Growth Rate of Escherichia coli During Human Urinary Tract Infection: Implications for Antibiotic Effect
Abstract
1. Introduction
2. Results
2.1. Study Population
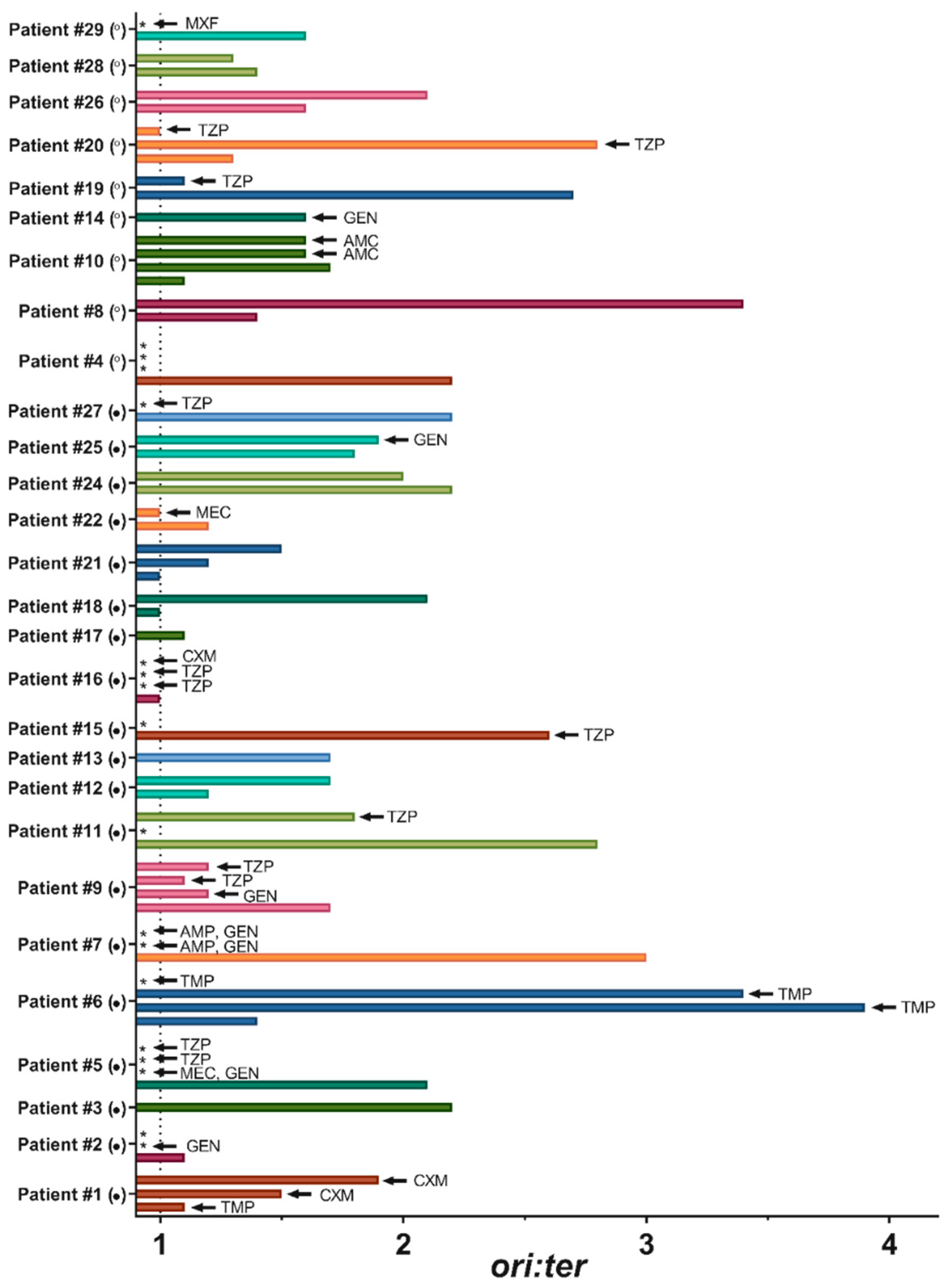
2.2. Bacterial Growth Rates in Human Urine
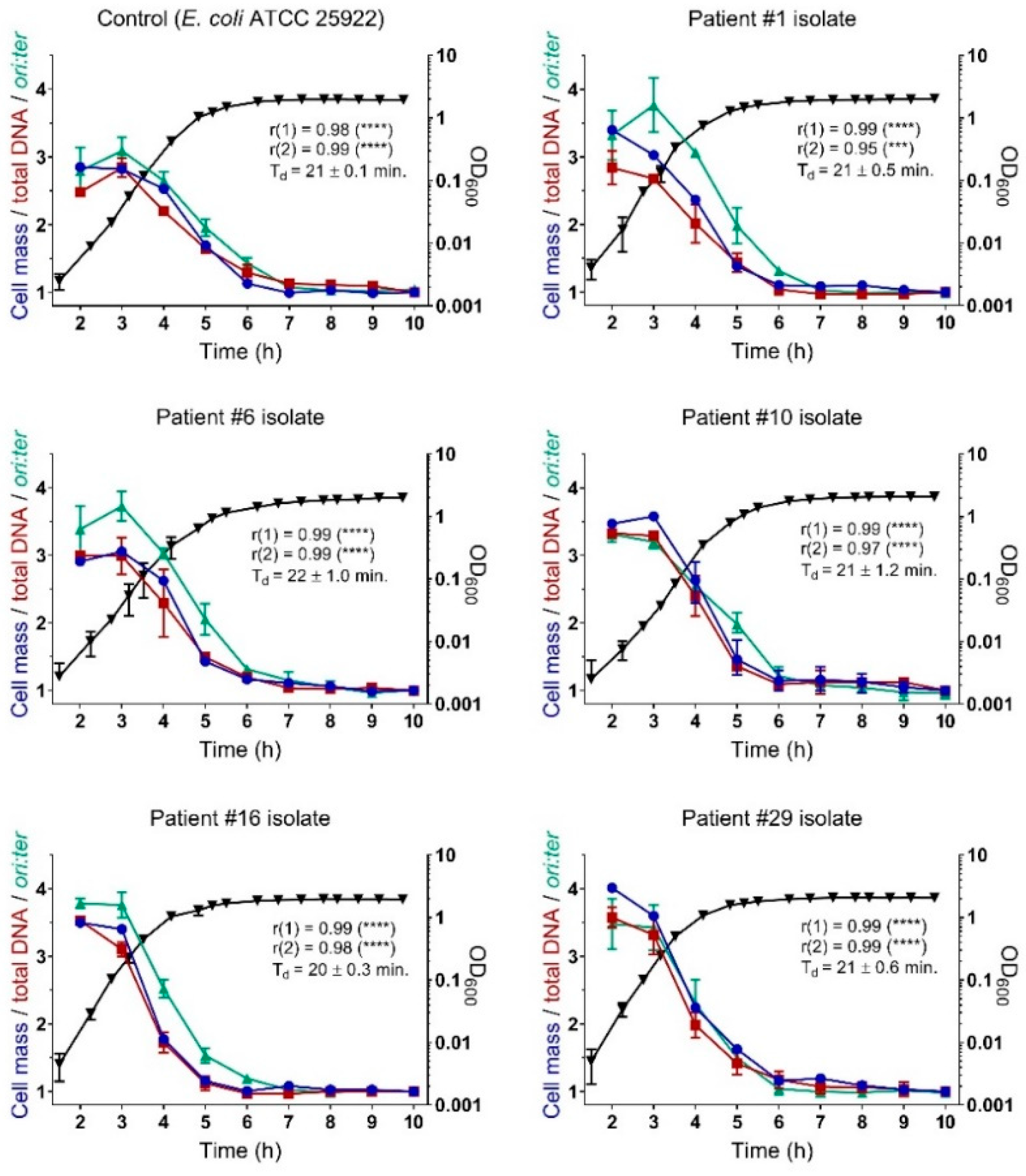
2.3. Bacterial Growth Rates During Controlled Propagation (in vitro)
3. Discussion
4. Materials and Methods
4.1. Study Population and Strain Collection
4.2. Research Ethical Approvals
4.3. Bacterial Identification and Susceptibility Testing
4.4. In Vitro Growth Experiment
4.5. Whole-Genome Sequencing and Data Analysis
4.6. Flow Cytometry
4.7. Real-Time Quantitative PCR (qPCR)
4.8. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hagan, E.C.; Donnenberg, M.S.; Mobley, H.L.T. Uropathogenic Escherichia coli. EcoSal Plus 2009, 3. [Google Scholar] [CrossRef]
- Johnson, J.R.; Russo, T.A. Extraintestinal pathogenic Escherichia coli: “The other bad E coli”. J. Lab. Clin. Med. 2002, 139, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Smith, H. Pathogenicity and the Microbe in vivo: The 1989 Fred Griffith Review Lecture. Microbiology 1990, 136, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Haugan, M.S.; Løbner-Olesen, A.; Frimodt-Møller, N. Comparative activity of ceftriaxone, ciprofloxacin and gentamicin as a function of bacterial growth rate probed by Escherichia coli chromosome replication in the mouse peritonitis model. Antimicrob. Agents Chemother. 2019, 63, e02133-e18. [Google Scholar] [CrossRef] [PubMed]
- Tuomanen, E.; Cozens, R.; Tosch, W.; Zak, O.; Tomasz, A. The rate of killing of Escherichia coli by β-lactam antibiotics is strictly proportional to the rate of bacterial growth. Microbiology 1986, 132, 1297–1304. [Google Scholar] [CrossRef]
- Lee, A.J.; Wang, S.; Meredith, H.R.; Zhuang, B.; Dai, Z.; You, L. Robust, linear correlations between growth rates and β-lactam–mediated lysis rates. Proc. Natl. Acad. Sci. USA 2018, 115, 4069–4074. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.R.; Collier, P.J.; Gilbert, P. Influence of growth rate on susceptibility to antimicrobial agents: Modification of the cell envelope and batch and continuous culture studies. Antimicrob. Agents Chemother. 1990, 34, 1623–1628. [Google Scholar] [CrossRef]
- Cozens, R.M.; Tuomanen, E.; Tosch, W.; Zak, O.; Suter, J.; Tomasz, A. Evaluation of the bactericidal activity of beta-lactam antibiotics on slowly growing bacteria cultured in the chemostat. Antimicrob. Agents Chemother. 1986, 29, 797–802. [Google Scholar] [CrossRef]
- Eng, R.H.; Padberg, F.T.; Smith, S.M.; Tan, E.N.; Cherubin, C.E. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 1991, 35, 1824–1828. [Google Scholar] [CrossRef]
- Fantin, B.; Leggett, J.; Ebert, S.; Craig, W.A. Correlation between in vitro and in vivo activity of antimicrobial agents against gram-negative bacilli in a murine infection model. Antimicrob. Agents Chemother. 1991, 35, 1413–1422. [Google Scholar] [CrossRef] [PubMed]
- Zeiler, H.J.; Voigt, W.H. Efficacy of ciprofloxacin in stationary-phase bacteria in vivo. Am. J. Med. 1987, 82, 87–90. [Google Scholar] [PubMed]
- Subashchandrabose, S.; Mobley, H.L.T. Virulence and Fitness Determinants of Uropathogenic Escherichia coli. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Ulett, G.C.; Totsika, M.; Schaale, K.; Carey, A.J.; Sweet, M.J.; Schembri, M.A. Uropathogenic Escherichia coli virulence and innate immune responses during urinary tract infection. Curr. Opin. Microbiol. 2013, 16, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Forsyth, V.S.; Armbruster, C.E.; Smith, S.N.; Pirani, A.; Springman, A.C.; Walters, M.S.; Nielubowicz, G.R.; Himpsl, S.D.; Snitkin, E.S.; Mobley, H.L.T. Rapid Growth of Uropathogenic Escherichia coli during Human Urinary Tract Infection. MBio 2018, 9, e00186-e18. [Google Scholar] [CrossRef] [PubMed]
- Korem, T.; Zeevi, D.; Suez, J.; Weinberger, A.; Avnit-Sagi, T.; Pompan-Lotan, M.; Matot, E.; Jona, G.; Harmelin, A.; Cohen, N.; et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 2015, 349, 1101–1106. [Google Scholar] [CrossRef]
- Brown, C.T.; Olm, M.R.; Thomas, B.C.; Banfield, J.F. Measurement of bacterial replication rates in microbial communities. Nat. Biotechnol. 2016, 34, 1256–1263. [Google Scholar] [CrossRef]
- Olm, M.R.; Brown, C.T.; Brooks, B.; Firek, B.; Baker, R.; Burstein, D.; Soenjoyo, K.; Thomas, B.C.; Morowitz, M.; Banfield, J.F. Identical bacterial populations colonize premature infant gut, skin, and oral microbiomes and exhibit different in situ growth rates. Genome Res. 2017, 27, 601–612. [Google Scholar] [CrossRef]
- Haugan, M.S.; Charbon, G.; Frimodt-Møller, N.; Løbner-Olesen, A. Chromosome replication as a measure of bacterial growth rate during Escherichia coli infection in the mouse peritonitis model. Sci. Rep. 2018, 8, 14961. [Google Scholar] [CrossRef]
- Cooper, S.; Helmstetter, C.E. Chromosome replication and the division cycle of Escherichia coli Br. J. Mol. Biol. 1968, 31, 519–540. [Google Scholar] [CrossRef]
- Donachie, W.D. Relationship between Cell Size and Time of Initiation of DNA Replication. Nature 1968, 219, 1077–1079. [Google Scholar] [CrossRef] [PubMed]
- Skarstad, K.; Boye, E.; Steen, H.B. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 1986, 5, 1711–1717. [Google Scholar] [CrossRef] [PubMed]
- Eng, R.H.; Smith, S.M.; Cherubin, C.E.; Tan, E.N. Evaluation of two methods for overcoming the antibiotic carry-over effect. Eur. J. Clin. Microbiol. Infect. Dis. 1991, 10, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Bushby, S.R.; Hitchings, G.H. Trimethoprim, a sulphonamide potentiator. Br. J. Pharmacol. Chemother. 1968, 33, 72–90. [Google Scholar] [CrossRef] [PubMed]
- Krogfelt, K.A.; Bergmans, H.; Klemm, P. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect. Immun. 1990, 58, 1995–1998. [Google Scholar]
- Sarkar, S.; Ulett, G.C.; Totsika, M.; Phan, M.D.; Schembri, M.A. Role of Capsule and O Antigen in the Virulence of Uropathogenic Escherichia coli. PLoS ONE 2014, 9, e94786. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S. Molecular epidemiology of uropathogenic Escherichia coli. J. Infect. Chemother. 2007, 13, 68–73. [Google Scholar] [CrossRef]
- Kornberg, A.; Baker, T.A. DNA Replication, 2nd ed.; University Science Books: Sausalito, CA, USA, 1992; Volume 1992. [Google Scholar]
- Sezonov, G.; Joseleau-Petit, D.; D’Ari, R. Escherichia coli Physiology in Luria-Bertani Broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef]
- Si, F.; Li, D.; Cox, S.E.; Sauls, J.T.; Azizi, O.; Sou, C.; Schwartz, A.B.; Erickstad, M.J.; Jun, Y.; Li, X.; et al. Invariance of Initiation Mass and Predictability of Cell Size in Escherichia coli. Curr. Biol. 2017, 27, 1278–1287. [Google Scholar] [CrossRef]
- Antimicrobial Susceptibility Testing EUCAST Disk Diffusion Method, Version 6.0. Available online: http://www.eucast.org/ast_of_bacteria/disk_diffusion_methodology/ (accessed on 3 December 2018).
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Pontén, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of Methods for Genomic Taxonomy. J. Clin. Microbiol. 2014, 52, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Løbner-Olesen, A.; Skarstad, K.; Hansen, F.G.; von Meyenburg, K.; Boye, E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell 1989, 57, 881–889. [Google Scholar] [CrossRef]
- Fujimitsu, K.; Su’etsugu, M.; Yamaguchi, Y.; Mazda, K.; Fu, N.; Kawakami, H.; Katayama, T. Modes of Overinitiation, dnaA Gene Expression, and Inhibition of Cell Division in a Novel Cold-Sensitive hda Mutant of Escherichia coli. J. Bacteriol. 2008, 190, 5368–5381. [Google Scholar] [CrossRef]
- Riber, L.; Olsson, J.A.; Jensen, R.B.; Skovgaard, O.; Dasgupta, S.; Marinus, M.G.; Løbner-Olesen, A. Hda-mediated inactivation of the DnaA protein and dnaA gene autoregulation act in concert to ensure homeostatic maintenance of the Escherichia coli chromosome. Genes Dev. 2006, 20, 2121–2134. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
| Characteristic | Value |
|---|---|
| Total number of participants | 29 |
| Median age, years (range) | 79 (29–99) |
| Sex, number female (%) | 22 (76) |
| Symptoms and/or other signs of urinary tract infection (UTI), number of participants (%) * | 20 (69) |
| Median duration of symptoms, days (range) † | 1 (1–14) |
| Urethral catheter before collection of day 0 urine specimen, number of participants (%) | 11 (38) |
| E. coli bacteraemia, number of participants (%) | 5 (17) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haugan, M.S.; Hertz, F.B.; Charbon, G.; Sahin, B.; Løbner-Olesen, A.; Frimodt-Møller, N. Growth Rate of Escherichia coli During Human Urinary Tract Infection: Implications for Antibiotic Effect. Antibiotics 2019, 8, 92. https://doi.org/10.3390/antibiotics8030092
Haugan MS, Hertz FB, Charbon G, Sahin B, Løbner-Olesen A, Frimodt-Møller N. Growth Rate of Escherichia coli During Human Urinary Tract Infection: Implications for Antibiotic Effect. Antibiotics. 2019; 8(3):92. https://doi.org/10.3390/antibiotics8030092
Chicago/Turabian StyleHaugan, Maria Schei, Frederik Boëtius Hertz, Godefroid Charbon, Berivan Sahin, Anders Løbner-Olesen, and Niels Frimodt-Møller. 2019. "Growth Rate of Escherichia coli During Human Urinary Tract Infection: Implications for Antibiotic Effect" Antibiotics 8, no. 3: 92. https://doi.org/10.3390/antibiotics8030092
APA StyleHaugan, M. S., Hertz, F. B., Charbon, G., Sahin, B., Løbner-Olesen, A., & Frimodt-Møller, N. (2019). Growth Rate of Escherichia coli During Human Urinary Tract Infection: Implications for Antibiotic Effect. Antibiotics, 8(3), 92. https://doi.org/10.3390/antibiotics8030092







