., Nyandoro, S. S., Lyantagaye, S. L., Kusari, S., & Hatti-Kaul, R. (2020). Omics for Bioprospecting and Drug Discovery from Bacteria and Microalgae. Antibiotics, 9(5), 229. https://doi.org/10.3390/antibiotics9050229







