Characterization of a Carbapenem-Resistant Kluyvera Cryocrescens Isolate Carrying Blandm-1 from Hospital Sewage
Abstract
1. Introduction
2. Results and Discussion
2.1. Antimicrobial Susceptibility of SCW13
2.2. Genome Characteristics of SCW13
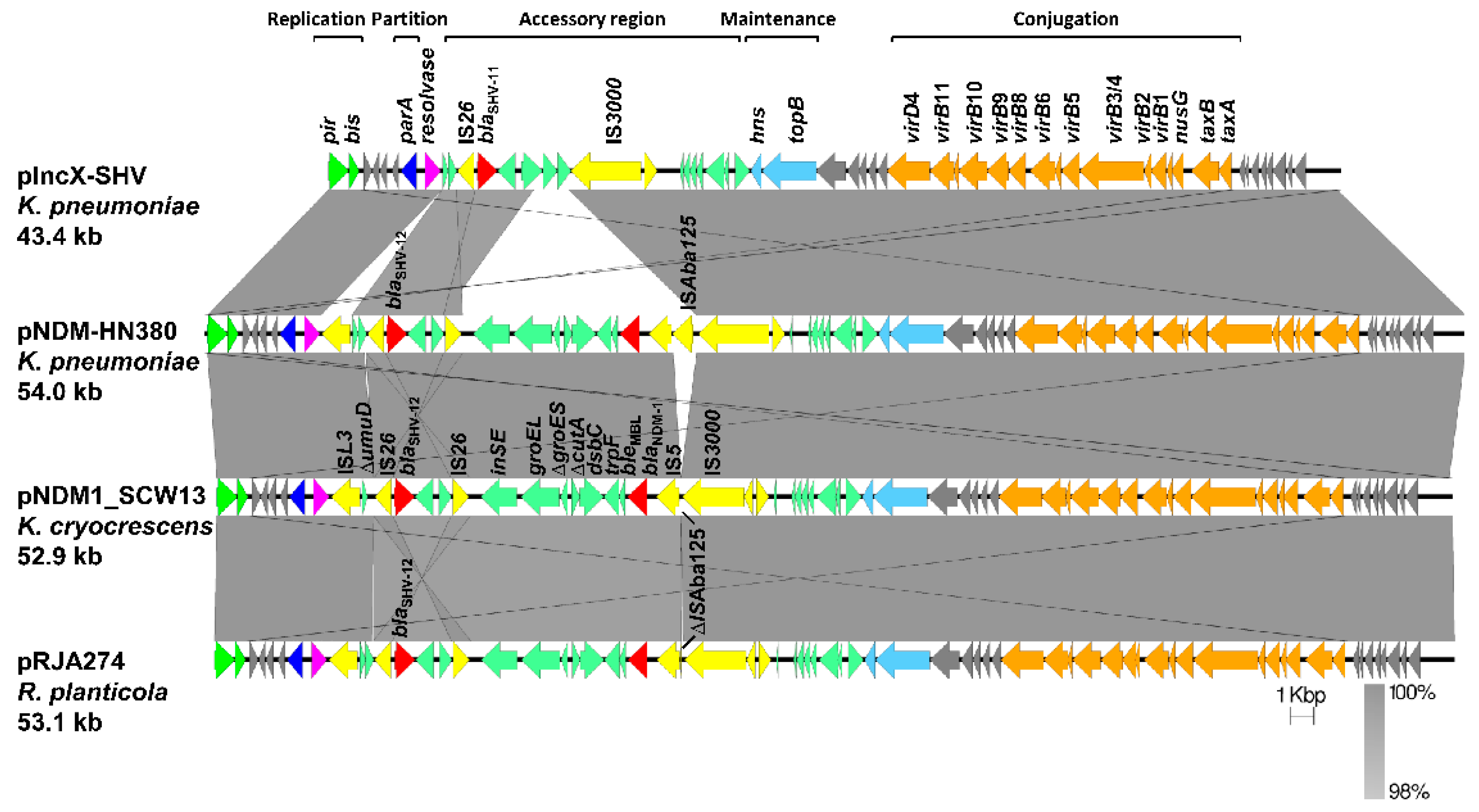
2.3. Analysis of the BlaNDM-1-Harboring Plasmid PNDM1_SCW13
2.4. Analysis of the Chromosomally-Encoded KLUC-2
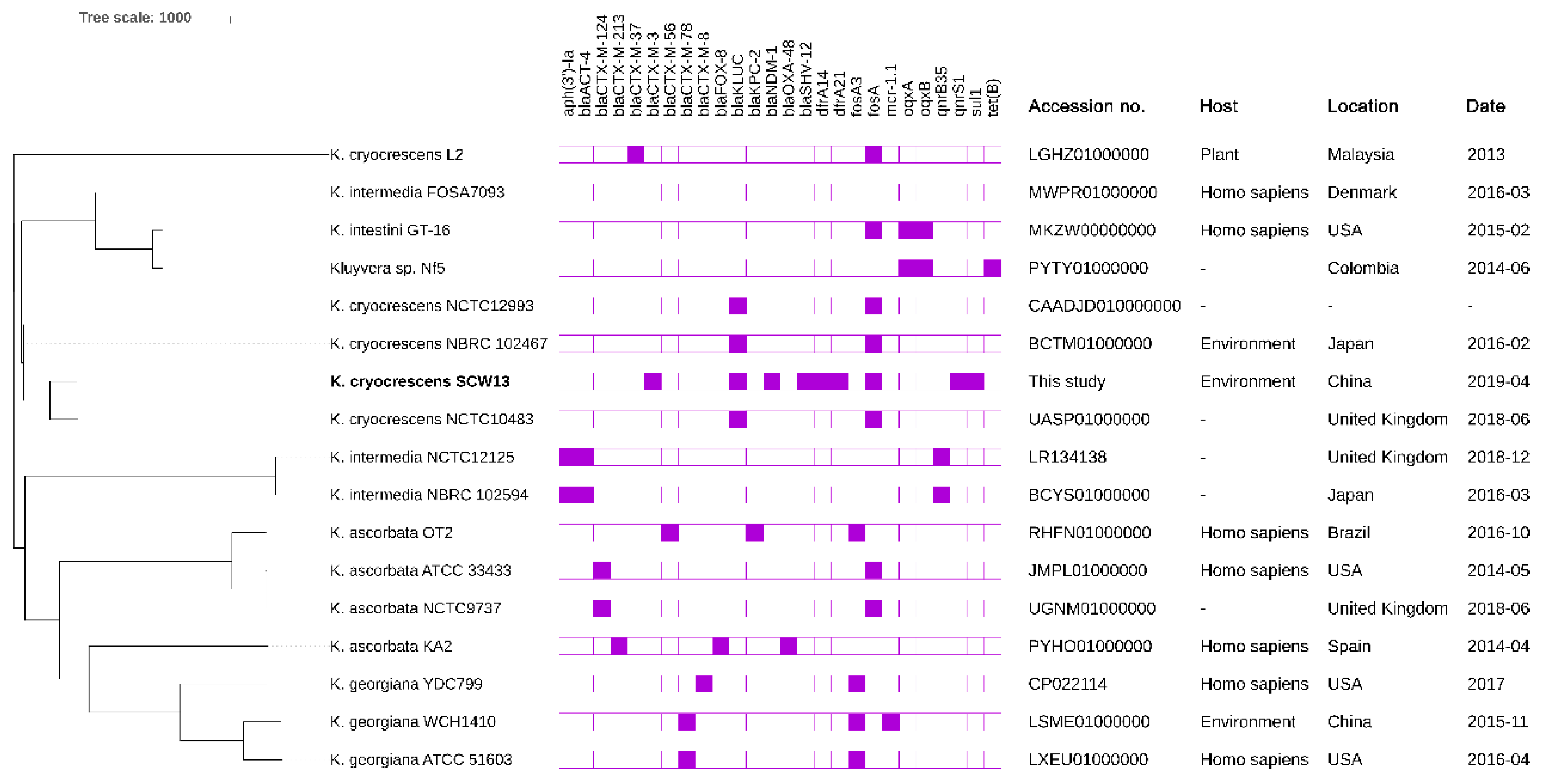
2.5. Phylogenetic Analysis of Different Kluyvera Species
3. Materials and Methods
3.1. Strain Identification
3.2. Antimicrobial Susceptibility Testing
3.3. Conjugation Assay
3.4. Genome Sequencing and Bioinformatic Analysis
3.5. Nucleotide Sequence Accession Numbers
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Perez, F.; Bonomo, R.A. Carbapenem-Resistant Enterobacteriaceae: Global Action Required. Lancet Infect. Dis. 2019, 19, 561–562. [Google Scholar] [CrossRef]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. Ndm Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef] [PubMed]
- Yong, D.; Toleman, M.A.; Giske, C.G.; Cho, H.S.; Sundman, K.; Lee, K.; Walsh, T.R. Characterization of a New Metallo-Beta-Lactamase Gene, Bla(Ndm-1), and a Novel Erythromycin Esterase Gene Carried on a Unique Genetic Structure in Klebsiella Pneumoniae Sequence Type 14 from India. Antimicrob. Agents Chemother. 2009, 53, 5046–5054. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, R.; Li, J.; Wu, Z.; Yin, W.; Schwarz, S.; Tyrrell, J.M.; Zheng, Y.; Wang, S.; Shen, Z.; et al. Comprehensive Resistome Analysis Reveals the Prevalence of Ndm and Mcr-1 in Chinese Poultry Production. Nat. Microbiol. 2017, 2, 16260. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, Genetics and Worldwide Spread of New Delhi Metallo-Beta-Lactamase (Ndm): A Threat to Public Health. BMC Microbiol. 2017, 17, 101. [Google Scholar] [CrossRef]
- Qu, H.; Wang, X.; Ni, Y.; Liu, J.; Tan, R.; Huang, J.; Li, L.; Sun, J. Ndm-1-Producing Enterobacteriaceae in a Teaching Hospital in Shanghai, China: Incx3-Type Plasmids May Contribute to the Dissemination of Blandm-1. Int. J. Infect. Dis. 2015, 34, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Sekizuka, T.; Matsui, M.; Takahashi, T.; Hayashi, M.; Suzuki, S.; Tokaji, A.; Kuroda, M. Complete Genome Sequence of BlaImp-6-Positive Metakosakonia Sp. Mry16-398 Isolate from the Ascites of a Diverticulitis Patient. Front. Microbiol. 2018, 9, 2853. [Google Scholar] [CrossRef] [PubMed]
- Tetz, G.; Tetz, V. Complete Genome Sequence of Kluyvera Intestini Sp. Nov., Isolated from the Stomach of a Patient with Gastric Cancer. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Karadag Oncel, E.; Ozsurekci, Y.; Akyon, Y.; Gur, D.; Cengiz, A.B.; Kara, A. Kluyvera Ascorbata Infections in Children: A Case Series. Turk Pediatri Ars. 2015, 50, 123–128. [Google Scholar] [CrossRef]
- Yusuke, Y.; Nakazawa, S.; Otani, S.; Sekizuka, E.; Ota, Y. Nosocomial Bacteremia Due to Kluyvera Cryocrescens: Case Report and Literature Review. IDCases 2016, 4, 24–26. [Google Scholar]
- Zhao, F.; Zong, Z. Kluyvera Ascorbata Strain from Hospital Sewage Carrying the Mcr-1 Colistin Resistance Gene. Antimicrob. Agents Chemother. 2016, 60, 7498–7501. [Google Scholar] [PubMed]
- Wang, L.; Jing, Y.; Lai, K.; An, J.; Yang, J. A Case of Biliary Tract Infection Caused by Kpc-2-Producing Kluyvera Ascorbata. Case Rep. Infect. Dis. 2018, 2018, 5745708. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, V.B.; Zavascki, A.P.; Nodari, C.S.; Sandri, A.M.; Silva, M.P.; Campos, J.C.; Sampaio, J.L.; Barth, A.L. Detection of Blakpc-2 in a Carbapenem-Resistant Kluyvera Georgiana. J. Antimicrob. Chemother. 2012, 67, 2776–2777. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, V.B.; Zavascki, A.P.; Rozales, F.P.; Pagano, M.; Magagnin, C.M.; Nodari, C.S.; da Silva, R.C.; Dalarosa, M.G.; Falci, D.R.; Barth, A.L. Detection of Bla(Ges-5) in Carbapenem-Resistant Kluyvera Intermedia Isolates Recovered from the Hospital Environment. Antimicrob. Agents Chemother. 2014, 58, 622–623. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, M.; Leon-Sampedro, R.; Perez-Viso, B.; Morosini, M.I.; Lopez-Fresnena, N.; Diaz-Agero, C.; Coque, T.M.; Ruiz-Garbajosa, P.; Canton, R. First Report of an Oxa-48- and Ctx-M-213-Producing Kluyvera Species Clone Recovered from Patients Admitted in a University Hospital in Madrid, Spain. Antimicrob. Agents Chemother. 2018, 62, e01238. [Google Scholar] [CrossRef] [PubMed]
- Paskova, V.; Medvecky, M.; Skalova, A.; Chudejova, K.; Bitar, I.; Jakubu, V.; Bergerova, T.; ZemLickova, H.; Papagiannitsis, C.C.; Hrabak, J. Characterization of Ndm-Encoding Plasmids from Enterobacteriaceae Recovered from Czech Hospitals. Front. Microbiol. 2018, 9, 1549. [Google Scholar] [CrossRef]
- D’Andrea, M.M.; Arena, F.; Pallecchi, L.; Rossolini, G.M. Ctx-M-Type Beta-Lactamases: A Successful Story of Antibiotic Resistance. Int. J. Med. Microbiol. 2013, 303, 305–317. [Google Scholar] [CrossRef]
- Li, P.; Shen, K.; Zhang, Y.; Ying, J.; Zhu, T.; Liu, Y.; Xu, L.; Lin, C.; Zhang, K.; Li, P.; et al. Characterization of a Novel Blakluc Variant with Reduced Beta-Lactam Resistance from an Inca/C Group Plasmid in a Clinical Klebsiella Pneumoniae Isolate. Front. Microbiol. 2018, 9, 1908. [Google Scholar] [CrossRef]
- Decousser, J.W.; Poirel, L.; Nordmann, P. Characterization of a Chromosomally Encoded Extended-Spectrum Class a Beta-Lactamase from Kluyvera Cryocrescens. Antimicrob. Agents Chemother. 2001, 45, 3595–3598. [Google Scholar] [CrossRef]
- Richter, M.; Rossello-Mora, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Ho, P.L.; Li, Z.; Lo, W.U.; Cheung, Y.Y.; Lin, C.H.; Sham, P.C.; Cheng, V.C.; Ng, T.K.; Que, T.L.; Chow, K.H. Identification and Characterization of a Novel Incompatibility Group X3 Plasmid Carrying Bla Ndm-1 in Enterobacteriaceae Isolates with Epidemiological Links to Multiple Geographical Areas in China. Emerg. Microbes Infect. 2012, 1, e39. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.-L.; Wang, Y.; Liu, M.C.-J.; Lai, E.L.-Y.; Law, P.Y.-T.; Ca, H.O. Incx3 Epidemic Plasmid Carrying Blandm-5 in Escherichia Coli from Swine in Multiple Geographic Areas in China. Antimicrob. Agents Chemother. 2017, 62, e02295. [Google Scholar]
- Hao, Y.; Shao, C.; Bai, Y.; Jin, Y. Genotypic and Phenotypic Characterization of Incx3 Plasmid Carrying Bla Ndm-7 in Escherichia Coli Sequence Type 167 Isolated from a Patient with Urinary Tract Infection. Front. Microbiol. 2018, 9, 2468. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Xie, Y.; Feng, P.; Zong, Z. Blandm-5 Carried by an Incx3 Plasmid in Escherichia Coli Sequence Type 167. Antimicrob. Agents Chemother. 2014, 58, 7548–7552. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Zhou, M.; Zhang, Q.; Tao, H.; Ye, Y.; Chen, H.; Xu, L.; Xu, H.; Wang, P.; Feng, X. First Identification of Ndm-4-Producing Escherichia Coli St410 in China. Emerg. Microbes Infect. 2016, 5, e118. [Google Scholar] [CrossRef]
- Petrella, S.; Ziental-Gelus, N.; Mayer, C.; Renard, M.; Jarlier, V.; Sougakoff, W. Genetic and Structural Insights into the Dissemination Potential of the Extremely Broad-Spectrum Class a Beta-Lactamase Kpc-2 Identified in an Escherichia Coli Strain and an Enterobacter Cloacae Strain Isolated from the Same Patient in France. Antimicrob. Agents Chemother. 2008, 52, 3725–3736. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nordmann, P.; Lartigue, M.; Poirel, L. β-Lactam Induction of Isecp1b-Mediated Mobilization of the Naturally Occurring Blactx-M-β-Lactamase Gene of Kluyvera Ascorbata. FEMS Microbiol. Lett. 2008, 288, 247–249. [Google Scholar] [CrossRef]
- Lartigue, M.F.; Poirel, L.; Aubert, D.; Nordmann, P. In Vitro Analysis of Isecp1b-Mediated Mobilization of Naturally Occurring β -Lactamase Gene Blactx-M of Kluyvera Ascorbata. Antimicrob. Agents Chemother. 2006, 50, 1282–1286. [Google Scholar] [CrossRef]
- Johnson, M.; Irena, Z.; Yan, R.; Yuri, M.; Scott, M.; Thomas, M. Ncbi Blast: A Better Web Interface. Nucl. Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Lane, D.J. 16s/23s Rrna Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Wiley: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Single-Cell Genomes and Mini-Metagenomes from Chimeric Mda Products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A Large-Scale Evaluation of Algorithms to Calculate Average Nucleotide Identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Aarestrup, M.F.; Hasman, H. In Silico Detection and Typing of Plasmids Using Plasmidfinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, M.F.; Larsen, V.M. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A Genome Comparison Visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Kaas, R.S.; Leekitcharoenphon, P.; Aarestrup, F.M.; Lund, O. Solving the Problem of Comparing Whole Bacterial Genomes across Different Sequencing Platforms. PloS ONE 2014, 9, e104984. [Google Scholar] [CrossRef]
- Croucher, N.J.; Page, A.J.; Connor, T.R.; Delaney, A.J.; Keane, J.A.; Bentley, S.D.; Parkhill, J.; Harris, S.R. Rapid Phylogenetic Analysis of Large Samples of Recombinant Bacterial Whole Genome Sequences Using Gubbins. Nucl. Acids Res. 2015, 43, e15. [Google Scholar] [CrossRef]

| Strain | MIC (μg/mL) a | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | FOS | GEN | CST | MEM | IMP | CEF | CFT | AZT | CIP | CTX | TGC | |
| SCW13 | 32 | 256 | ≤4 | ≤4 | 128 | 256 | >512 | 256 | 512 | ≤4 | >512 | ≤4 |
| SCW13 b | 16 | 64 | ≤4 | ≤4 | 128 | 256 | >512 | 512 | 512 | ≤4 | >512 | ≤4 |
| E. coli J53 | 16 | 32 | ≤4 | ≤4 | 0.5 | 1 | ≤4 | 4 | 8 | ≤4 | 8 | ≤4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Luo, L.; Xiao, Z.; Wang, G.; Li, C.; Zhang, Z.; Zhou, Y.; Zhang, L. Characterization of a Carbapenem-Resistant Kluyvera Cryocrescens Isolate Carrying Blandm-1 from Hospital Sewage. Antibiotics 2019, 8, 149. https://doi.org/10.3390/antibiotics8030149
Li Y, Luo L, Xiao Z, Wang G, Li C, Zhang Z, Zhou Y, Zhang L. Characterization of a Carbapenem-Resistant Kluyvera Cryocrescens Isolate Carrying Blandm-1 from Hospital Sewage. Antibiotics. 2019; 8(3):149. https://doi.org/10.3390/antibiotics8030149
Chicago/Turabian StyleLi, Ying, Li Luo, Zhijiao Xiao, Guangxi Wang, Chengwen Li, Zhikun Zhang, Yingshun Zhou, and Luhua Zhang. 2019. "Characterization of a Carbapenem-Resistant Kluyvera Cryocrescens Isolate Carrying Blandm-1 from Hospital Sewage" Antibiotics 8, no. 3: 149. https://doi.org/10.3390/antibiotics8030149
APA StyleLi, Y., Luo, L., Xiao, Z., Wang, G., Li, C., Zhang, Z., Zhou, Y., & Zhang, L. (2019). Characterization of a Carbapenem-Resistant Kluyvera Cryocrescens Isolate Carrying Blandm-1 from Hospital Sewage. Antibiotics, 8(3), 149. https://doi.org/10.3390/antibiotics8030149





