The Cell Envelope Stress Response of Bacillus subtilis towards Laspartomycin C
Abstract
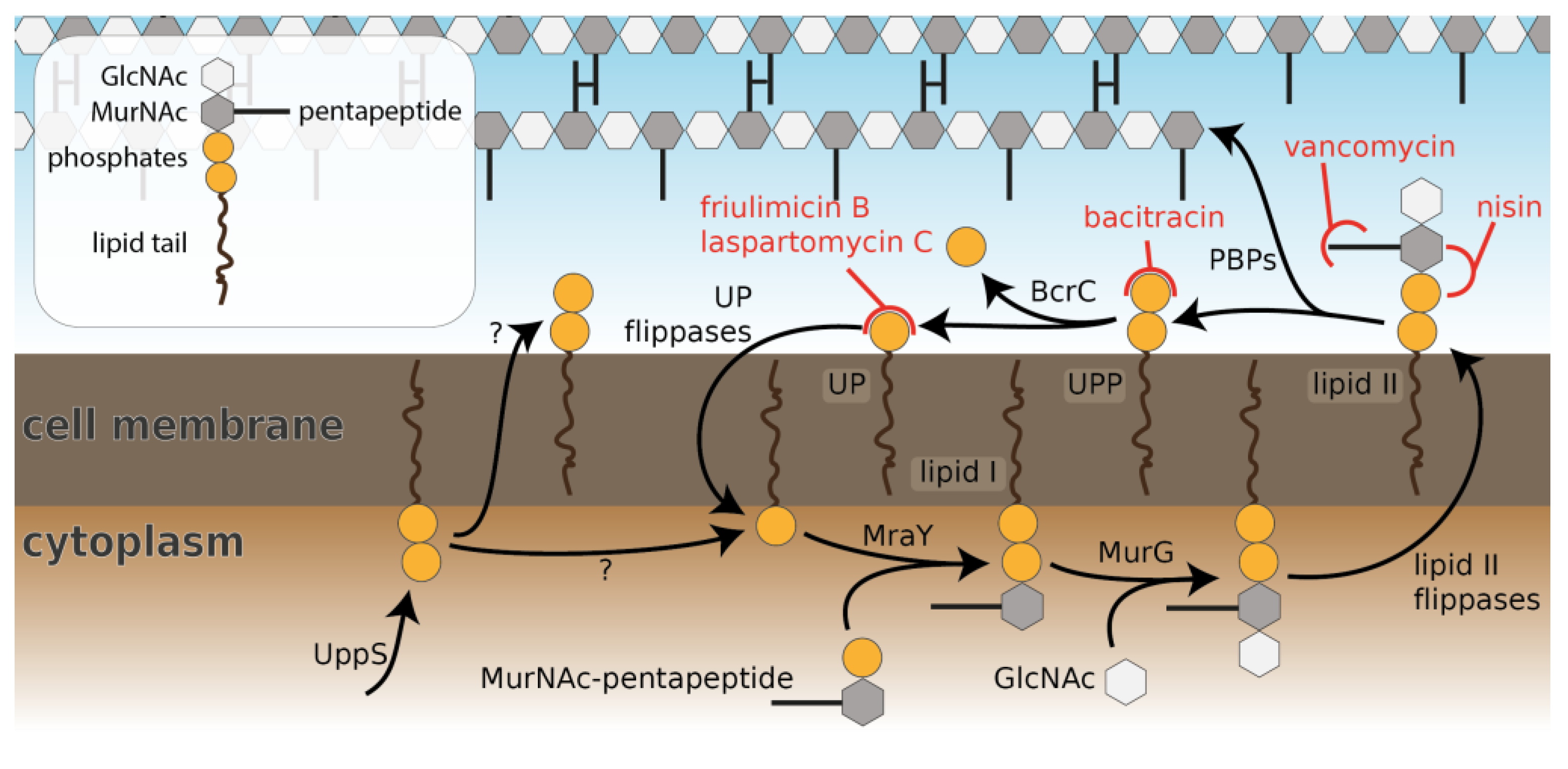
1. Introduction
2. Results
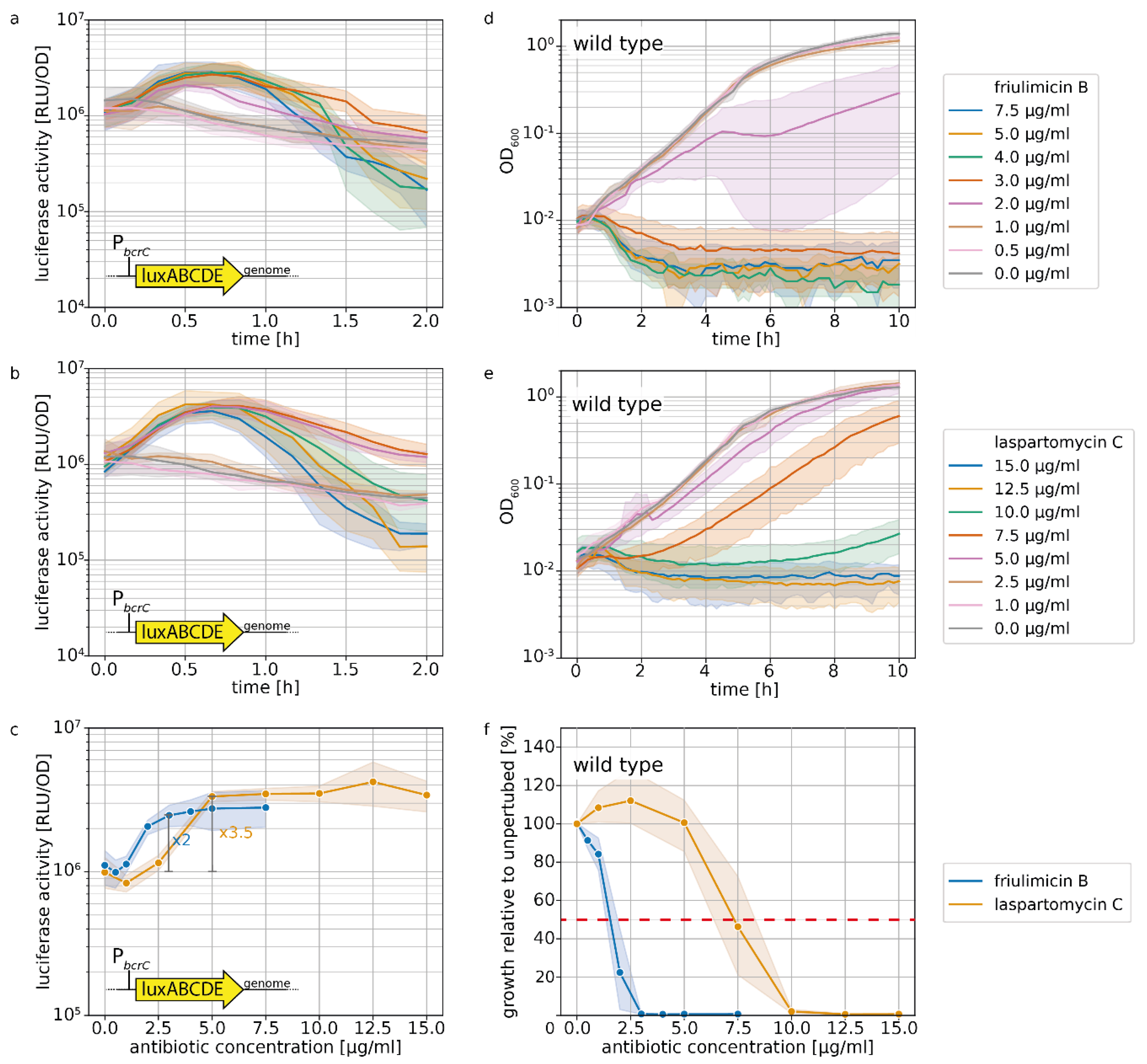
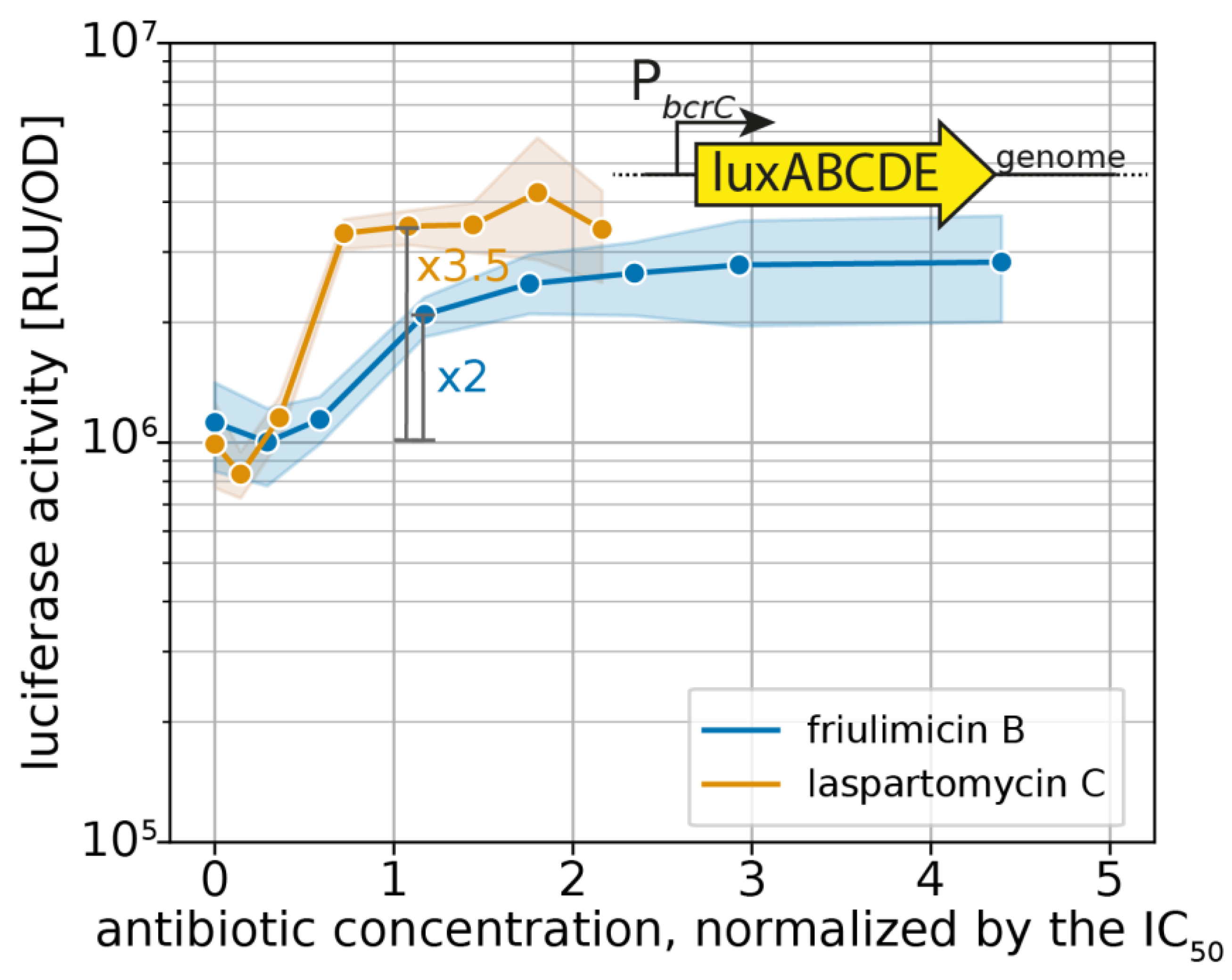
2.1. Laspartomycin C Induces a Slightly Stronger Response of the σM Regulon than Friulimicin B
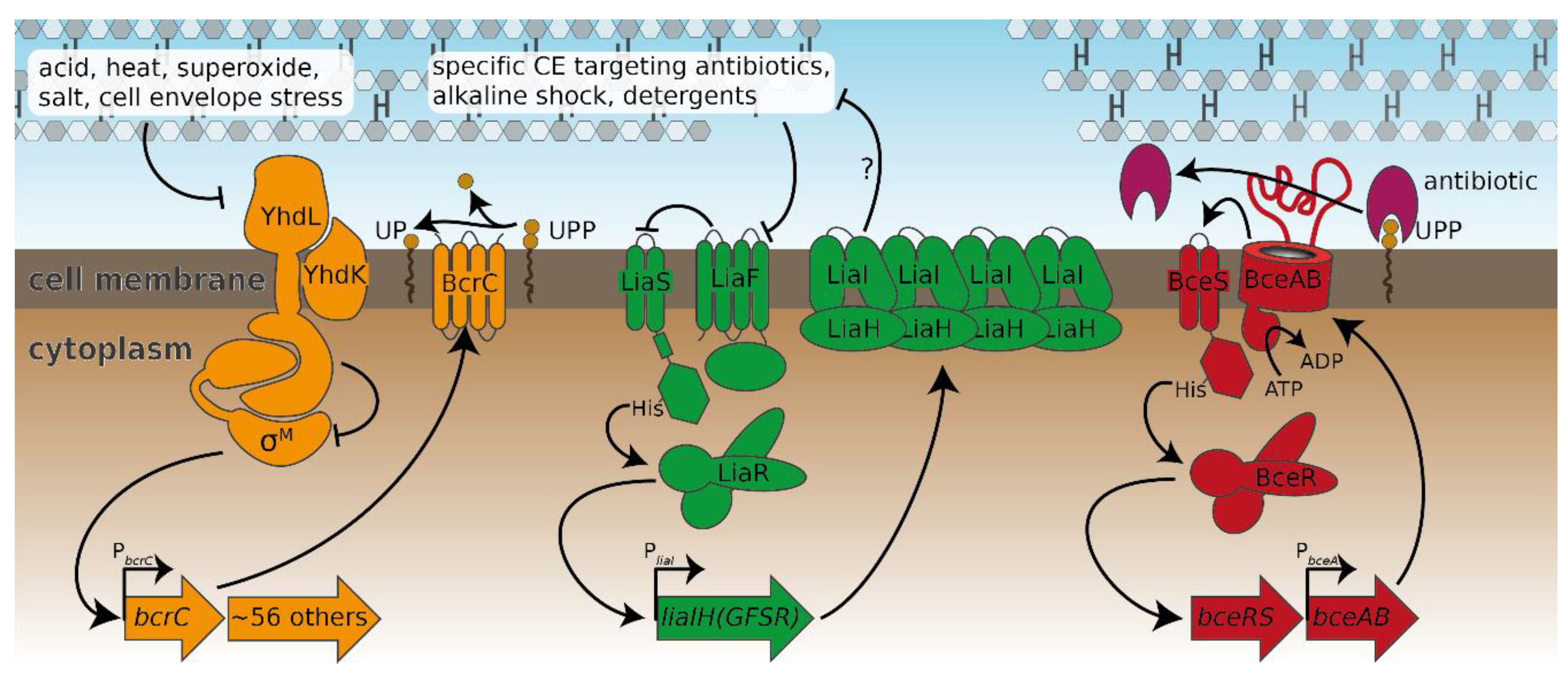
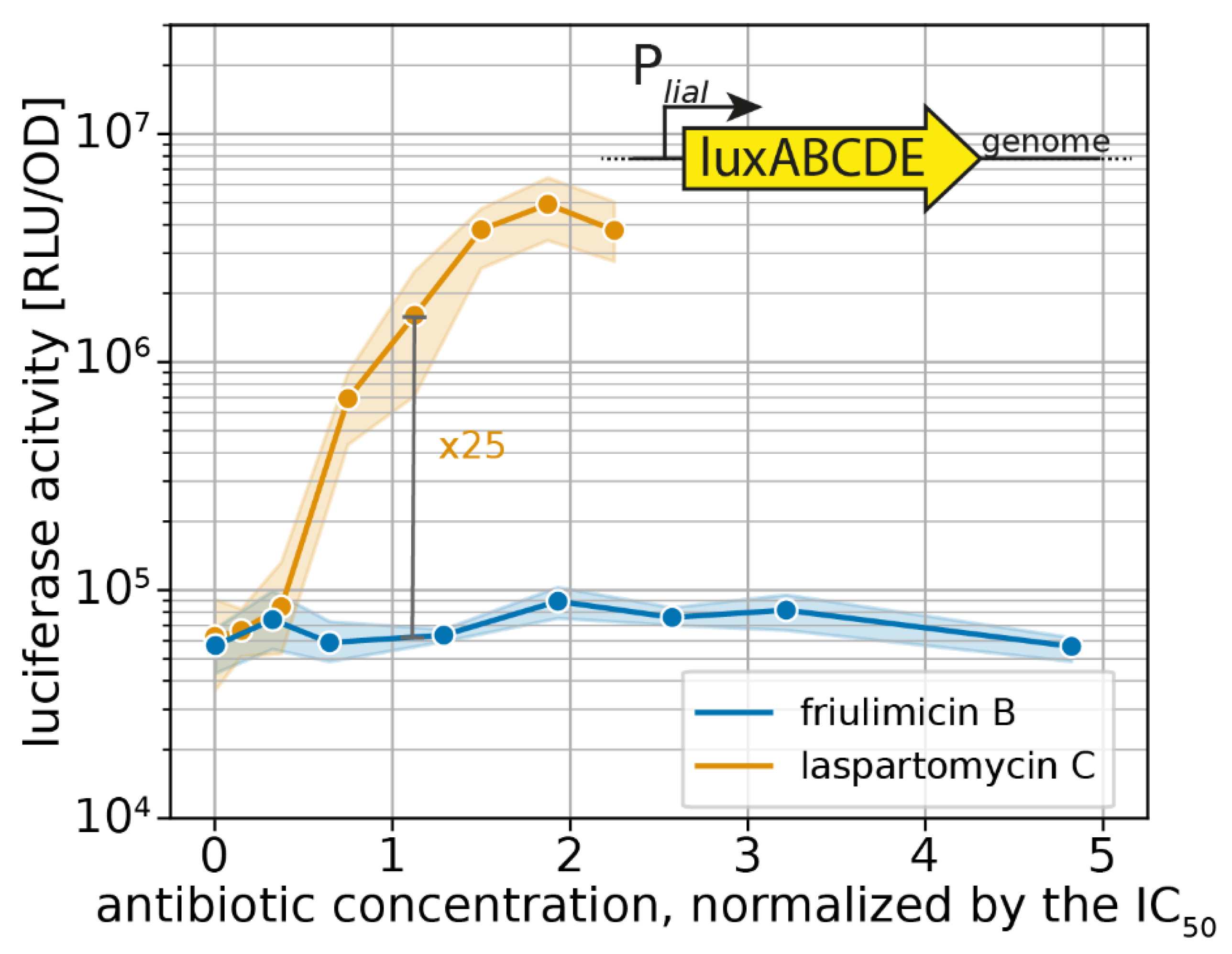
2.2. Probing the Broader CESR against Laspartomycin C Shows Induction of the LiaFSR Module
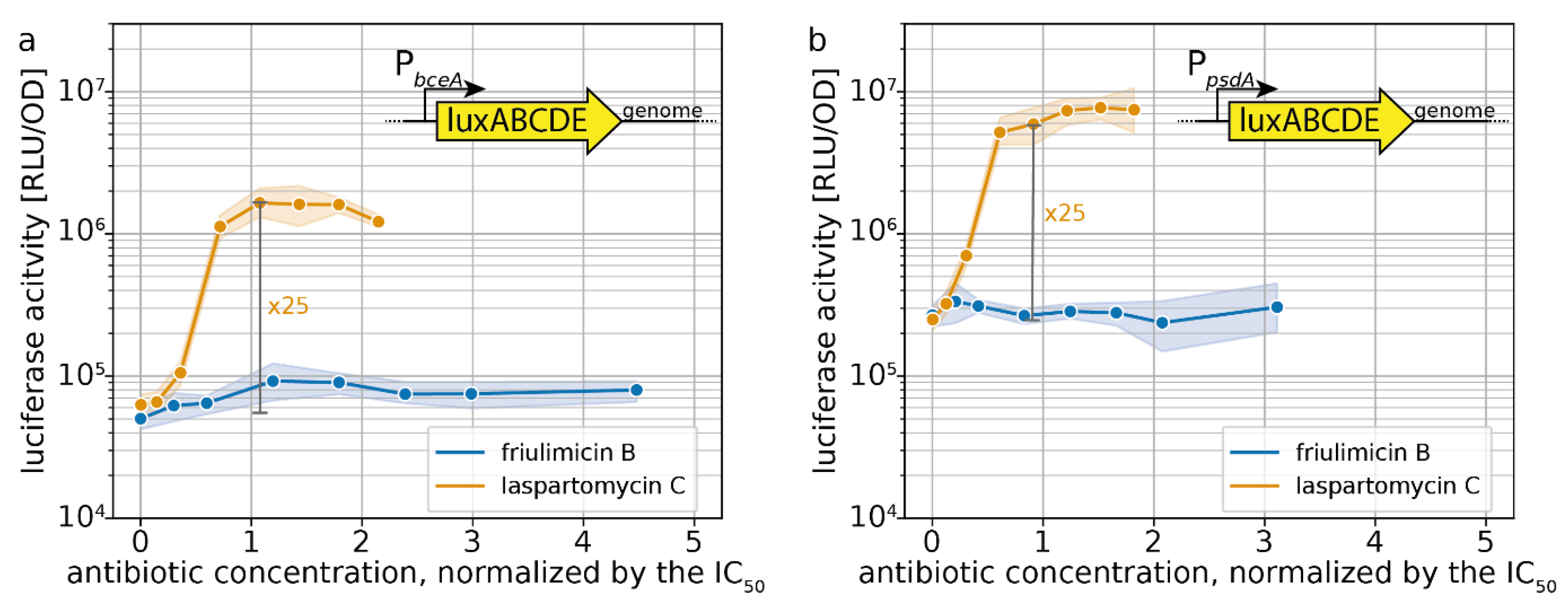
2.3. Induction of Specific CESR Modules by Laspartomycin C
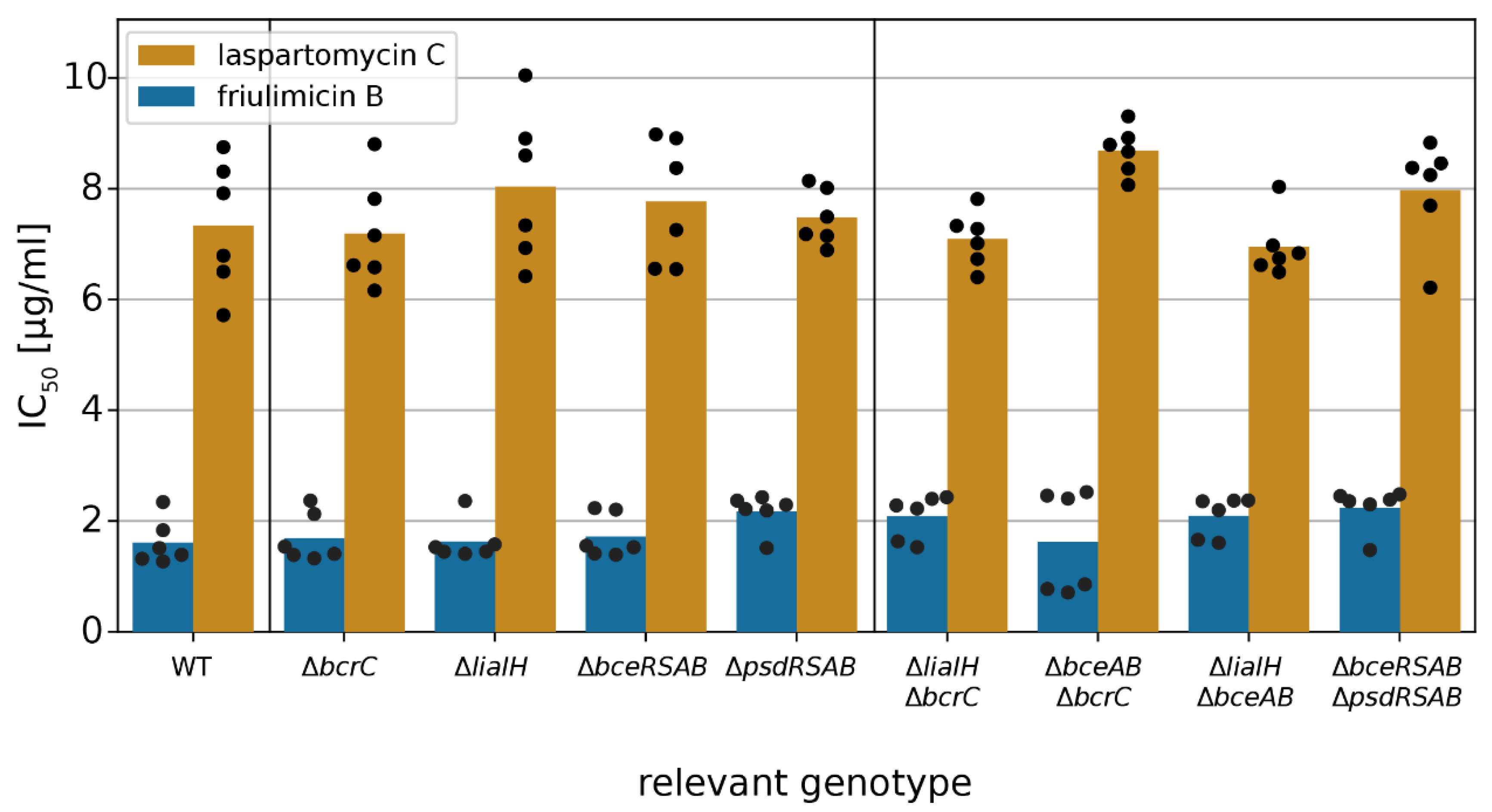
2.4. Acitvated Resistance Modules do not Protect against Laspartomycin C Attack
3. Discussion
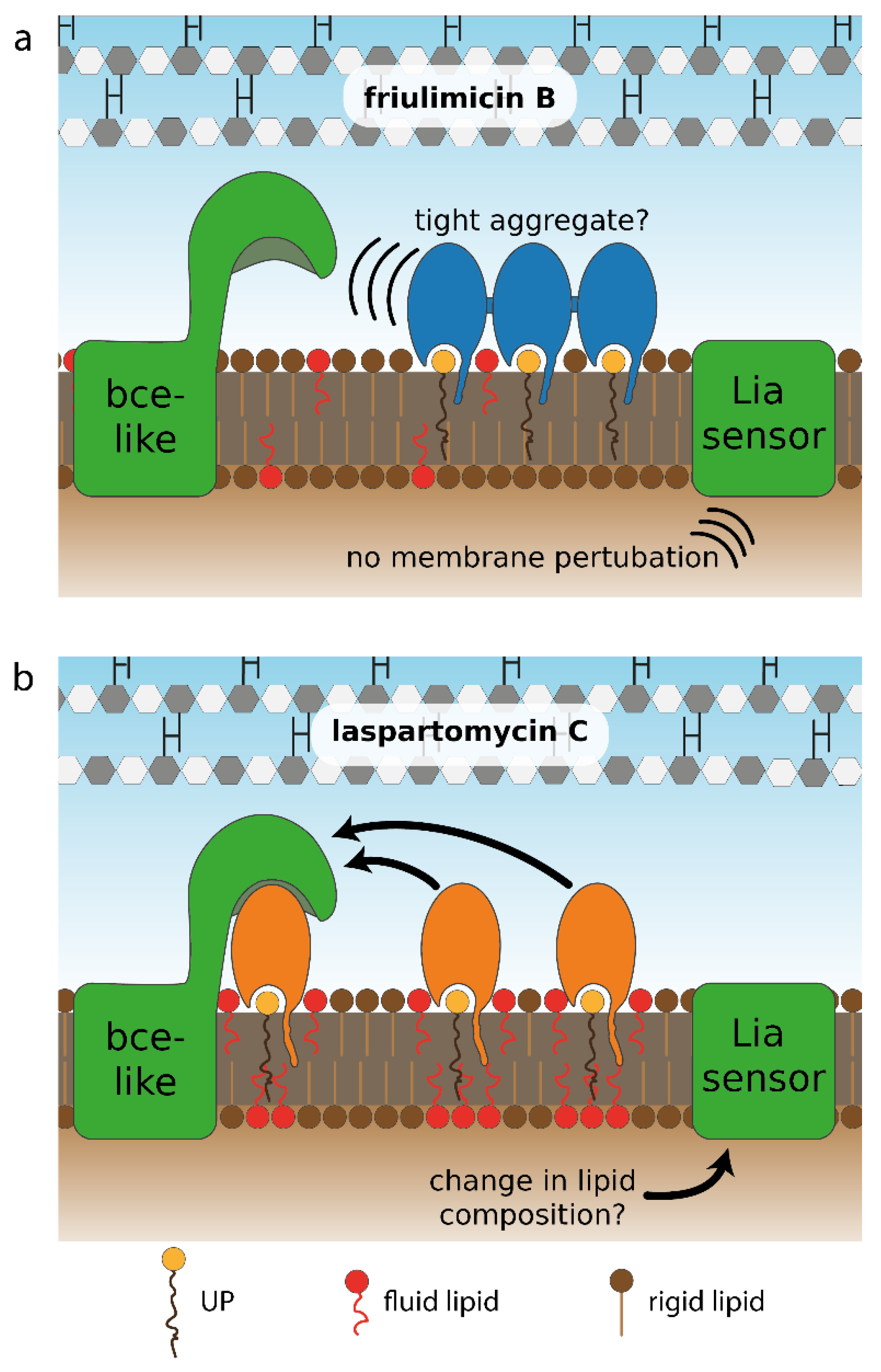
3.1. Novel Clues on CESR Signal Perception
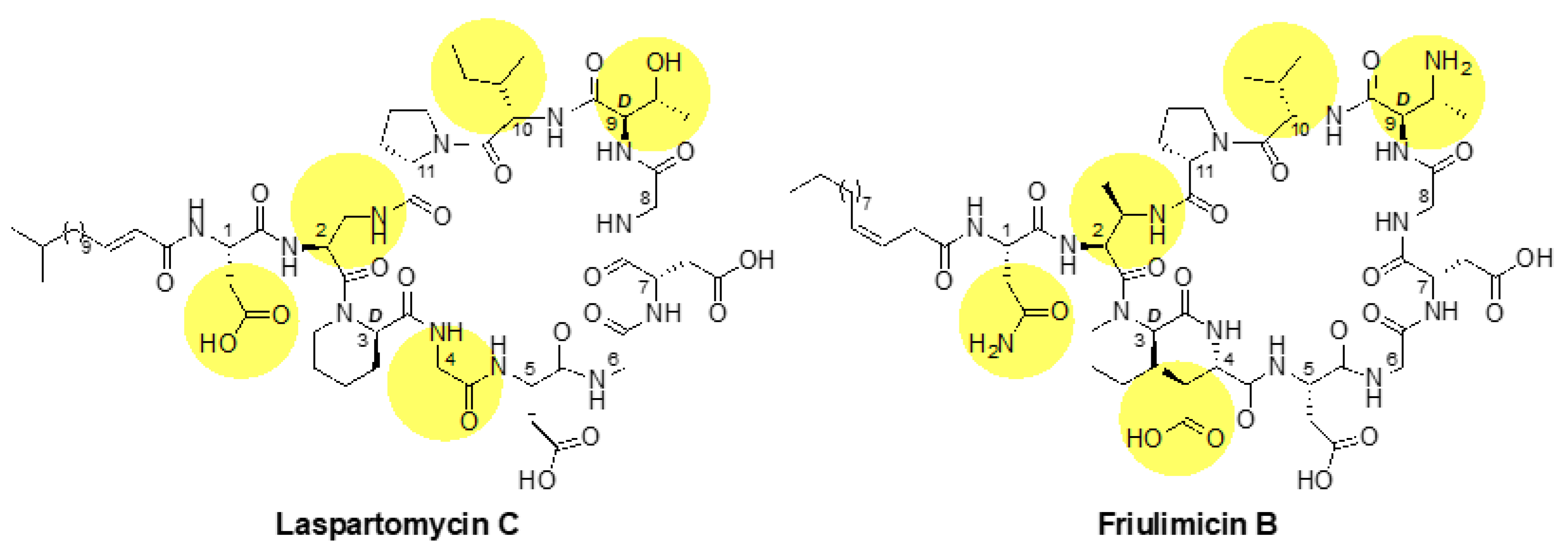
3.2. Differential Sensing of Friulimicin B and Laspartomycin C
3.3. Lacking Antibiotic Protection is Widespread in Bce-like Resistance Modules and may Precede the Spontaneous Evolution of Resistance
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Luciferase Reporter and IC50 Determination Assay
4.3. Data Processing
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Piepenbreier, H.; Diehl, A.; Fritz, G. Minimal exposure of lipid II cycle intermediates triggers cell wall antibiotic resistance. Nat. Commun. 2019, 10, 2733. [Google Scholar] [CrossRef] [PubMed]
- Breukink, E.; de Kruijff, B. Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 2006, 5, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Gries, K.; Josten, M.; Wiedemann, I.; Pelzer, S.; Labischinski, H.; Sahl, H.G. The lipopeptide antibiotic Friulimicin B inhibits cell wall biosynthesis through complex formation with bactoprenol phosphate. Antimicrob. Agents Chemother. 2009, 53, 1610–1618. [Google Scholar] [CrossRef] [PubMed]
- Wood, T.M.; Martin, N.I. The calcium-dependent lipopeptide antibiotics: Structure, mechanism, & medicinal chemistry. Medchemcomm 2019, 10, 634–646. [Google Scholar]
- Kleijn, L.H.J.; Oppedijk, S.F.; ’t Hart, P.; van Harten, R.M.; Martin-Visscher, L.A.; Kemmink, J.; Breukink, E.; Martin, N.I. Total Synthesis of Laspartomycin C and Characterization of Its Antibacterial Mechanism of Action. J. Med. Chem. 2016, 59, 3569–3574. [Google Scholar] [CrossRef]
- Corcilius, L.; Elias, N.T.; Ochoa, J.L.; Linington, R.G.; Payne, R.J. Total Synthesis of Glycinocins A-C. J. Org. Chem. 2017, 82, 12778–12785. [Google Scholar] [CrossRef]
- Wood, T.M.; Bertheussen, K.; Martin, N.I. The contribution of achiral residues in the laspartomycin family of calcium-dependent lipopeptide antibiotics. Org. Biomol. Chem. 2019. [Google Scholar] [CrossRef]
- Kleijn, L.H.J.; Vlieg, H.C.; Wood, T.M.; Sastre Toraño, J.; Janssen, B.J.C.; Martin, N.I. High-resolution crystal structure reveals molecular details of target recognition by the calcium-dependent lipopeptide antibiotic laspartomycin C. Angew. Chem. Int. Ed Engl. 2017. [Google Scholar] [CrossRef]
- Wecke, T.; Zühlke, D.; Mäder, U.; Jordan, S.; Voigt, B.; Pelzer, S.; Labischinski, H.; Homuth, G.; Hecker, M.; Mascher, T. Daptomycin versus Friulimicin B: In-depth profiling of Bacillus subtilis cell envelope stress responses. Antimicrob. Agents Chemother. 2009, 53, 1619–1623. [Google Scholar] [CrossRef]
- Yoshimura, M.; Asai, K.; Sadaie, Y.; Yoshikawa, H. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology 2004, 150, 591–599. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cao, M.; Helmann, J.D. Regulation of the Bacillus subtilis bcrC bacitracin resistance gene by two extracytoplasmic function sigma factors. J. Bacteriol. 2002, 184, 6123–6129. [Google Scholar] [CrossRef] [PubMed]
- Jordan, S.; Junker, A.; Helmann, J.D.; Mascher, T. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: Identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 2006, 188, 5153–5166. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Escobar, J.; Wolf, D.; Fritz, G.; Höfler, C.; Wedlich-Söldner, R.; Mascher, T. Subcellular localization, interactions and dynamics of the phage-shock protein-like Lia response in Bacillus subtilis. Mol. Microbiol. 2014, 92, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Wecke, T.; Bauer, T.; Harth, H.; Mäder, U.; Mascher, T. The rhamnolipid stress response of Bacillus subtilis. FEMS Microbiol. Lett. 2011, 323, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.; Kalamorz, F.; Wecke, T.; Juszczak, A.; Mäder, U.; Homuth, G.; Jordan, S.; Kirstein, J.; Hoppert, M.; Voigt, B.; et al. In-depth profiling of the LiaR response of Bacillus subtilis. J. Bacteriol. 2010, 192, 4680–4693. [Google Scholar] [CrossRef] [PubMed]
- McAuley, S.; Vadia, S.; Jani, C.; Huynh, A.; Yang, Z.; Levin, P.A.; Nodwell, J.R. A Chemical Inhibitor of Cell Growth Reduces Cell Size in Bacillus subtilis. ACS Chem. Biol. 2019, 14, 688–695. [Google Scholar] [CrossRef]
- Radeck, J.; Fritz, G.; Mascher, T. The cell envelope stress response of Bacillus subtilis: From static signaling devices to dynamic regulatory network. Curr. Genet. 2017, 63, 79–90. [Google Scholar] [CrossRef]
- Kobras, C.M.; Piepenbreier, H.; Emenegger, J.; Sim, A.; Fritz, G.; Gebhard, S. BceAB-type antibiotic resistance transporters appear to act by target protection of cell wall synthesis. Antimicrob. Agents Chemother. 2019, 2019/12/25, 835702. [Google Scholar] [CrossRef]
- Koh, A.; Gibbon, M.J.; van der Kamp, M.W.; Pudney, C.R.; Gebhard, S. Conformation control of the histidine kinase BceS of Bacillus subtilis by its cognate ABC-transporter facilitates need-based activation of antibiotic resistance. Mol. Microbiol. 2020. [Google Scholar] [CrossRef]
- Fritz, G.; Dintner, S.; Treichel, N.S.; Radeck, J.; Gerland, U.; Mascher, T.; Gebhard, S. A new way of sensing: Need-based activation of antibiotic resistance by a flux-sensing mechanism. MBio 2015, 6, e00975. [Google Scholar] [CrossRef]
- Rietkötter, E.; Hoyer, D.; Mascher, T. Bacitracin sensing in Bacillus subtilis. Mol. Microbiol. 2008, 68, 768–785. [Google Scholar] [CrossRef]
- Staroń, A.; Finkeisen, D.E.; Mascher, T. Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob. Agents Chemother. 2011, 55, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Wolf, D.; Gutzeit, H.O. The black soldier fly, Hermetia illucens—A promising source for sustainable production of proteins, lipids and bioactive substances. Z. Naturforsch. C 2017, 72, 351–363. [Google Scholar] [CrossRef] [PubMed]
- Dintner, S.; Staroń, A.; Berchtold, E.; Petri, T.; Mascher, T.; Gebhard, S. Coevolution of ABC transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes Bacteria. J. Bacteriol. 2011, 193, 3851–3862. [Google Scholar] [CrossRef]
- Joseph, P.; Fichant, G.; Quentin, Y.; Denizot, F. Regulatory relationship of two-component and ABC transport systems and clustering of their genes in the Bacillus/Clostridium group, suggest a functional link between them. J. Mol. Microbiol. Biotechnol. 2002, 4, 503–513. [Google Scholar] [PubMed]
- Dintner, S.; Heermann, R.; Fang, C.; Jung, K.; Gebhard, S. A sensory complex consisting of an ATP-binding cassette transporter and a two-component regulatory system controls bacitracin resistance in Bacillus subtilis. J. Biol. Chem. 2014, 289, 27899–27910. [Google Scholar] [CrossRef]
- Wilson, D.N.; Hauryliuk, V.; Atkinson, G.C.; O’Neill, A.J. Target protection as a key antibiotic resistance mechanism. Nat. Rev. Microbiol. 2020, 18, 637–648. [Google Scholar] [CrossRef]
- Mascher, T.; Margulis, N.G.; Wang, T.; Ye, R.W.; Helmann, J.D. Cell wall stress responses in Bacillus subtilis: The regulatory network of the bacitracin stimulon. Mol. Microbiol. 2003, 50, 1591–1604. [Google Scholar] [CrossRef]
- Mascher, T.; Zimmer, S.L.; Smith, T.-A.; Helmann, J.D. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 2004, 48, 2888–2896. [Google Scholar] [CrossRef]
- Kepplinger, B.; Morton-Laing, S.; Seistrup, K.H.; Marrs, E.C.L.; Hopkins, A.P.; Perry, J.D.; Strahl, H.; Hall, M.J.; Errington, J.; Allenby, N.E.E. Mode of Action and Heterologous Expression of the Natural Product Antibiotic Vancoresmycin. ACS Chem. Biol. 2018, 13, 207–214. [Google Scholar] [CrossRef]
- Radeck, J.; Gebhard, S.; Orchard, P.S.; Kirchner, M.; Bauer, S.; Mascher, T.; Fritz, G. Anatomy of the bacitracin resistance network in Bacillus subtilis. Mol. Microbiol. 2016, 100, 607–620. [Google Scholar] [CrossRef]
- Gebhard, S.; Mascher, T. Antimicrobial peptide sensing and detoxification modules: Unravelling the regulatory circuitry of Staphylococcus aureus. Mol. Microbiol. 2011, 81, 581–587. [Google Scholar] [CrossRef]
- Höfler, C.; Heckmann, J.; Fritsch, A.; Popp, P.; Gebhard, S.; Fritz, G.; Mascher, T. Cannibalism stress response in Bacillus subtilis. Microbiology 2016, 162, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Chang, G. Multidrug resistance ABC transporters. FEBS Lett. 2003, 555, 102–105. [Google Scholar] [CrossRef]
- Asai, K. Anti-sigma factor-mediated cell surface stress responses in Bacillus subtilis. Genes Genet. Syst. 2018, 92, 223–234. [Google Scholar] [CrossRef]
- Müller, A.; Wenzel, M.; Strahl, H.; Grein, F.; Saaki, T.N.V.; Kohl, B.; Siersma, T.; Bandow, J.E.; Sahl, H.-G.; Schneider, T.; et al. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc. Natl. Acad. Sci. USA 2016, 113, E7077–E7086. [Google Scholar] [CrossRef] [PubMed]
- Grein, F.; Müller, A.; Scherer, K.M.; Liu, X.; Ludwig, K.C.; Klöckner, A.; Strach, M.; Sahl, H.-G.; Kubitscheck, U.; Schneider, T. Ca2+-Daptomycin targets cell wall biosynthesis by forming a tripartite complex with undecaprenyl-coupled intermediates and membrane lipids. Nat. Commun. 2020, 11, 1455. [Google Scholar] [CrossRef] [PubMed]
- Mescola, A.; Ragazzini, G.; Alessandrini, A. Daptomycin Strongly Affects the Phase Behavior of Model Lipid Bilayers. J. Phys. Chem. B 2020. [Google Scholar] [CrossRef] [PubMed]
- Crow, A.; Greene, N.P.; Kaplan, E.; Koronakis, V. Structure and mechanotransmission mechanism of the MacB ABC transporter superfamily. Proc. Natl. Acad. Sci. 2017, 114, 12572–12577. [Google Scholar] [CrossRef] [PubMed]
- Mavrici, D.; Marakalala, M.J.; Holton, J.M.; Prigozhin, D.M.; Gee, C.L.; Zhang, Y.J.; Rubin, E.J.; Alber, T. Mycobacterium tuberculosis FtsX extracellular domain activates the peptidoglycan hydrolase, RipC. Proc. Natl. Acad. Sci. USA 2014, 111, 8037–8042. [Google Scholar] [CrossRef] [PubMed]
- Neidhardt, F.C.; Bloch, P.L.; Smith, D.F. Culture Medium for Enterobacteria. J. Bacteriol. 1974, 119, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, S.; Fang, C.; Shaaly, A.; Leslie, D.J.; Weimar, M.R.; Kalamorz, F.; Carne, A.; Cook, G.M. Identification and characterization of a bacitracin resistance network in Enterococcus faecalis. Antimicrob. Agents Chemother. 2014, 58, 1425–1433. [Google Scholar] [CrossRef] [PubMed]
| B. subtilis Strain | Source/Reference |
|---|---|
| W168 trpC2 | Laboratory stock |
| W168 PbcrC-lux | [31] |
| W168 PliaI-lux | [31] |
| W168 PbceA-lux | [31] |
| W168 PpsdA-lux | [33] |
| W168 bcrC::kan | [31] |
| W168 ΔliaIH | [31] |
| W168 ΔbceRSAB | [20] |
| W168 ΔpsdRSAB | Intermediate strain to produce TMB1518 [42] |
| W168 ΔliaIH bceAB::kan | [31] |
| W168 ΔliaIH bcrC::tet | [31] |
| W168 bceAB::kan bcrC::tet PliaI-lux | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diehl, A.; Wood, T.M.; Gebhard, S.; Martin, N.I.; Fritz, G. The Cell Envelope Stress Response of Bacillus subtilis towards Laspartomycin C. Antibiotics 2020, 9, 729. https://doi.org/10.3390/antibiotics9110729
Diehl A, Wood TM, Gebhard S, Martin NI, Fritz G. The Cell Envelope Stress Response of Bacillus subtilis towards Laspartomycin C. Antibiotics. 2020; 9(11):729. https://doi.org/10.3390/antibiotics9110729
Chicago/Turabian StyleDiehl, Angelika, Thomas M. Wood, Susanne Gebhard, Nathaniel I. Martin, and Georg Fritz. 2020. "The Cell Envelope Stress Response of Bacillus subtilis towards Laspartomycin C" Antibiotics 9, no. 11: 729. https://doi.org/10.3390/antibiotics9110729
APA StyleDiehl, A., Wood, T. M., Gebhard, S., Martin, N. I., & Fritz, G. (2020). The Cell Envelope Stress Response of Bacillus subtilis towards Laspartomycin C. Antibiotics, 9(11), 729. https://doi.org/10.3390/antibiotics9110729





