Genotypic and Phenotypic Evaluation of Biofilm Production and Antimicrobial Resistance in Staphylococcus aureus Isolated from Milk, North West Province, South Africa
Abstract
1. Introduction
2. Results
2.1. Isolation of S. aureus
2.2. Phenotypic Biofilm Production
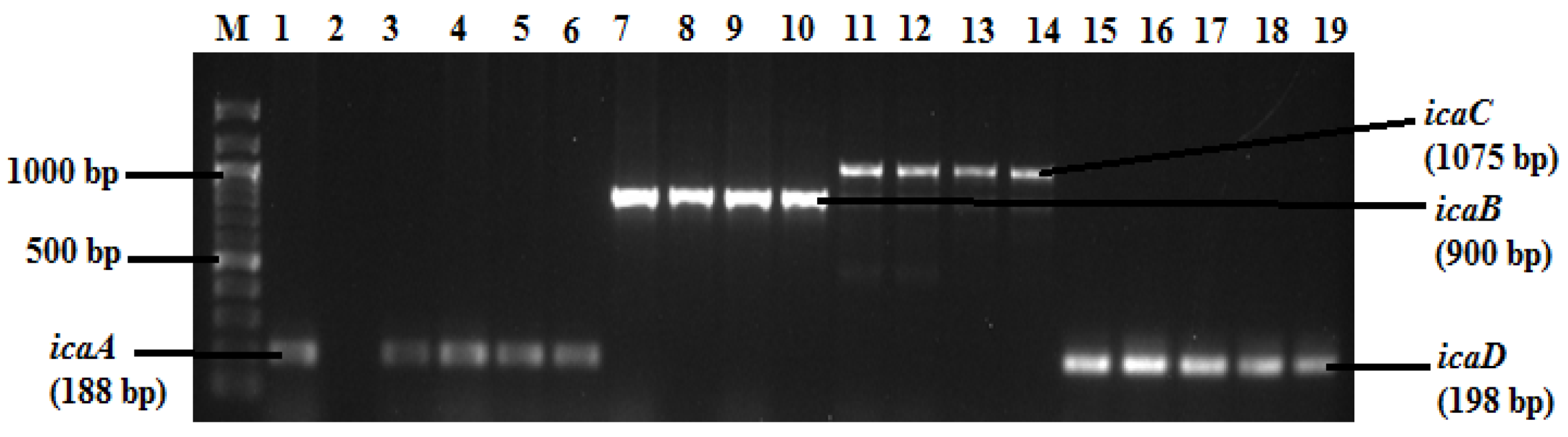
2.3. Detection of Biofilm Genes
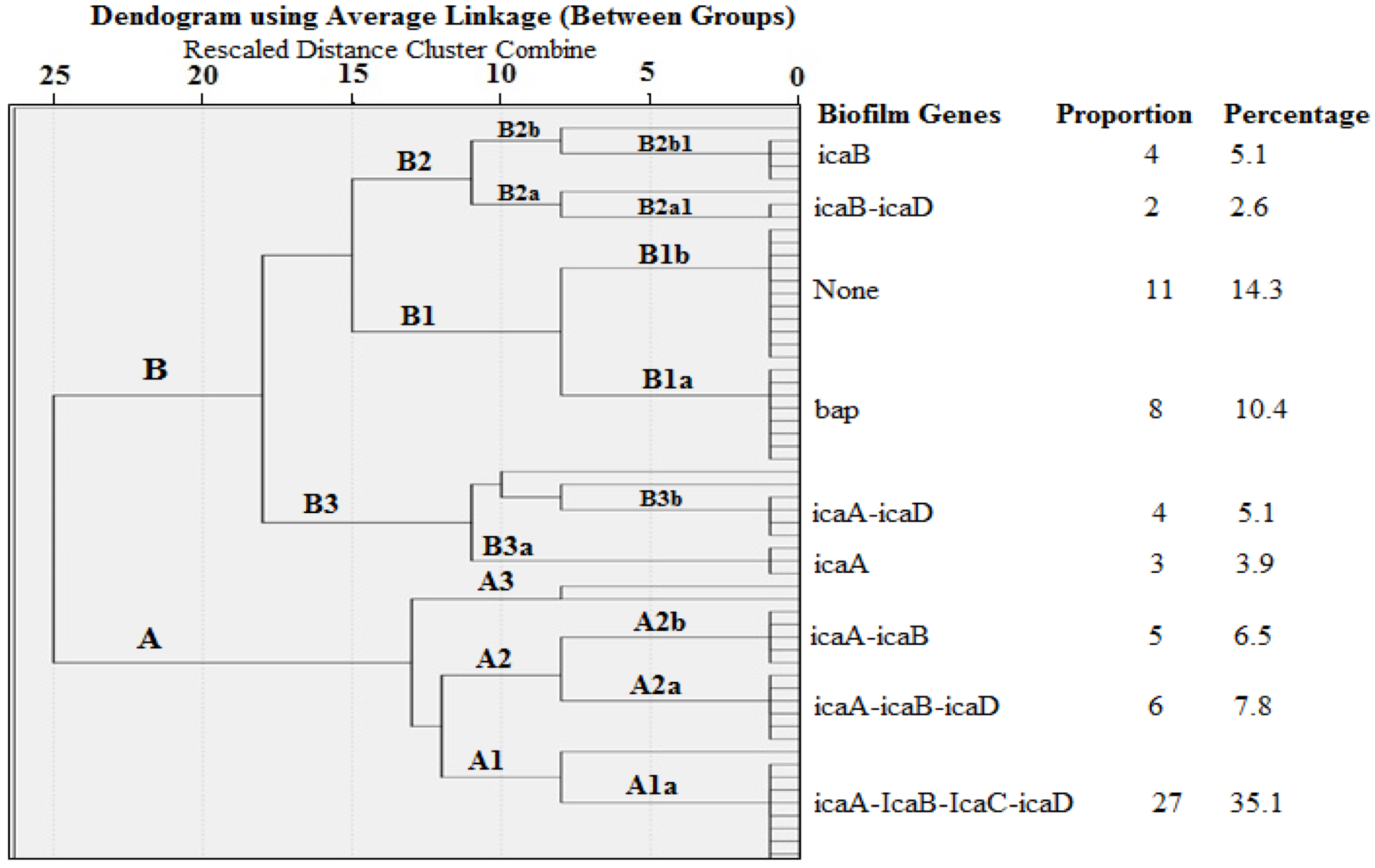
2.4. Cluster Analysis of Biofilm Genes in S. aureus Isolated from Milk
2.5. Relationship between Biofilm Formation and Biofilm Genes in S. aureus Isolates
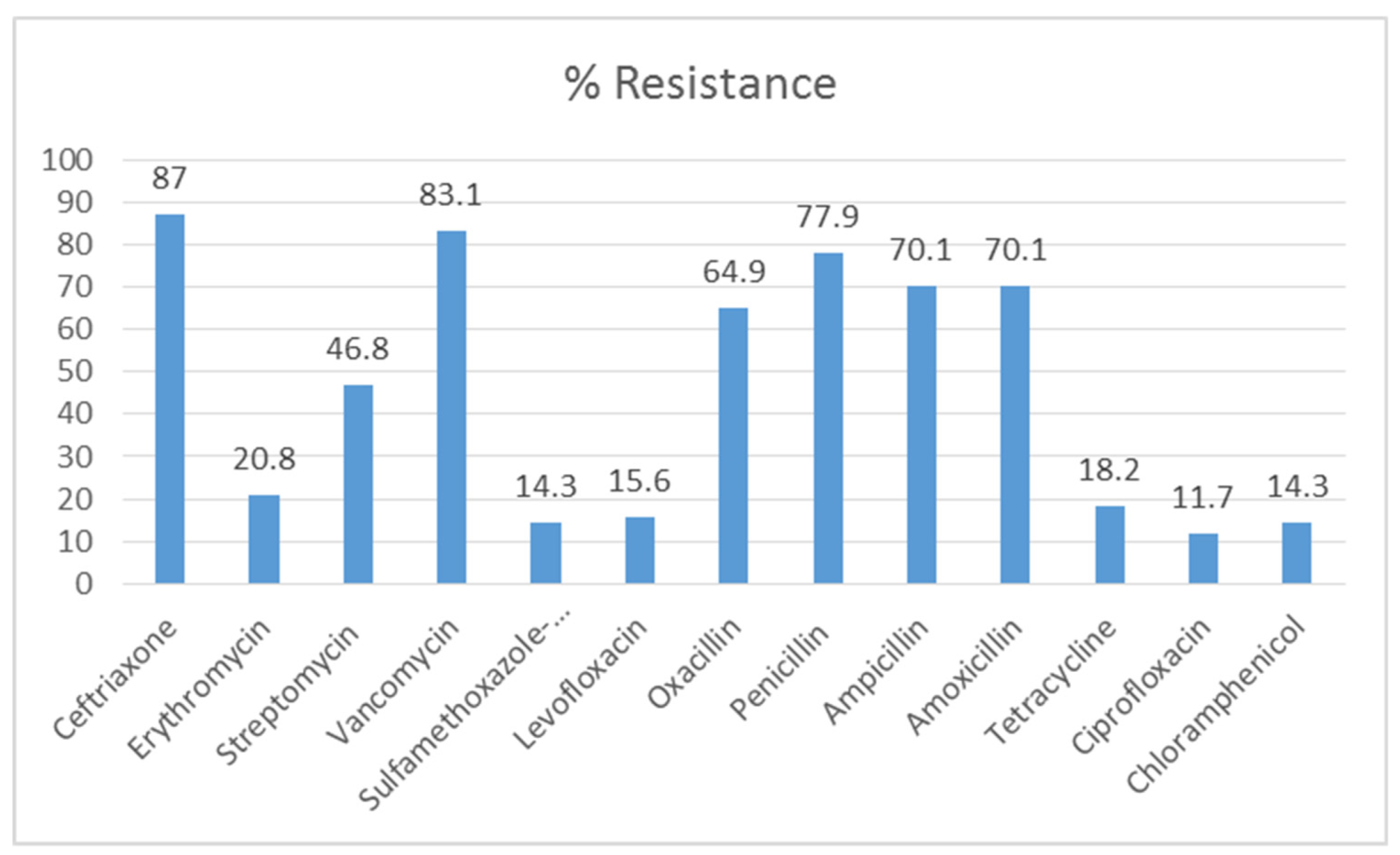
2.6. Antibiotic Susceptibility Test (AST)
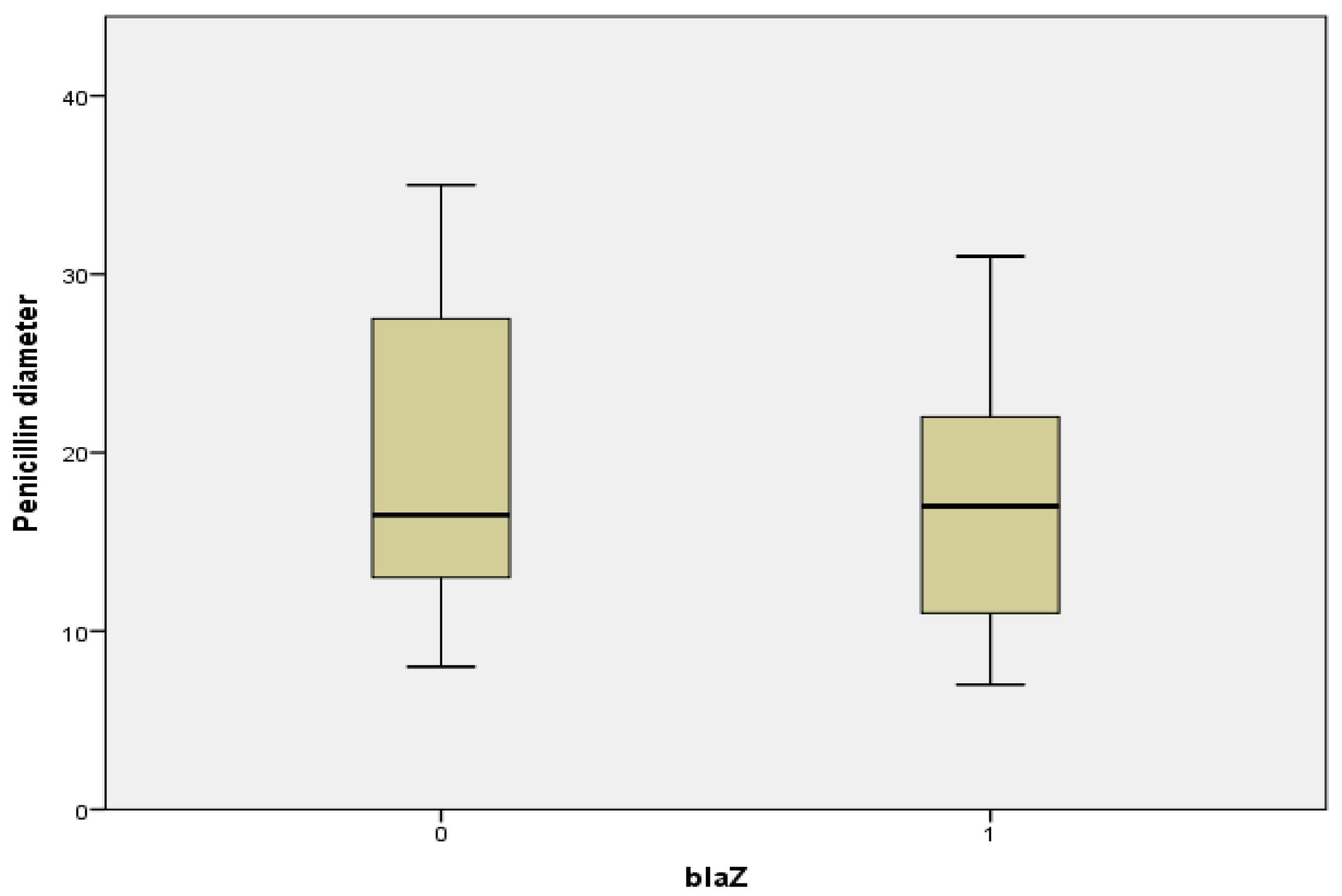
2.7. Relationship between Penicillin Diameter, blaZ Genes, and Beta Lactamase Production
2.8. Profiling of Biofilm Formation and Resistance Patterns of MDR S. aureus Isolates
3. Discussion
4. Materials and Methods
4.1. Study Site
4.2. Sample Collection
4.3. Isolation and Identification of Staphylococcus aureus Isolates
4.4. PCR Detection of Biofilm and Antimicrobial Resistance Genes
4.5. Phenotypic Biofilm Production
4.6. Antibiotic Susceptibility Testing (AST)
4.7. Phenotypic Evaluation of Beta-Lactamase Production and Inducible Clindamycin Resistance
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kloos, W.E. Natural populations of the genus Staphylococcus. Annu. Rev. Microbiol. 1980, 34, 559–592. [Google Scholar] [CrossRef]
- Foster, T.J. Staphylococcus aureus. In Molecular Medical Microbiology, 2nd ed.; Sussmann, M., Newcastle, U.T., Eds.; Academic Press: Cambridge, MA, USA, 2002; pp. 839–888. [Google Scholar]
- Hermans, K.; Devriese, L.A.; Haesebrouck, F. Staphylococcus. In Pathogenesis of Bacterial Infections in Animals, 3rd ed.; Gyles, C.L., Songer, J.G., Thoen, C.O., Eds.; Blackwell Publishing: Hoboken, NJ, USA, 2004; pp. 43–56. [Google Scholar]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Le Loir, Y.; Baron, F.; Gautier, M. Staphylococcus aureus and food poisoning. Gen. Mol. Res. 2003, 2, 63–76. [Google Scholar]
- Ferry, T.; Perpoint, T.; Vandenesch, F.; Etienne, J. Virulence determinants in Staphylococcus aureus and their involvement in clinical syndromes. Curr. Infect. Dis. Rep. 2005, 7, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 207–228. [Google Scholar] [PubMed]
- Periasamy, S.; Joo, H.-S.; Duong, A.C.; Bach, T.-H.L.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.C.; Otto, M. How Staphylococcus aureus biofilms develop their characteristic structure. PNAS 2012, 109, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- McKenney, D.; Ubner, J.H.; Muller, E.; Wang, Y.; Goldmann, D.A.; Pier, G.B. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesion. Infect. Immun. 1998, 66, 4711–4720. [Google Scholar] [CrossRef]
- Atshan, S.S.; Nor Shamsudin, M.; Sekawi, Z.; Lung, L.; Hamat, R.A.; Karunanidhi, A.; Ali, A.M.; Ghaznavi-Rad, E.; Moghaddam, H.G.; Seng, J.S.C.; et al. Prevalence of adhesion and regulation of biofilm-related genes in different clones of Staphylococcus aureus. J. Biomed. Biotech. 2012, 2012, 1–10. [Google Scholar]
- Cucarella, C.; Tormo, M.A.; Ubeda, C.; Trotonda, M.P.; Monzón, M.; Peris, C.; Amorena, B.; Lasa, I.; Penadés, J.R. Role of biofilm—Associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect. Immun. 2004, 72, 2177–2185. [Google Scholar] [CrossRef]
- Lassa, I.; Penades, J.R. A family of surface proteins involved in biofilm formation. Res. Microbiol. 2006, 157, 99–107. [Google Scholar] [CrossRef]
- Garcés, L. The Detrimental Impacts of Industrial Animal Agriculture: A Case for Humane and Sustainable Agriculture, Compassion in World Farming Trust. 2002. Available online: http://www.ciwf.org.uk/includes/documents/cm_docs/2008/d/detrimental_impact_industrial_animal_agriculture_2002.pdf (accessed on 12 January 2020).
- Girardini, L.K.; Paim, D.S.; Ausani, T.C.; Lopes, G.V.; Pellegrini, D.C.P.; Brito, M.A.V.P.; Cardoso, M. Antimicrobial resistance profiles of Staphylococcus aureus clusters on small dairy farms in southern Brazil. Pesq. Vet. Bras. 2016, 36, 951–956. [Google Scholar] [CrossRef][Green Version]
- Neopane, P.; Nepal, H.P.; Hrestha, R.; Uehara, O.; Abiko, Y. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int. J. Gen. Med. 2018, 11, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Henton, M.M.; Eagar, H.A.; Swan, G.E.; van Vuuren, M. Antibiotic management and resistance in livestock production. S. Afr. Med. J. 2011, 101, 583–586. [Google Scholar] [PubMed]
- Lim, D.; Strynadka, N.C. Structural basis for the ß-lactam resistance of PBP2a from methicillin—Resistant Staphylococcus aureus. Nat. Struct. Mol. Biol. 2002, 9, 870–876. [Google Scholar]
- Juhász-Kaszanyitzky, E.; Jánosi, S.; Somogyi, P.; Dán, A.; Bloois, L.G.; Van Duijkeren, E.; Wagenaar, J.A. MRSA transmission between cows and humans. Emerg. Infect. Dis. 2007, 13, 630–632. [Google Scholar] [CrossRef]
- Cosgrove, S.E. The relationship between antimicrobial resistance and patient outcomes: Mortality, length of hospital stay, and health care costs. Genet. Mol. Res. 2006, 42, S82–S89. [Google Scholar] [CrossRef]
- Lowy, F. Staphylococcal infections. In Harrison’s Principles of Internal Medicine; Fauci, A., Braunwald, E., Casper, D., Hauser, S., Longo, D., Jameson, J., Eds.; The McGraw-Hill Companies Inc.: New York, NY, USA, 2013; pp. 386–399. [Google Scholar]
- Akindolire, M.A.; Babalola, O.O.; Ateba, C.N. Detection of antibiotic resistant Staphylococcus aureus from milk: A public health implication. Int. J. Environ. Res. Public Health. 2015, 12, 10254–10275. [Google Scholar] [CrossRef]
- Ateba, C.N.; Mbewe, M.; Moneoang, M.S.; Bezuidenhout, C.C. Antibiotic-resistant Staphylococcus aureus isolated from milk in the Mafikeng Area, North West province, South Africa. S. Afr. J. Sci. 2010, 106. [Google Scholar] [CrossRef]
- Schmidt, T.; Kock, M.M.; Ehlers, M.M. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in south african dairy herds: Genetic diversity and inter-species host transmission. Front. Microbiol. 2017, 8, 1–15. [Google Scholar] [CrossRef]
- Lochem, S.V.; Thompson, P.N.; Annandale, C.H. Prevalence of MRSA among large commercial pig herds in South Africa. Onderstepoort J. Vet. Res. 2018, 85, 1–4. [Google Scholar]
- Akanbi, O.E.; Njom, H.A.; Fri, J.; Otigbu, A.C.; Clarke, A.M. Antimicrobial susceptibility of Staphylococcus aureus isolated from recreational waters and beach sand in eastern cape province of South Africa. Int. J. Environ. Res. Public Health. 2017, 14, 1001. [Google Scholar] [CrossRef] [PubMed]
- Fortuin-de Smidt, M.C.; Singh-Moodley, A.; Badat, R.; Quan, V.; Kularatne, R.; Nana, T.; Lekalaka, R.; Govender, N.P.; Perovic, O. Staphylococcus aureus bacteraemia in Gauteng academic hospitals, South Africa. Int. J. Infect. Dis. 2015, 30, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Naicker, P.R.; Karayem, K.; Hoek, K.G.; Harvey, J.; Wasserman, E. Biofilm formation in invasive Staphylococcus aureus isolates is associated with the clonal lineage. Microb. Pathog. 2016, 90, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, S.; Meng, L.; Dong, L.; Zhao, S.; Lan, X.; Wang, J.; Zheng, N. Prevalence, antimicrobial susceptibility, and molecular characterization of Staphylococcus aureus isolated from dairy herds in northern China. J. Dairy Sci. 2017, 100, 8796–8803. [Google Scholar] [CrossRef] [PubMed]
- Avila-Novoa, M.G.; Iñıguez-Moreno, M.; Solıs-Velazquez, O.A.; Gonzalez-Gomez, J.P.; Guerrero-Medina, P.J.; Gutierrez-Lomel, M. Biofilm Formation by Staphylococcus aureus isolated from Food Contact Surfaces in the Dairy Industry of Jalisco, Mexico. J. Food Qual. 2018, 1746139. [Google Scholar] [CrossRef]
- Nourbakhsh, F.; Namvar, A.E. Detection of genes involved in biofilm formation in Staphylococcus aureus isolates. GMS Hyg. Infect. Control 2016, 11. [Google Scholar] [CrossRef]
- Szweda, P.; Schielmann, M.; Milewski, S.; Jakubczak, A.; Frankowska, A. Biofilm production and presence of ica and bap genes in Staphylococcus aureus strains isolated from cows with Mastitis in the Eastern Poland. Pol. J. Microbiol. 2012, 61, 65–69. [Google Scholar] [CrossRef]
- Begum, H.A.; Uddin, M.S.; Islam, M.J.; Nazir, K.H.; Islam, M.A.; Rahman, M.T. Detection of biofilm producing coagulase positive Staphylococcus aureus from bovine mastitis, their pigment production, hemolytic activity and antibiotic sensitivity pattern. J. Bangladesh Soc. Agric. Sci. Technol. 2007, 4, 97–100. [Google Scholar]
- Marques, V.F.; Motta, C.C.; Soares, B.S.; Melo, D.A.; Coelho, S.M.O.; Coelho, I.S.; Barbosa, H.S. Biofilm production and beta-lactamic resistance in Brazilian Staphylococcus aureus isolates from bovine mastitis. Braz. J. Microbiol. 2017, 48, 118–124. [Google Scholar] [CrossRef]
- Wang, W.; Lin, X.; Jiang, T.; Peng, Z.; Xu, J.; Yi, L.; Li, F.; Fanning, S.; Baloch, Z. Prevalence and Characterization of Staphylococcus aureus Cultured From Raw Milk Taken From Dairy Cows With Mastitis in Beijing, China. Front. Microbiol. 2018, 9, 11–23. [Google Scholar] [CrossRef]
- Gowrishankar, S.; Kamaladevi, A.; Balamurugan, K.; Pandian, S.K. In Vitro and In Vivo Biofilm Characterization of Methicillin-Resistant Staphylococcus aureus from Patients Associated with Pharyngitis Infection. BioMed Res. Int. 2016, 2016, 1289157. [Google Scholar] [CrossRef]
- Li, L.; Yang, H.J.; Liu, D.C.; He, H.B.; Wang, C.F.; Zhang, J.F.; Gao, Y.D.; Yangjun, Z. Analysis of biofilm formation and associated genes detected in Staphylococcus isolates from bovine mastitis. Intern. J. Appl. Res. Vet. Med. 2012, 10, 62–68. [Google Scholar]
- Eagar, H.; Swan, G.; Van Vuuren, M. A survey of antimicrobial usage in animals in South Africa with specific reference to food animals. J. S. Afr. Vet Assoc. 2012, 83, 8. [Google Scholar] [CrossRef] [PubMed]
- Gardete, S.; Tomasz, A. Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Investig. 2014, 124, 2836–2840. [Google Scholar] [CrossRef] [PubMed]
- Bissong, M.E.A.; Wirgham, T.; Enekegbe, M.A.; Niba, P.T.N.; Foka, F.E.T. Prevalence and antibiotic susceptibility patterns of methicillin resistant Staphylococcus aureus in patients attending the Laquintinie Hospital Douala, Cameroon. Eur. J. Clin. Biomed. Sci. 2016, 2, 92–96. [Google Scholar] [CrossRef]
- Zehra, A.; Singh, R.; Kaur, S.; Gill, J.P.S. Molecular characterization of antibiotic-resistant Staphylococcus aureus from livestock (bovine and swine). Vet. World 2017, 10, 598–604. [Google Scholar] [CrossRef]
- Akpaka, P.E.; Roberts, R.; Monecke, S. Molecular characterization of antimicrobial resistance genes against Staphylococcus aureus isolates from Trinidad and Tobago. J. Infect. Public Health. 2017, 10, 316–323. [Google Scholar] [CrossRef]
- Ferreira, A.M.; Martins, K.B.; da Silva, V.R.; Mondelli, A.L.; de Souza da Cunha, M.L.R. Correlation of phenotypic tests with the presence of the blaZ gene for detection of beta-lactamase. Braz. J. Microbiol. 2017, 48, 159–162. [Google Scholar] [CrossRef]
- Kaase, M.; Lenga, S.; Friedrich, S.; Szabados, F.; Sakinc, T.; Kleine, B.; Gatermann, S.G. Comparison of phenotypic methods for penicillinase detection in Staphylococcus aureus. Clin. Microbiol. Infect. 2008, 4, 614–616. [Google Scholar] [CrossRef]
- El Feghaly, R.E.; Stamm, J.E.; Fritz, S.A.; Burnham, C.D. Presence of the blaZ beta-lactamase gene in isolates of Staphylococcus aureus that appear penicillin susceptible by conventional phenotypic methods. Diagn. Microbiol. Infect. Dis. 2012, 74, 388–393. [Google Scholar] [CrossRef]
- Brakstad, O.G.; Aasbakk, K.; Maeland, J.A. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 1992, 30, 1654–1660. [Google Scholar] [CrossRef] [PubMed]
- Alfatemi, S.M.H.; Motamedifar, M.; Hadi, N.; Ebrahim-Saraie, H.S. Analysis of virulence genes among methicillin resistant Staphylococcus aureus (MRSA) strains. Jundishapur J. Microbiol. 2014, 7, e10741. [Google Scholar]
- Strommenger, B.; Kettlitz, C.; Werner, G.; Witte, W. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 2003, 41, 4089–4094. [Google Scholar] [CrossRef] [PubMed]
- Huys, G.; D’Haene, K.; Van Eldere, J.; von Holy, A.; Swings, J. Molecular diversity and characterization of tetracycline-resistant Staphylococcus aureus isolates from a poultry processing plant. Appl. Environ. Microbiol. 2005, 71, 574–579. [Google Scholar] [CrossRef][Green Version]
- Ding, Z.F.; Zhang, H.; Tang, W.; Tong, C.Y.; Li, R.T.; Chen, L.X.; Pu, L.J.; Zhu, Z.B.; Cui, Y.D. Methylase genes-mediated erythromycin resistance in Staphylococcus aureus from bovine mastitis in China. Israel J. Vet. Med. 2012, 67, 170–179. [Google Scholar]
- Saha, B.; Singh, A.K.; Ghosh, A.; Bal, M. Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). J. Med. Microbiol. 2008, 57, 72–79. [Google Scholar] [CrossRef]
- Saadat, S.; Solhjoo, K.; Norooz-Nejad, M.J.; Kazemi, A. VanA and vanB positive vancomycin-resistant Staphylococcus aureus among clinical isolates in Shiraz, South of Iran. Oman Med. J. 2014, 29, 335. [Google Scholar] [CrossRef] [PubMed]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; CLSI supplement M100S; Wayne, P.A., Ed.; Clinical and Laboratory Standards Institute: West Valley Road, PA, USA, 2016. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 3rd ed.; CLSI document M31-A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008; ISBN 1-56238-659-X. [Google Scholar]
- Ghanbari, F.; Ghajavand, H.; Havaei, R.; Jami, M.-S.; Khademi, F.; Heydari, L.; Shahin, M.; Havaei, S.A. Distribution of erm genes among Staphylococcus aureus isolates with inducible resistance to clindamycin in Isfahan, Iran. Adv. Biomed. Res. 2016, 5, 62. [Google Scholar] [CrossRef]
| Proportion of Isolates in the Different Biofilm Forming Categories | Location | Total | P-Value | ||||
|---|---|---|---|---|---|---|---|
| Mafikeng | Rooidepand | Rooigrond | Zeerust | ||||
| Biofilm Formation | Moderate | 3 | 1 | 0 | 0 | 4 | |
| None | 3 | 1 | 2 | 1 | 7 | 0.526 | |
| Strong | 27 | 13 | 25 | 1 | 66 | ||
| Total | 33 (42.9) | 15 (19.5) | 27 (35.1) | 2 (2.5) | 77 | ||
| Biofilm Formation | Number (%) of Isolates Having the Biofilm Gene | ||||
|---|---|---|---|---|---|
| icaA | icaB | icaC | icaD | bap | |
| None (n = 7) | 4 (57.1) | 0 (0.0) | 0 (0.0) | 3 (42.9) | 1 (14.3) |
| Moderate (n = 4) | 3 (75.0) | 2 (50) | 3 (75.0) | 4 (100) | 1 (25) |
| Strong (n = 66) | 42 (63.6) | 46 (69.7) | 27 (40.9) | 36 (54.5) | 10 (15.1) |
| Total (n = 77) | 49 (63.6) | 48 (62.3) | 30 (38.9) | 43 (55.8) | 12 (15.6) |
| P-value | 0.777 | 0.027 | 0.259 | 0.674 | 0.084 |
| Phenotypic Characteristic | blaZ Gene (% *) | Total | ||
|---|---|---|---|---|
| Negative | Positive | |||
| Beta Lactamase Production | Negative | 17 (35.4) | 4 (13.8) | 21 |
| Positive | 31 (64.6) | 25 (86.2) | 56 | |
| Total | 48 | 29 | 77 | |
| MDR Isolate | Resistance Phenotype | Biofilm Production | Resistance Genes | Biofilm Genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bla Z | van C | tet K | tet L | Msr A/B | Ica Genes | bap | ||||||
| A | B | C | D | |||||||||
| Sum 1 | cef-str-van-oxa | strong | + | - | - | - | - | + | + | + | + | - |
| Ctrl 2 | cef-str-van-oxa | strong | + | + | + | - | - | + | + | + | + | - |
| XWL2 | cef-str-van-oxa | strong | + | + | + | + | - | + | + | + | + | - |
| RPN4L1 | cef-van-oxa | strong | + | - | - | - | - | - | - | - | - | + |
| RPN4L2 | cef-van-oxa | strong | + | + | - | - | - | + | + | + | - | - |
| RPN21 | cef-oxa-cip-chl | none | - | + | + | + | - | - | - | - | - | + |
| RPN3 | str-van-oxa-tet | strong | + | + | + | + | - | + | + | + | + | - |
| SUM 2 | cef-str-van-oxa | strong | + | - | + | - | - | + | + | + | + | - |
| RPN2L | cef-van-oxa-tet-chl | strong | - | - | - | - | - | + | + | + | + | - |
| RPN1L | cef-van-oxa | strong | + | - | - | - | - | + | - | - | - | - |
| RPN30 | cef-ery-str-van-lev-oxa-tet | strong | + | + | + | + | + | - | + | - | - | - |
| Fa1L | cef-ery-van-oxa-tet | moderate | - | - | + | + | - | - | - | - | - | + |
| Fa2S | cef-str-van-sul-lev-oxa-cip-chl | strong | - | - | - | - | - | - | + | - | - | - |
| Bb1LY2 | cef-van-oxa | strong | - | - | - | - | - | - | - | - | - | - |
| LIC3M | cef-van-oxa-cip-chl | strong | + | + | - | - | - | + | + | - | + | - |
| XY | cef-van-oxa | strong | + | + | + | + | - | - | - | - | - | + |
| LIC 1 | str-van-oxa-tet | strong | - | + | + | + | - | - | - | - | - | + |
| LIC 2M | cef-str-van | strong | - | - | - | - | - | + | + | - | + | - |
| KE18 | cef-oxa-tet | strong | - | - | + | + | + | - | - | - | - | - |
| K5 | cef-van-oxa | strong | + | + | + | + | - | + | + | - | + | - |
| K9 | cef-van-oxa | strong | - | - | - | - | + | + | + | - | + | - |
| K10 | cef-van-oxa | strong | + | - | - | - | - | + | + | - | + | - |
| K11 | cef-van-oxa | strong | + | + | + | + | - | - | + | - | + | - |
| K85 | cef-van-oxa | strong | + | + | - | - | - | + | + | + | + | - |
| K87 | cef-van-oxa | moderate | + | + | - | - | + | + | - | - | + | - |
| Gene | Primer Sequence | Amplicon Size (bp) | Reference |
|---|---|---|---|
| nuc | F-GCGATTGATGGTGGATACGGT R-AGCCAAGCCTTGACGAACTAAAGC | 279 | [45] |
| icaA | F-ACACTTGCTGGCGCAGTCAA R-TCTGGAACCAACATCCAACA | 188 | [46] |
| icaB | F-AGAATCGTGAAGTATAGAAAATT R-TCTAATCTTTTTCATGGAATCCGT | 900 | [46] |
| icaC | F-ATGGGACGGATTCCATGAAAAAGA R-TAATAAGCATTAATGTTCAATT | 1075 | [46] |
| icaD | F-ATGGTCAAGCCCAGACAGAG R-AGTATTTTCAATGTTTAAAGCAA | 198 | [46] |
| bap | F-CCCTATATCGAAGGTGTAGAATTGCAC R-GCTGTTGAAGTTAATACTGTACCTGC | 971 | [11] |
| blaZ | F-CAAAGATGATATAGTTGCTTATTCTCC R-TGCTTGACCACTTTTATCAGC | 421 | [43] |
| tetK | F-GTAGCGACAATAGGTAATAGT R-GTAGTGACAATAAACCTCCTA | 360 | [47] |
| tetL | F-GTCGTTGCGCGCTATATTCC R-GTGAACGGTAGCCCACCTAA | 696 | [48] |
| mefA | F-AGTATCATTAATCACTAGTGC R-TTCTTCTGGTACAAAAGTGG | 367 | [49] |
| msrA | F-CGATGAAGGAGGATTAAAATG R-CATGAATAGATTGTCCTGTTAATT | 1733 | [49] |
| vanA | F-ATGAATAGAATAAAAGTTGC R-TCACCCCTTTAACGCTAATA | 1032 | [50] |
| vanB | F-GTGACAAACCGGAGGCGAGGA R-CCGCCATCCTCCTGCAAAAAA | 430 | [51] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bissong, M.E.A.; Ateba, C.N. Genotypic and Phenotypic Evaluation of Biofilm Production and Antimicrobial Resistance in Staphylococcus aureus Isolated from Milk, North West Province, South Africa. Antibiotics 2020, 9, 156. https://doi.org/10.3390/antibiotics9040156
Bissong MEA, Ateba CN. Genotypic and Phenotypic Evaluation of Biofilm Production and Antimicrobial Resistance in Staphylococcus aureus Isolated from Milk, North West Province, South Africa. Antibiotics. 2020; 9(4):156. https://doi.org/10.3390/antibiotics9040156
Chicago/Turabian StyleBissong, Marie Ebob Agbortabot, and Collins Njie Ateba. 2020. "Genotypic and Phenotypic Evaluation of Biofilm Production and Antimicrobial Resistance in Staphylococcus aureus Isolated from Milk, North West Province, South Africa" Antibiotics 9, no. 4: 156. https://doi.org/10.3390/antibiotics9040156
APA StyleBissong, M. E. A., & Ateba, C. N. (2020). Genotypic and Phenotypic Evaluation of Biofilm Production and Antimicrobial Resistance in Staphylococcus aureus Isolated from Milk, North West Province, South Africa. Antibiotics, 9(4), 156. https://doi.org/10.3390/antibiotics9040156






