Antibiotic and Metal Resistance in Escherichia coli Isolated from Pig Slaughterhouses in the United Kingdom
Abstract
1. Introduction
2. Results
2.1. Isolate Recovery and EAggEC Genes Re-Confirmation
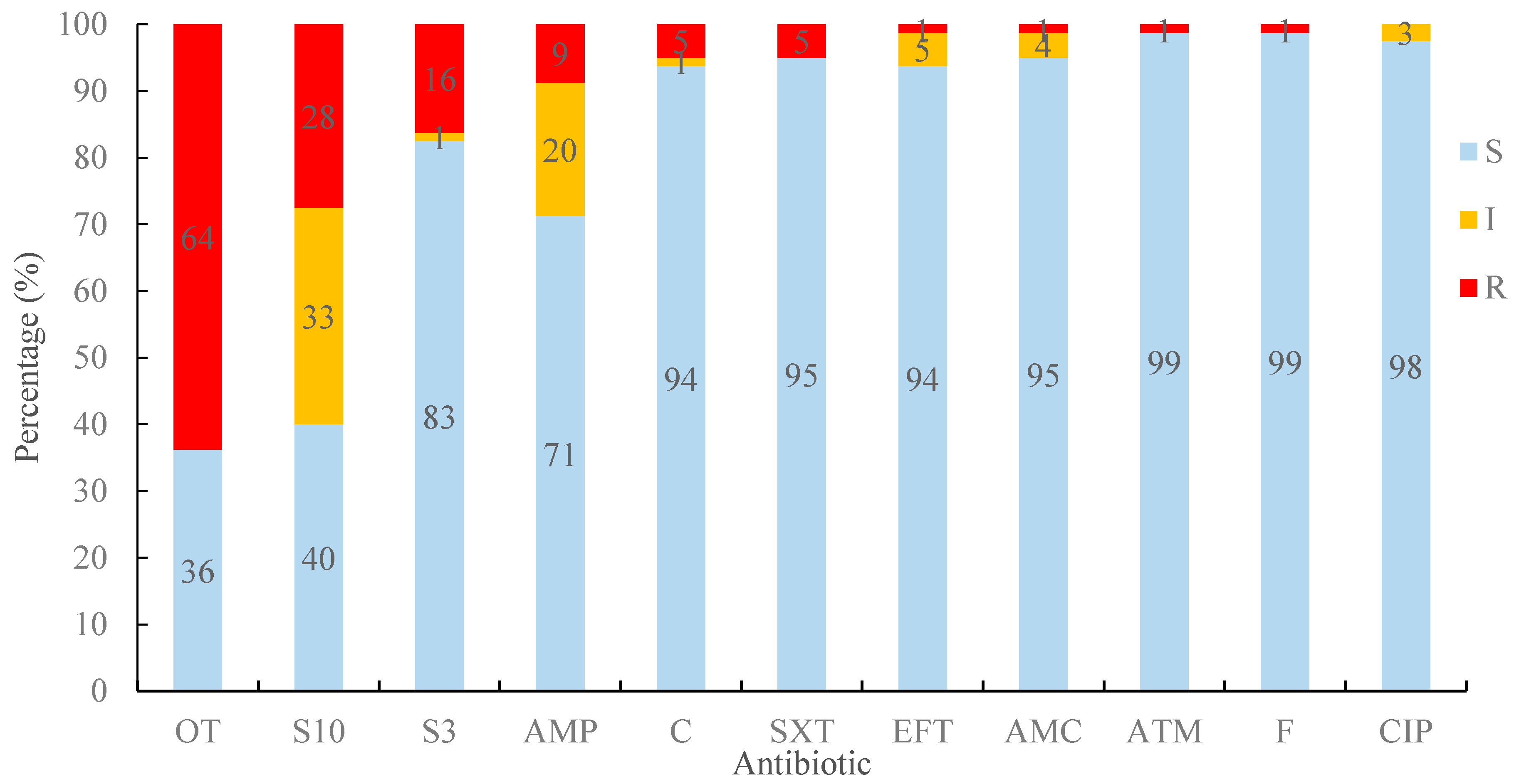
2.2. Resistance Prevalence to Antimicrobials
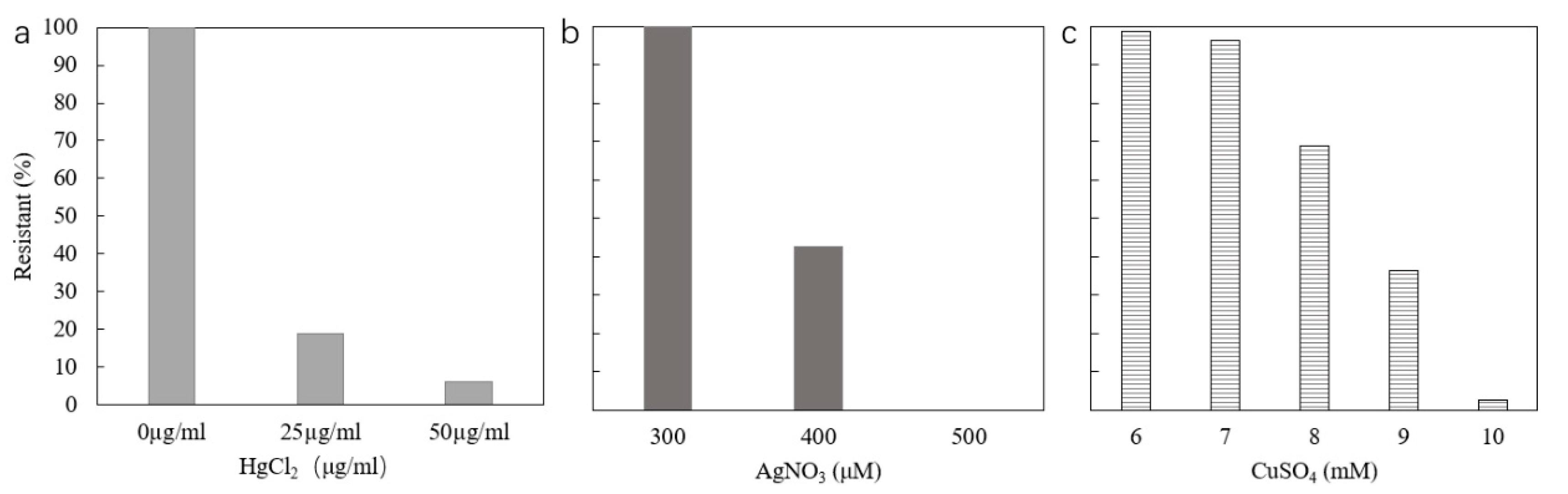
2.3. Resistance to Mercury
2.4. Resistance to Silver
2.5. Resistance to Copper
3. Discussion
4. Materials and Methods
4.1. Isolate Recovery and Re-confirmation
4.2. Antibiotic Sensitivity Tests
4.3. Metal Sensitivity Tests
4.4. PCR Detection of Genetic Elements Carrying Metal and Antibiotic Resistance Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hudson, J.A.; Frewer, L.J.; Jones, G.; Brereton, P.A.; Whittingham, M.J.; Stewart, G. The agri-food chain and antimicrobial resistance: A review. Trends Food Sci. Technol. 2017, 69, 131–147. [Google Scholar] [CrossRef]
- Zhou, B.; Wang, C.; Zhao, Q.; Wang, Y.; Huo, M.; Wang, J.; Wang, S. Prevalence and dissemination of antibiotic resistance genes and coselection of heavy metals in Chinese dairy farms. J. Hazard. Mater. 2016, 320, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Teuber, M. Veterinary use and antibiotic resistance. Curr. Opin. Microbiol. 2001, 4, 493–499. [Google Scholar] [CrossRef]
- Wright, G.D. Antibiotic resistance in the environment: A link to the clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef]
- Qiao, M.; Ying, G.G.; Singer, A.C.; Zhu, Y.G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef]
- Hobman, J.L.; Crossman, L.C. Bacterial antimicrobial metal ion resistance. J. Med. Microbiol. 2015, 64, 471–497. [Google Scholar] [CrossRef]
- He, X.; Xu, Y.; Chen, J.; Ling, J.; Li, Y.; Huang, L.; Zhou, X.; Zheng, L.; Xie, G. Evolution of corresponding resistance genes in the water of fish tanks with multiple stresses of antibiotics and heavy metals. Water Res. 2017, 124, 39–48. [Google Scholar] [CrossRef]
- Nicholson, F.A.; Smith, S.R.; Alloway, B.J.; Carlton-Smith, C.; Chambers, B.J. An inventory of heavy metals inputs to agricultural soils in England and Wales. Sci. Total. Environ. 2003, 311, 205–219. [Google Scholar] [CrossRef]
- Li, Y.X.; Li, W.; Wu, J.; Xu, L.C.; Su, Q.H.; Xiong, X. Contribution of additives Cu to its accumulation in pig feces: Study in Beijing and Fuxin of China. J. Environ. Sci. 2007, 19, 610–615. [Google Scholar] [CrossRef]
- Li, L.; Xu, Z.; Wu, J.; Tian, G. Bioaccumulation of heavy metals in the earthworm Eisenia fetida in relation to bioavailable metal concentrations in pig manure. Bioresour. Technol. 2010, 101, 3430–3436. [Google Scholar] [CrossRef]
- Holzel, C.S.; Muller, C.; Harms, K.S.; Mikolajewski, S.; Schafer, S.; Schwaiger, K.; Bauer, J. Heavy metals in liquid pig manure in light of bacterial antimicrobial resistance. Environ. Res. 2012, 113, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Parkhill, J.; Dougan, G.; James, K.D.; Thomson, N.R.; Pickard, D.; Wain, J.; Churcher, C.; Mungall, K.L.; Bentley, S.D.; Holden, M.T.G. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 2001, 413, 848–852. [Google Scholar] [CrossRef]
- Akinbowale, O.L.; Peng, H.; Grant, P.; Barton, M.D. Antibiotic and heavy metal resistance in motile aeromonads and pseudomonads from rainbow trout (Oncorhynchus mykiss) farms in Australia. Int. J. Antimicrob. Agents 2007, 30, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Najiah, M.; Lee, S.W.; Wendy, W.; Tee, L.W.; Nadirah, M.; Faizah, S.H. Antibiotic Resistance and Heavy Metals Tolerance in Gram-Negative Bacteria from Diseased American Bullfrog (Rana catesbeiana) Cultured in Malaysia. Agric. Sci. China 2009, 8, 1270–1275. [Google Scholar] [CrossRef]
- Alonso, A.; Sanchez, P.; Martinez, J.L. Environmental selection of antibiotic resistance genes. Environ. Microbiol. 2001, 3, 1–9. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, L.; Ye, C.; Yu, X. Co-selection of antibiotic resistance via copper shock loading on bacteria from a drinking water bio-filter. Environ. Pollut. 2018, 233, 132–141. [Google Scholar] [CrossRef]
- McAuliffe, G.A.; Takahashi, T.; Mogensen, L.; Hermansen, J.E.; Sage, C.L.; Chapman, D.V.; Lee, M.R.F. Environmental trade-offs of pig production systems under varied operational efficiencies. J. Clean Prod. 2017, 165, 1163–1173. [Google Scholar] [CrossRef]
- Sheridan, J.J. Sources of contamination during slaughter and measures for control. J. Food Saf. 1998, 18, 321–339. [Google Scholar] [CrossRef]
- Wheatley, P.; Giotis, E.S.; McKevitt, A.I. Effects of slaughtering operations on carcass contamination in an Irish pork production plant. Ir. Vet. J. 2014, 67, 1. [Google Scholar] [CrossRef]
- Martins, B.T.F.; Botelho, C.V.; Silva, D.A.L.; Lanna, F.; Grossi, J.L.; Campos-Galvao, M.E.M.; Yamatogi, R.S.; Falcao, J.P.; Bersot, L.D.S.; Nero, L.A. Yersinia enterocolitica in a Brazilian pork production chain: Tracking of contamination routes, virulence and antimicrobial resistance. Int. J. Food Microbiol. 2018, 276, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Warriner, K.; Aldsworth, T.G.; Kaur, S.; Dodd, C.E. Cross-contamination of carcasses and equipment during pork processing. J. Appl. Microbiol. 2002, 93, 169–177. [Google Scholar] [CrossRef]
- Bouvet, J.; Bavai, C.; Rossel, R.; Le Roux, A.; Montet, M.P.; Ray-Gueniot, S.; Mazuy, C.; Arquillière, C.; Vernozy-Rozand, C. Prevalence of verotoxin-producing Escherichia coli and E. coli O157:H7 in pig carcasses from three French slaughterhouses. Int. J. Food Microbiol. 2001, 71, 249–255. [Google Scholar] [CrossRef]
- Morales-Partera, A.M.; Cardoso-Toset, F.; Luque, I.; Astorga, R.J.; Maldonado, A.; Herrera-León, S.; Hernández, M.; Gómez-Laguna, J.; Tarradas, C. Prevalence and diversity of Salmonella spp., Campylobacter spp., and Listeria monocytogenes in two free-range pig slaughterhouses. Food Control 2018, 92, 208–215. [Google Scholar] [CrossRef]
- Eblen, D.R.; Annous, B.A.; Sapers, G.M. Studies to select appropriate nonpathogenic surrogate Escherichia coli strains for potential use in place of Escherichia coli O157:H7 and salmonella in pilot plant studiest. J. Food Prot. 2005, 68, 282–291. [Google Scholar] [CrossRef]
- Hartl, D.L.; Dykhuizen, D. The Population Genetics of Escherichia Coli. Annu. Rev. Genet. 1984, 18, 31–68. [Google Scholar] [CrossRef] [PubMed]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 403. [Google Scholar] [CrossRef]
- Botteldoorn, N.; Heyndrickx, M.; Rijpens, N.; Herman, L. Detection and characterization of verotoxigenic Escherichia coli by a VTEC/EHEC multiplex PCR in porcine faeces and pig carcass swabs. Res. Microbiol. 2003, 154, 97–104. [Google Scholar] [CrossRef]
- Ojo, O.E.; Ajuwape, A.T.; Otesile, E.B.; Owoade, A.A.; Oyekunle, M.A.; Adetosoye, A.I. Potentially zoonotic shiga toxin-producing Escherichia coli serogroups in the faeces and meat of food-producing animals in Ibadan, Nigeria. Int. J. Food Microbiol. 2010, 142, 214–221. [Google Scholar] [CrossRef]
- Martinez-Vazquez, A.V.; Rivera-Sanchez, G.; Lira Mendez, K.; Reyes-Lopez, M.A.; Bocanegra-Garcia, V. Prevalence, antimicrobial resistance and virulence genes in Escherichia coli isolated from retail meats in Tamaulipas, Mexico. J. Glob. Antimicrob. Resist. 2018, 14, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Burger, R. EHEC O104:H4 in Germany 2011: Large Outbreak of Bloody Diarrhea and Haemolytic Uraemic Syndrome by Shiga Toxin-Producing E. coli via Contaminated Food; Robert Koch-Institut: Berlin, Germany, 2012. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Van Noten, N.; Gorissen, L.; De Smet, S. Assistance Update SLR: Copper and antibiotic resistance in pigs. EFSA Supporting Publ. 2016, 13, 1005E. [Google Scholar] [CrossRef]
- Halik, K.A. Preservation of some extremely thermophilic chemolithoautotrophic bacteria by deep-freezing and liquid-drying methods. J. Microbiol. Methods 1999, 35, 177–182. [Google Scholar] [CrossRef]
- Zhang, H.; Cai, L.; Zhang, F.; Ge, C.; Yang, F. Vacuum lyophilization preservation and rejuvenation performance of anammox bacteria. J. Biosci. Bioeng. 2020, 129, 519–527. [Google Scholar] [CrossRef]
- Lakshman, D.K.; Singh, V.; Camacho, M.E. Long-term cryopreservation of non-spore-forming fungi in Microbank™ beads for plant pathological investigations. J. Microbiol. Methods 2018, 148, 120–126. [Google Scholar] [CrossRef]
- Wei, S.H. Escherichia coli Contamination of Pork Carcasses in UK Slaughterhouses; The University of Nottingham: Nottingham, UK, 2012.
- Ibrahim, D.R.; Dodd, C.E.; Stekel, D.J.; Ramsden, S.J.; Hobman, J.L. Multidrug resistant, extended spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from a dairy farm. FEMS Microbiol. Ecol. 2016, 92, 1–12. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; CLSI standard M07; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th ed.; CLSI standard VET01; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests, 13th ed.; CLSI standard M02; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Colom, K.; Pérez, J.; Alonso, R.; Fernández-Aranguiz, A.; Lariño, E.; Cisterna, R. Simple and reliable multiplex PCR assay for detection of blaTEM, blaSHV and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 2003, 223, 147–151. [Google Scholar] [CrossRef]
- World Health Organization. Global Acton Plan on Antimicrobial Resistance; WHO: Geneva, Switzerland, 2015. [Google Scholar]
- Hwang, A.Y.; Gums, J.G. The emergence and evolution of antimicrobial resistance: Impact on a global scale. Bioorganic Med. Chem. 2016, 24, 6440–6445. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef]
- Rankin, S.; Roberts, S.; O’Shea, K.; Maloney, D.; Lorenzo, M.; Benson, C.E. Panton valentine leukocidin (PVL) toxin positive MRSA strains isolated from companion animals. Vet. Microbiol. 2005, 108, 145–148. [Google Scholar] [CrossRef]
- García-González, P.; García-Lamas, N.; Fuentes Edfuf, C.; Santos, Y. Development of a PCR method for the specific identification of the marine fish pathogen Tenacibaculum soleae. Aquaculture 2011, 319, 1–4. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, A.A.; Checkley, S.; Avery, B.; Chalmers, G.; Bohaychuk, V.; Boerlin, P.; Reid-Smith, R.; Aslam, M. Antimicrobial resistance and resistance genes in Escherichia coli isolated from retail meat purchased in Alberta, Canada. Foodborne Pathog Dis. 2012, 9, 625–631. [Google Scholar] [CrossRef]
- Llorente, P.; Barnech, L.; Irino, K.; Rumi, M.V.; Bentancor, A. Characterization of Shiga toxin-producing Escherichia coli isolated from ground beef collected in different socioeconomic strata markets in Buenos Aires, Argentina. Biomed. Res. Int. 2014, 2014, 795104. [Google Scholar] [CrossRef]
- Tadesse, D.A.; Zhao, S.; Tong, E.; Ayers, S.; Singh, A.; Bartholomew, M.J.; McDermott, P.F. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg. Infect Dis. 2012, 18, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Munir, M.; Xagoraraki, I. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 2012, 421, 173–183. [Google Scholar] [CrossRef]
- Harja, M.; Ciobanu, G. Studies on adsorption of oxytetracycline from aqueous solutions onto hydroxyapatite. Sci. Total Environ. 2018, 628, 36–43. [Google Scholar] [CrossRef]
- Zhang, S.; Qu, Q.; Zhang, J.; Lai, Z.; Zhu, X. Prevalence, genetic diversity, and antibiotic resistance of enterotoxigenic Escherichia coli in retail ready-to-eat foods in China. Food Control 2016, 68, 236–243. [Google Scholar] [CrossRef]
- Skočková, A.; Koláčková, I.; Bogdanovičová, K.; Karpíšková, R. Characteristic and antimicrobial resistance in Escherichia coli from retail meats purchased in the Czech Republic. Food Control 2015, 47, 401–406. [Google Scholar] [CrossRef]
- EMEA. Antibiotic Resistance in the European Union Associated with the Therapeutic Use of Veterinary Medicines. European Agency for the Evaluation of Medicinal Products; Report EMEA/CVMP/342/99-Final; Veterinary Medicines Evaluation Unit: London, UK, 1999.
- Abo-Amer, A.E.; Shobrak, M.Y.; Altalhi, A.D. Isolation and antimicrobial resistance among Escherichia coli isolated from farm chickens in Taif province, Saudi Arabia. J. Glob. Antimicrob. Resist. 2018. [Google Scholar] [CrossRef]
- Aragaw, K.; Molla, B.; Muckle, A.; Cole, L.; Wilkie, E.; Poppe, C.; Kleer, J.; Hildebrandt, G. The characterization of Salmonella serovars isolated from apparently healthy slaughtered pigs at Addis Ababa abattoir, Ethiopia. Prev. Vet. Med. 2007, 82, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Bonardi, S.; Alpigiani, I.; Bruini, I.; Barilli, E.; Brindani, F.; Morganti, M.; Cavallini, P.; Bolzoni, L.; Pongolini, S. Detection of Salmonella enterica in pigs at slaughter and comparison with human isolates in Italy. Int. J. Food Microbiol. 2016, 218, 44–50. [Google Scholar] [CrossRef]
- Wu, S.; Dalsgaard, A.; Vieira, A.R.; Emborg, H.-D.; Jensen, L.B. Prevalence of tetracycline resistance and genotypic analysis of populations of Escherichia coli from animals, carcasses and cuts processed at a pig slaughterhouse. Int. J. Food Microbiol. 2009, 135, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Bonardi, S.; Bruini, I.; D’Incau, M.; Van Damme, I.; Carniel, E.; Bremont, S.; Cavallini, P.; Tagliabue, S.; Brindani, F. Detection, seroprevalence and antimicrobial resistance of Yersinia enterocolitica and Yersinia pseudotuberculosis in pig tonsils in Northern Italy. Int. J. Food Microbiol. 2016, 235, 125–132. [Google Scholar] [CrossRef] [PubMed]
- de Jong, A.; Simjee, S.; Garch, F.E.; Moyaert, H.; Rose, M.; Youala, M.; Dry, M.; Group, E.S. Antimicrobial susceptibility of enterococci recovered from healthy cattle, pigs and chickens in nine EU countries (EASSA Study) to critically important antibiotics. Vet. Microbiol. 2018, 216, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Ozgumus, O.B.; Celik-Sevim, E.; Alpay-Karaoglu, S.; Sandalli, C.; Sevim, A. Molecular characterization of antibiotic resistant Escherichia coli strains isolated from tap and spring waters in a coastal region in turkey. J. Microbiol. 2007, 45, 379–387. [Google Scholar]
- DANMAP. DANMAP 2005-Use of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Bacteria from Food Animals, Foods, and Humans in Denmark; Danish Institute for Food and Veterinary Research: Copenhagen, Denmark, 2006. [Google Scholar]
- Schwaiger, K.; Huther, S.; Holzel, C.; Kampf, P.; Bauer, J. Prevalence of antibiotic-resistant Enterobacteriaceae isolated from chicken and pork meat purchased at the slaughterhouse and at retail in Bavaria, Germany. Int. J. Food Microbiol. 2012, 154, 206–211. [Google Scholar] [CrossRef]
- Sudarwanto, M.B.; Lukman, D.W.; Purnawarman, T.; Latif, H.; Pisestyani, H.; Sukmawinata, E. Multidrug resistance extended spectrum β-lactamase and AmpC producing Escherichia coli isolated from the environment of Bogor Slaughterhouse, Indonesia. Asian Pac. J. Trop. Biomed. 2017, 7, 708–711. [Google Scholar] [CrossRef]
- Ishihara, K.; Hosokawa, Y.; Makita, K.; Noda, J.; Ueno, H.; Muramatsu, Y.; Ueno, H.; Mukai, T.; Yamamoto, H.; Ito, M.; et al. Factors associated with antimicrobial-resistant Escherichia coli in zoo animals. Res. Vet. Sci. 2012, 93, 574–580. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Laupland, K.B. Extended-spectrum β-lactamase-producting Enterobacteriaceae: An emerging public-health concern. Lancet Infect. Dis. 2008, 8, 159–166. [Google Scholar] [CrossRef]
- Bradford, P.A. Extended-spectrum β-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Reinthaler, F.F.; Feierl, G.; Galler, H.; Haas, D.; Leitner, E.; Mascher, F.; Melkes, A.; Posch, J.; Winter, I.; Zarfel, G.; et al. ESBL-producing E. coli in Austrian sewage sludge. Water Res. 2010, 44, 1981–1985. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.; Vinué, L.; Poeta, P.; Coelho, A.C.; Matos, M.; Sáenz, Y.; Somalo, S.; Zarazaga, M.; Rodrigues, J.; Torres, C. Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli isolates in faecal samples of broilers. Vet. Microbiol. 2009, 138, 339–344. [Google Scholar] [CrossRef]
- Ojer-Usoz, E.; Gonzalez, D.; Vitas, A.I.; Leiva, J.; Garcia-Jalon, I.; Febles-Casquero, A.; Escolano Mde, L. Prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in meat products sold in Navarra, Spain. Meat Sci. 2013, 93, 316–321. [Google Scholar] [CrossRef]
- Peter, S.; Polsfuss, S.; Poledica, M.; Hombach, M.; Giger, J.; Böttger, E.; Zbinden, R.; Bloemberg, G. Detection of AmpC Beta-Lactamase in Escherichia coli: Comparison of Three Phenotypic Confirmation Assays and Genetic Analysis. J. Clin. Microbiol. 2011, 49, 2924–2932. [Google Scholar] [CrossRef] [PubMed]
- Philippon, A.; Arlet, G.; Lagrange, P.H. Origin and impact of plasmid-mediated extended-spectrum beta-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 1994, 13, S17–S29. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Alarcon, C.; Singer, R.S.; Johnson, T.J. Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS ONE 2011, 6, e23415. [Google Scholar] [CrossRef]
- Yu, Z.; Gunn, L.; Wall, P.; Fanning, S. Antimicrobial resistance and its association with tolerance to heavy metals in agriculture production. Food Microbiol. 2017, 64, 23–32. [Google Scholar] [CrossRef]
- Barkay, T.; Miller, S.M.; Summers, A.O. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 2003, 27, 355–384. [Google Scholar] [CrossRef]
- Pal, C.; Asiani, K.; Arya, S.; Rensing, C.; Stekel, D.J.; Larsson, D.G.J.; Hobman, J.L. Chapter Seven—Metal Resistance and Its Association With Antibiotic Resistance. In Advances in Microbial Physiology; Poole, R.K., Ed.; Academic Press: Cambridge, MA, USA, 2017; Volume 70, pp. 261–313. [Google Scholar]
- Olson, B.H.; Lester, J.N.; Cayless, S.M.; Ford, S. Distribution of mercury resistance determinants in bacterial communities of river sediments. Water Res. 1989, 23, 1209–1217. [Google Scholar] [CrossRef]
- Mazel, D.; Dychinco, B.; Webb, V.A.; Davies, J. Antibiotic resistance in the ECOR collection: Integrons and identification of a novel aad gene. Antimicrob. Agents Chemother. 2000, 44, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Maynes, M.; Silver, S. Effects of halides on plasmid-mediated silver resistance in Escherichia coli. Appl. Environ. Microbiol. 1998, 64, 5042–5045. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Matsui, K.; Lo, J.-F.; Silver, S. Molecular basis for resistance to silver cations in Salmonella. Nat. Med. 1999, 5, 183. [Google Scholar] [CrossRef] [PubMed]
- Silver, S. Bacterial silver resistance: Molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 2003, 27, 341–353. [Google Scholar] [CrossRef]
- Solioz, M.; Abicht, H.K.; Mermod, M.; Mancini, S. Response of Gram-positive bacteria to copper stress. J. Biol. Inorg. Chem. 2010, 15, 3–14. [Google Scholar] [CrossRef]
- Debski, B. Supplementation of pigs diet with zinc and copper as alternative to conventional antimicrobials. Pol. J. Vet. Sci. 2016, 19, 917–924. [Google Scholar] [CrossRef]
- Agga, G.E.; Scott, H.M.; Amachawadi, R.G.; Nagaraja, T.G.; Vinasco, J.; Bai, J.; Norby, B.; Renter, D.G.; Drita, S.S.; Nelssen, J.L.; et al. Effects of chlortetracycline and copper supplementation on antimicrobial resistance of fecal Escherichia coli from weaned pigs. Prev. Vet. Med. 2014, 114, 231–246. [Google Scholar] [CrossRef]
- Marazzato, M.; Aleandri, M.; Massaro, M.R.; Vitanza, L.; Conte, A.L.; Conte, M.P.; Nicoletti, M.; Comanducci, A.; Goldoni, P.; Maurizi, L.; et al. Escherichia coli strains of chicken and human origin: Characterization of antibiotic and heavy-metal resistance profiles, phylogenetic grouping, and presence of virulence genetic markers. Res. Vet. Sci. 2020, 132, 150–155. [Google Scholar] [CrossRef]
- Najar, I.N.; Sherpa, M.T.; Das, S.; Das, S.; Thakur, N. Diversity analysis and metagenomic insights into antibiotic and metal resistance among Himalayan hot spring bacteriobiome insinuating inherent environmental baseline levels of antibiotic and metal tolerance. J. Glob. Antimicrob. Resist. 2020, 21, 342–352. [Google Scholar] [CrossRef]
- Veterinary Medicines Directorate. UK-VARSS 2018 Highlights Report. Available online: https://www.gov.uk/government/publications/veterinary-antimicrobial-resistance-and-sales-surveillance-2018 (accessed on 7 September 2020).
- Kempf, I.; Jouy, E.; Chauvin, C. Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 2016, 48, 598–606. [Google Scholar] [CrossRef]
- Gupta, A.; Phung, L.T.; Taylor, D.E.; Silver, S. Diversity of silver resistance genes in IncH incompatibility group plasmids. Microbiology 2001, 147, 3393–3402. [Google Scholar] [CrossRef] [PubMed]
- Mourão, J.; Novais, C.; Machado, J.; Peixe, L.; Antunes, P. Metal tolerance in emerging clinically relevant multidrug-resistant Salmonella enterica serotype 4,[5],12:i:−clones circulating in Europe. Int. J. Antimicrob. Agents 2015, 45, 610–616. [Google Scholar] [CrossRef]
- Borck, K.; Beggs, J.D.; Brammar, W.J.; Hopkins, A.S.; Murray, N.E. The construction in vitro of transducing derivatives of phage lambda. Mol. Gen. Genet. MGG 1976, 146, 199–207. [Google Scholar] [CrossRef]
- Tetaz, T.J.; Luke, R.K. Plasmid-controlled resistance to copper in Escherichia coli. J. Bacteriol. 1983, 154, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Yokota, T. Plasmids of enterotoxigenic Escherichia coli H10407: Evidence for two heat-stable enterotoxin genes and a conjugal transfer system. J. Bacteriol. 1983, 153, 1352. [Google Scholar] [CrossRef]
- Dierikx, C.M.; van Duijkeren, E.; Schoormans, A.H.; van Essen-Zandbergen, A.; Veldman, K.; Kant, A.; Huijsdens, X.W.; van der Zwaluw, K.; Wagenaar, J.A.; Mevius, D.J. Occurrence and characteristics of extended-spectrum-beta-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J. Antimicrob. Chemother. 2012, 67, 1368–1374. [Google Scholar] [CrossRef]
- Asiani, K. Biochemical and Biophysical Studies on SilE from the Sil Silver Resistance Locus; The University of Nottingham: Nottingham, UK, 2016. [Google Scholar]
- Jenkins, C.; Tembo, M.; Chart, H.; Cheasty, T.; Willshaw, G.A.; Phillips, A.D.; Tompkins, D.; Smith, H. Detection of enteroaggregative Escherichia coli in faecal samples from patients in the community with diarrhoea. J. Med. Microbiol. 2006, 55, 1493–1497. [Google Scholar] [CrossRef] [PubMed]
- Liebert, C.A.; Wireman, J.; Smith, T.; Summers, A.O. Phylogeny of mercury resistance (mer) operons of gram-negative bacteria isolated from the fecal flora of primates. Appl. Environ. Microbiol. 1997, 63, 1066–1076. [Google Scholar] [CrossRef]
- Goldstein, C.; Lee, M.D.; Sanchez, S.; Hudson, C.; Phillips, B.; Register, B.; Grady, M.; Liebert, C.; Summers, A.O.; White, D.G.; et al. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 2001, 45, 723–726. [Google Scholar] [CrossRef]
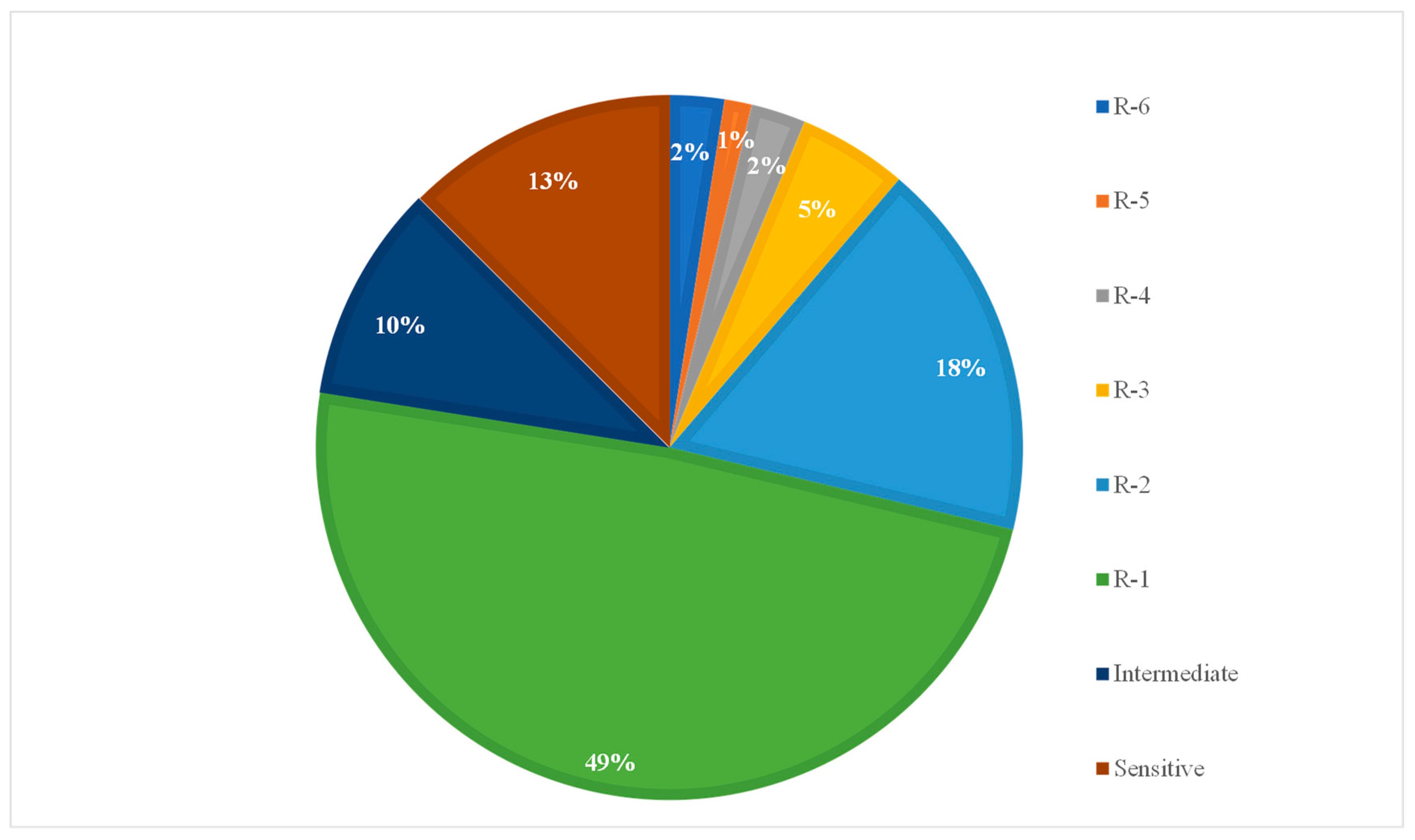
| Number of Resistance Carried | Total Number of Isolates | Number of Isolates Showing Each Phenotype | Combination of Resistances |
|---|---|---|---|
| 6 | 2 | 2 | AMP *, S10, OT, SXT, S300, C |
| 5 | 1 | 1 | S10, OT, SXT, S300, C |
| 4 | 2 | 1 | OT, SXT, S300, C |
| 1 | OT, F, S300, C | ||
| 3 | 4 | 1 | AMP, S10, EFT |
| 1 | AMP, S10, OT | ||
| 1 | S10, OT, S300 | ||
| 1 | AMP, ATM, OT | ||
| 2 | 14 | 9 | S10, OT |
| 4 | OT, S300 | ||
| 1 | S10, S300 | ||
| 1 | 39 | 30 | OT |
| 7 | S10 | ||
| 2 | S300 | ||
| Intermediate | 8 | 3 | AMP |
| 3 | S10 | ||
| 1 | EFT | ||
| 1 | EFT, S10 | ||
| 0 | 10 | 10 | Sensitive to all |
| Total | 80 | 80 |
| Strain | Silver Resistance | Mercury Resistance | Integron | Copper Resistance | Antibiotic Resistance | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Phenotype (400 μM) | Genotype | Phenotype (≥25 μg/mL) | Genotype pcoE | Phenotype (≥8 mM) | Phenotype | β-Lactamase Genes | ||||||
| ID | Original ID in Reference [37] | silA | silB | silE | merA | merC | intl1 | ||||||
| 4 | EAggEC7BP4 | + ◊ | R∇ (9 mM) | AMP *, S10, OT, SXT, S300, C | blaTEM | ||||||||
| 7 | EAggEC7BP5 | + | R (9 mM) | S10, OT, SXT, S300, C | blaTEM | ||||||||
| 9 | EAggEC9AP1 | + | + | + | R | + | + | R(25 μg/mL) | R (9 mM) | OT | |||
| 10 | EAggEC8BP5 | R | + | + | R(50 μg/mL) | + | R (8 mM) | AMP(I⊗), AMC(I), S10(I), OT, S300, SXT, C | |||||
| 12 | EAggEC10BP1 | + | + | + | R | R (8 mM) | S10(I), OT, S300, C, F | ||||||
| 13 | EAggEC9BP1 | + | + | + | R | AMP, S10, OT | |||||||
| 14 | EAggEC5AP4 | + | + | + | R | + | + | R(25 μg/mL) | + | + | S10(I), OT, S300, C(I) | ||
| 15 | EAggEC9AP5 | + | + | + | R | + | + | R(25 μg/mL) | R (9 mM) | OT, CIP(I) | |||
| 17 | EAggEC8AP4 | + | + | + | R | R (9 mM) | EFT(I), S10(I) | ||||||
| 21 | EAggEC10AP1 | + | + | + | R | + | + | R(25 μg/mL) | R (9 mM) | S10(I), OT | |||
| 23 | EAggEC 11AP3 | + | + | + | R | + | + | R(25 μg/mL) | R (9 mM) | S10(I), OT | |||
| 24 | EAggEC 11AP4 | + | + | + | R | + | + | R(25 μg/mL) | R (9 mM) | S10(I), OT | |||
| 25 | EAggEC9AP2 | + | + | + | R | + | + | R(25 μg/mL) | R (9 mM) | S10(I), OT | |||
| 27 | EAggEC10AP2 | + | + | + | R | S10, OT | |||||||
| 38 | EAggEC5AO2 | + | + | R(50 μg/mL) | R (9 mM) | OT | |||||||
| 41 | EAggEC5AO1 | + | + | R(50 μg/mL) | R (9 mM) | OT | |||||||
| 48 | EAggEC5AO3 | + | + | R(50 μg/mL) | R (9 mM) | OT | |||||||
| 50 | EAggEC10AO1 | + | + | + | R | + | R (8 mM) | S10(I), S300 | |||||
| 57 | EAggEC5AS2 | + | + | R(50 μg/mL) | R (9 mM) | AMP, ATM, OT | |||||||
| 59 | EAggEC5AS1 | + | + | R(25 μg/mL) | R (9 mM) | AMP(I), OT | |||||||
| 61 | EAggEC10AO3 | AMP, AMC, OT, S300, SXT | blaTEM | ||||||||||
| 62 | EAggEC12AO3 | + | + | + | R | S10(I), OT | |||||||
| 63 | EAggEC12AO4 | + | + | + | R | + | |||||||
| 70 | EAggEC11AS5 | + | + | + | R | + | + | R(25μg/mL) | R (9 mM) | OT | |||
| 74 | EAggEC14AS2 | + | + | + | R | + | |||||||
| 75 | EAggEC14AS1 | + | + | + | R | + | |||||||
| 77 | EAggEC15AS1 | + | + | + | R | + | R (9 mM) | S10(I), OT | |||||
| 78 | EAggEC13AO3 | + | + | + | R | + | R (10 mM) | ||||||
| Antibiotic Discs | Content | Antimicrobial Group |
|---|---|---|
| β-Lactams | ||
| Ampicillin (AMP) | 10 μg | Penicillins |
| Amoxicillin–clavulanic acid (AMC) | 20/10 μg | Penicillin Combination with Beta-lactamase Inhibitor |
| Cefotaxime (CTX) | 30 μg | Third Generation Cephalosporin |
| Ceftazidime (CAZ) | 30 μg | Third Generation Cephalosporin |
| Ceftiofur (EFT) | 30 μg | Third Generation Cephalosporin |
| Cefquinome (CFQ) | 30 μg | Fourth Generation Cephalosporin |
| Aztreonam (ATM) | 30 μg | Monobactam |
| Imipenem (IPM) | 10 μg | Carbapenem |
| Aminoglycoside | ||
| Streptomycin (S10) | 10 μg | Amino-glycosides |
| Quinolones | ||
| Ciprofloxacin (CIP) | 5 μg | Fluoro-quinolones |
| Enrofloxacin (ENR) | 5 μg | Fluoro-quinolones |
| Nalidixic acid (NA) | 30 μg | Quinolones |
| Sulphonamide/complex | ||
| Trimethoprim–sulfamethoxazole (SXT) | 1.25/23.75 μg | Folate Pathway Inhibitors |
| Sulfonamide (S300) | 300 μg | Folate Pathway Inhibitors |
| Phenicol | ||
| Chloramphenicol (C) | 30 μg | Phenicols |
| Tetracycline | ||
| Oxytetracycline (OT) | 30 μg | Tetracyclines |
| Nitrofuran derivative | ||
| Nitrofurantoin (F) | 300 μg | Nitrofurans |
| Oligonucleotide Name | Sequence a | Product Size (Bp) | Reference |
|---|---|---|---|
| astA-F | CCATCAACACAGTATATCCGA | 111 | [98] |
| astA-R | GGTCGCGAGTGACGGCTTTGT | ||
| CTX-M-F | ATGTGCAGYACCAGTAARGTKATGGC | 529 | [96] |
| CTX-M-R | TGGGTRAARTARGTSACCAGAAYSAGCGG | ||
| TEM-F | GCGGAACCCCTATTTG | 964 | |
| TEM-R | ACCAATGCTTAATCAGTGAG | ||
| SHV-F | TTATCTCCCTGTTAGCCACC | 796 | |
| SHV-R | GATTTGCTGATTTCGCTCGG | ||
| OXA-1-F | ATGAAAAACACAATACATATCAACTTCGC | 820 | |
| OXA-1-R | GTGTGTTTAGAATGGTGATCGCATT | ||
| mcr-1-F | CGGTCAGTCCGTTTGTTC | 309 | [32] |
| mcr-1-R | CTTGGTCGGTCTGTAGGG | ||
| merA-F | ACCATCGGCACCTGCGT | 1237 | [99] |
| merA-R | ACCATCGTCAGGTAGGGGAACAA | ||
| merC-F | CATCGGGCTGGGCTGGGCTTCTTGAG | 364 | |
| merC-R | CATCGTTCCTTATTCGTGTGG | ||
| intl1-F | CCTCCCGCACGATGATC | 280 | [100] |
| intl1-R | TCCACGCATCGTCAGGC | ||
| silA-F | ATGATTGAATGGATTATCCG | 3147 | [97] |
| silA-R | TTATGACACGCTTTTTTTAT | ||
| silB-F | ATGGCTTCTTTAAAGATAAA | 1293 | |
| silB-R | TCAGTGCCCTGAATGCATAT | ||
| silE-F | ATGAAAAATATCGTATTAGC | 432 | |
| silE-R | TCAGCCTGCACTGAGCATGC | ||
| pcoE-F | ATGAATATATTAATCACGAC | 450 | |
| pcoE-R | TTACCTGGTCGAATACAGCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Wei, S.-H.; Hobman, J.L.; Dodd, C.E.R. Antibiotic and Metal Resistance in Escherichia coli Isolated from Pig Slaughterhouses in the United Kingdom. Antibiotics 2020, 9, 746. https://doi.org/10.3390/antibiotics9110746
Yang H, Wei S-H, Hobman JL, Dodd CER. Antibiotic and Metal Resistance in Escherichia coli Isolated from Pig Slaughterhouses in the United Kingdom. Antibiotics. 2020; 9(11):746. https://doi.org/10.3390/antibiotics9110746
Chicago/Turabian StyleYang, Hongyan, Shao-Hung Wei, Jon L. Hobman, and Christine E. R. Dodd. 2020. "Antibiotic and Metal Resistance in Escherichia coli Isolated from Pig Slaughterhouses in the United Kingdom" Antibiotics 9, no. 11: 746. https://doi.org/10.3390/antibiotics9110746
APA StyleYang, H., Wei, S.-H., Hobman, J. L., & Dodd, C. E. R. (2020). Antibiotic and Metal Resistance in Escherichia coli Isolated from Pig Slaughterhouses in the United Kingdom. Antibiotics, 9(11), 746. https://doi.org/10.3390/antibiotics9110746




