Carbapenemase Genes and Multidrug Resistance of Acinetobacter Baumannii: A Cross Sectional Study of Patients with Pneumonia in Southern Vietnam
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Sample Collection
2.2. Bacterial Isolates and Antimicrobial Susceptibility Testing
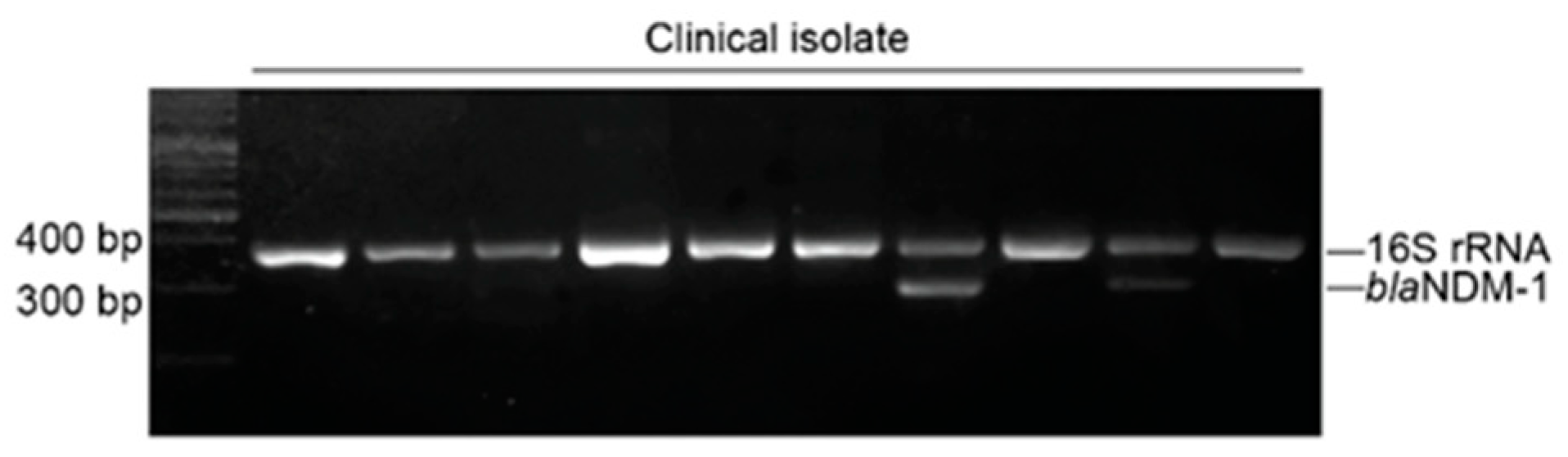
2.3. Identification of Carbapenemase Genes
2.4. Data Analysis
3. Results
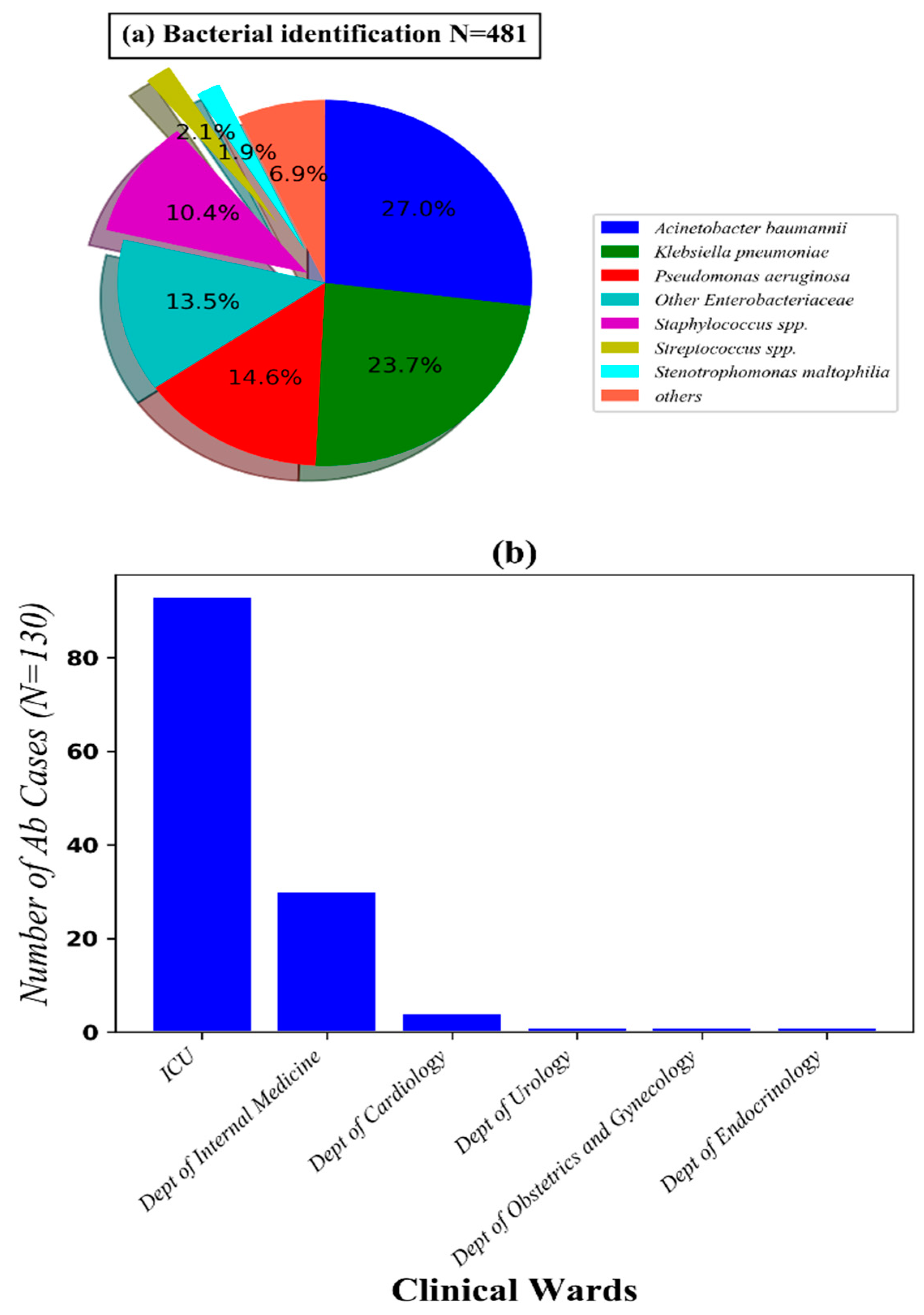
3.1. Characteristics of Ab Infected Patients with Pneumonia Admitted to Thong Nhat Dong Nai General Hospital from Septmeber 2017 to March 2018
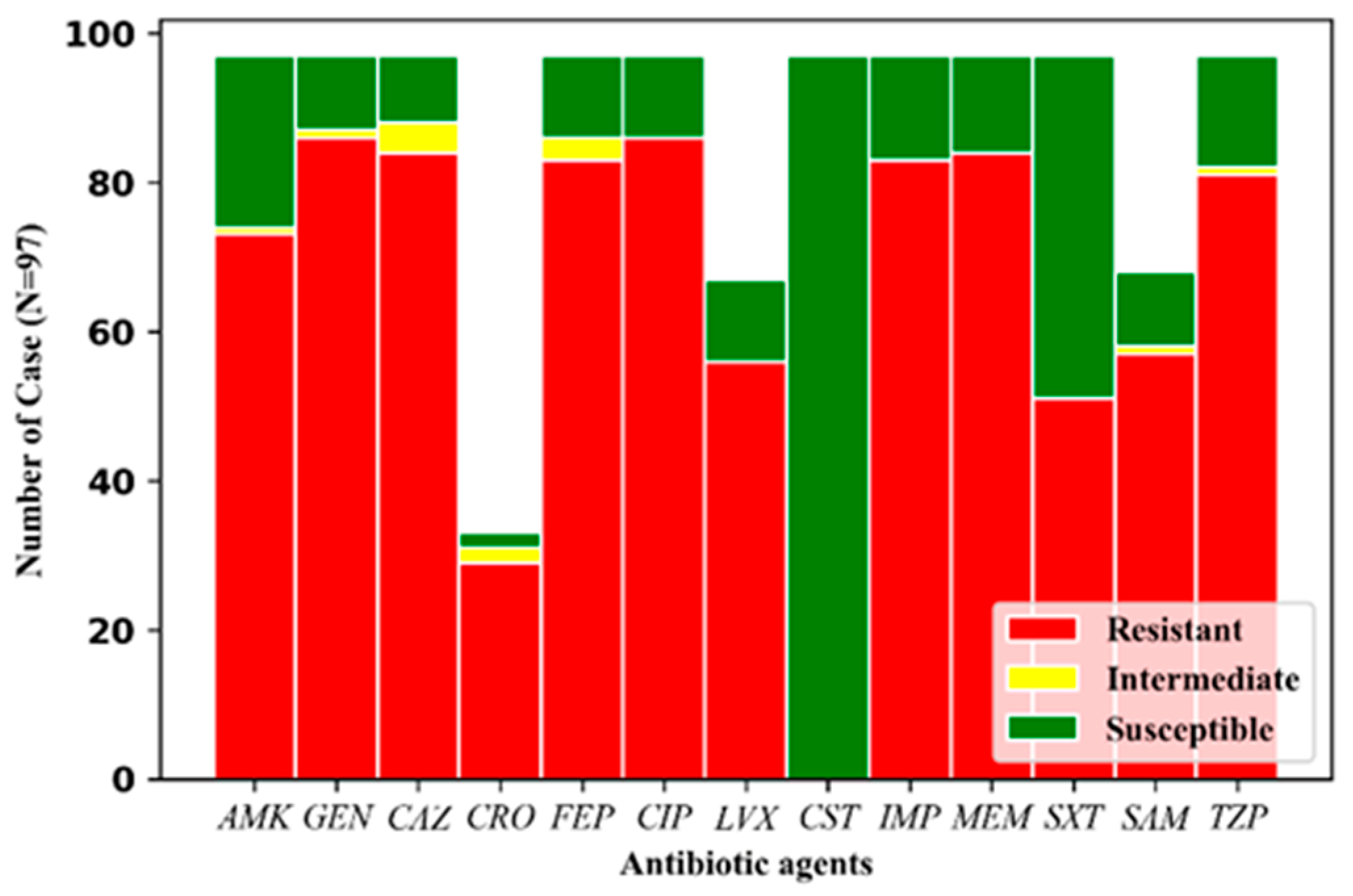
3.2. Drug Resistance Profile of 97 Ab Strains Recovered from Infected Patients
3.3. Genotypes and Relationship between Genotypes and Drug Resistant Profile of the Bacteria
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sultan, A.M.; Seliem, W.A. Identifying Risk Factors for Healthcare-Associated Infections Caused by Carbapenem-Resistant Acinetobacter baumannii in a Neonatal Intensive Care Unit. Sultan Qaboos Univ. Med. J. 2018, 18, e75–e80. [Google Scholar] [CrossRef]
- Maragakis, L.L.; Perl, T.M. Acinetobacter baumannii: Epidemiology, antimicrobial resistance, and treatment options. Clin. Infect. Dis. 2008, 46, 1254–1263. [Google Scholar] [CrossRef] [PubMed]
- Özgür, E.S.; Horasan, E.S.; Karaca, K.; Ersöz, G.; Atış, S.N.; Kaya, A. Ventilator-associated pneumonia due to extensive drug-resistant Acinetobacter baumannii: Risk factors, clinical features, and outcomes. Am. J. Infect. Control 2014, 42, 206–208. [Google Scholar] [CrossRef] [PubMed]
- Curcio, D.; Fernández, F.; Vergara, J.; Vazquez, W.; Luna, C. Late onset ventilator-associated pneumonia due to multidrug-resistant Acinetobacter spp.: Experience with tigecycline. J. Chemother. 2009, 21, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Inchai, J.; Liwsrisakun, C.; Theerakittikul, T.; Chaiwarith, R.; Khositsakulchai, W.; Pothirat, C. Risk factors of multidrug-resistant, extensively drug-resistant and pandrug-resistant Acinetobacter baumannii ventilator-associated pneumonia in a Medical Intensive Care Unit of University Hospital in Thailand. J. Infect. Chemother. 2015, 21, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Hasanin, A.; Mukhtar, A.; El-adawy, A.; Elazizi, H.; Lotfy, A.; Nassar, H.; Ghaith, D. Ventilator associated pneumonia caused by extensive-drug resistant Acinetobacter species: Colistin is the remaining choice. Egypt. J. Anaesth. 2016, 32, 409–413. [Google Scholar] [CrossRef]
- Moffatt, J.H.; Harper, M.; Harrison, P.; Hale, J.D.; Vinogradov, E.; Seemann, T.; Henry, R.; Crane, B.; Michael, F.S.; Cox, A.D.; et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 2010, 54, 4971–4977. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Liu, L.; Fang, Y.; Shi, Q.; Li, X.; Chen, Q.; Shi, K.; Jiang, Y.; Zhou, H.; Yu, Y. Colistin resistance in Acinetobacter baumannii MDR-ZJ06 revealed by a multiomics approach. Front. Cell. Infect. Microbiol. 2017, 7, 45. [Google Scholar] [CrossRef]
- Qureshi, Z.A.; Hittle, L.E.; O’Hara, J.A.; Rivera, J.I.; Syed, A.; Shields, R.K.; Pasculle, A.W.; Ernst, R.K.; Doi, Y. Colistin-resistant Acinetobacter baumannii: Beyond carbapenem resistance. Clin. Infect. Dis. 2015, 60, 1295–1303. [Google Scholar] [CrossRef]
- World Health Organization (WHO). WHO Publishes List of Bacteria for which New Antibiotics are Urgently Needed; WHO: Geneva, Switzerland, 2017. [Google Scholar]
- Da Silva, K.E.; Maciel, W.G.; Croda, J.; Cayô, R.; Ramos, A.C.; de Sales, R.O.; Kurihara, M.N.L.; Vasconcelos, N.G.; Gales, A.C.; Simionatto, S. A high mortality rate associated with multidrug-resistant Acinetobacter baumannii ST79 and ST25 carrying OXA-23 in a Brazilian intensive care unit. PLoS ONE 2018, 13, e0209367. [Google Scholar] [CrossRef]
- Yang, S.; Sun, J.; Wu, X.; Zhang, L. Determinants of Mortality in Patients with Nosocomial Acinetobacter baumannii Bacteremia in Southwest China: A Five-Year Case-Control Study. Can. J. Infect. Dis. Med. Microbiol. 2018, 2018, 3150965. [Google Scholar] [CrossRef]
- Hernández-Torres, A.; García-Vázquez, E.; Gómez, J.; Canteras, M.; Ruiz, J.; Yagüe, G. Multidrug and carbapenem-resistant Acinetobacter baumannii infections: Factors associated with mortality. Med. Clin. 2012, 138, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Fattouh, M.; El-din, A.N. Emergence of carbapenemresistant acinetobacter baumannii in the intensive care unit in Sohag University Hospital, Egypt. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 732–744. [Google Scholar]
- Poirel, L.; Naas, T.; Nordmann, P. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob. Agents Chemother. 2010, 54, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Yang, F. Drug-resistant gene of blaOXA-23, blaOXA-24, blaOXA-51 and blaOXA-58 in Acinetobacter baumannii. Int. J. Clin. Exp. Med. 2015, 8, 13859. [Google Scholar] [PubMed]
- Kim, D.H.; Choi, J.-Y.; Kim, H.W.; Kim, S.H.; Chung, D.R.; Peck, K.R.; Thamlikitkul, V.; So, T.M.-K.; Yasin, R.M.D.; Hsueh, P.-R.; et al. Spread of carbapenem-resistant Acinetobacter baumannii global clone 2 in Asia and AbaR-type resistance islands. Antimicrob. Agents Chemother. 2013, 57, 5239–5246. [Google Scholar] [CrossRef]
- Elbrolosy, A.M.; Labeeb, A.Z.; Hassan, D.M. new Delhi metallo-β-lactamase-producing Acinetobacter isolates among late-onset VaP patients: Multidrug-resistant pathogen and poor outcome. Infect. Drug Resist. 2019, 12, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, L.; Bakour, S.; Rolain, J.-M. Co-occurrence of carbapenemase encoding genes in Acinetobacter baumannii, a dream or reality? BMC Microbiol. 2018, 18, 107. [Google Scholar] [CrossRef]
- Isler, B.; Doi, Y.; Bonomo, R.A.; Paterson, D.L. New treatment options against carbapenem-resistant Acinetobacter baumannii infections. Antimicrob. Agents Chemother. 2019, 63, e01110-18. [Google Scholar] [CrossRef]
- Dortet, L.; Potron, A.; Bonnin, R.A.; Plesiat, P.; Naas, T.; Filloux, A.; Larrouy-Maumus, G. Rapid detection of colistin resistance in Acinetobacter baumannii using MALDI-TOF-based lipidomics on intact bacteria. Sci. Rep. 2018, 8, 16910. [Google Scholar] [CrossRef]
- Nhu, N.T.K.; Riordan, D.W.; Nhu, T.D.H.; Thanh, D.P.; Thwaites, G.; Lan, N.P.H.; Wren, B.W.; Baker, S.; Stabler, R.A. The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 2016, 6, 28291. [Google Scholar]
- Tuan Anh, N.; Nga, T.; Tuan, H.M.; Tuan, N.S.; Chau, N.; Baker, S.; Duong, H.H. The molecular epidemiology and antimicrobial resistance phenotypes of acinetobacter baumannii isolated from patients in three hospitals in Southern Vietnam. J. Med. Microbiol. 2016, 66. [Google Scholar] [CrossRef] [PubMed]
- Si-Tuan, N.; Thanh, N.N.; Hang, P.T.T.; Van Dung, P.; Huong, N.T. Antimicrobial resistance patterns among Acinetobacter baumannii isolated from Thong Nhat Dong Nai General Hospital. Orthopedics 2016, 2, 61–75. [Google Scholar]
- Xie, R.; Zhang, X.D.; Zhao, Q.; Peng, B.; Zheng, J. Analysis of global prevalence of antibiotic resistance in Acinetobacter baumannii infections disclosed a faster increase in OECD countries. Emerg. Microbes Infect. 2018, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Si-Tuan, N.; Ngoc, H.M.; Hang, P.T.T.; Nguyen, C.; Van, P.H.; Huong, N.T. New eight genes identified at the clinical multidrug-resistant Acinetobacter baumannii DMS06669 strain in a Vietnam hospital. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 74. [Google Scholar] [CrossRef] [PubMed]
- Trinh, L.P.; Luu, T.N.H.; Nguyen, M.P.; Dinh, T.H.; Doan, T.H.; Pham, B.Y. Studies of common antibiotic resistance-associated genes of Acinetobacter baumannii. Vietnam J. Sci. Technol. Eng. 2019, 61, 58–61. [Google Scholar] [CrossRef]
- Beidas, S.O. Evaluation of sputum gram stain. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 1992, 15, 1048–1049. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Tuan Anh, N.; Minh Tuan, H.; Huyen Nguyen, K. Development of a Taq-man real-time PCR assay for the detection of several blaOXA genes in Acinetobacter baumannii. Ho Chi Minh J. Med. 2014, 16, 107–201. [Google Scholar]
- CaLS I. Performance Standards for aNtimicrobial Susceptibility Testing; M100 S28; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Opazo-Capurro, A.; Martín, I.; Quezada-Aguiluz, M.; Morales-León, F.; Domínguez-Yévenes, M.; Lima, C.A.; Esposito, F.; Cerdeira, L.; Bello-Toledo, H.; Lincopan, N.; et al. Evolutionary dynamics of carbapenem-resistant Acinetobacter baumannii circulating in Chilean hospitals. Infect. Genet. Evol. 2019, 73, 93–97. [Google Scholar] [CrossRef]
- Shrestha, S.; Tada, T.; Miyoshi-Akiyama, T.; Ohara, H.; Shimada, K.; Satou, K.; Teruya, K.; Nakano, K.; Shiroma, A.; Sherchand, J.B.; et al. Molecular epidemiology of multidrug-resistant Acinetobacter baumannii isolates in a university hospital in Nepal reveals the emergence of a novel epidemic clonal lineage. Int. J. Antimicrob. Agents 2015, 46, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.R.; Acharya, M.; Kakshapati, T.; Leungtongkam, U.; Thummeepak, R.; Sitthisak, S. Co-existence of bla OXA-23 and bla NDM-1 genes of Acinetobacter baumannii isolated from Nepal: Antimicrobial resistance and clinical significance. Antimicrob. Resist. Infect. Control 2017, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, G.J.; Domingues, S. Insights on the Horizontal Gene Transfer of Carbapenemase Determinants in the Opportunistic Pathogen Acinetobacter baumannii. Microorganisms 2016, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Viana, G.F.; Zago, M.C.B.; Moreira, R.R.B.; Zarpellon, M.N.; Menegucci, T.C.; Cardoso, C.L.; Tognim, M.C.B. ISAba1/blaOXA-23: A serious obstacle to controlling the spread and treatment of Acinetobacter baumannii strains. Am. J. Infect. Control 2016, 44, 593–595. [Google Scholar] [CrossRef] [PubMed]
- Yazdansetad, S.; Najari, E.; Ghaemi, E.A.; Javid, N.; Hashemi, A.; Ardebili, A. Carbapenem-resistant Acinetobacter baumannii isolates carrying blaOXA genes with upstream ISAba1: First report of a novel OXA subclass from Iran. J. Glob. Antimicrob. Resist. 2019, 18, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Mugnier, P.D.; Poirel, L.; Naas, T.; Nordmann, P. Worldwide dissemination of the blaOXA-23 carbapenemase gene of Acinetobacter baumannii. Emerg. Infect. Dis. 2010, 16, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.L.; Chang, W.C.; Kuo, S.C.; Lee, Y.T.; Chen, C.P.; Siu, L.K.; Cho, W.L.; Fung, C.P. Contribution of a Plasmid-Borne blaOXA-58 Gene with Its Hybrid Promoter Provided by IS1006 and an ISAba3-Like Element to β-Lactam Resistance in Acinetobacter Genomic Species 13TU. Antimicrob. Agents Chemother. 2010, 54, 3107–3112. [Google Scholar] [CrossRef]
- Kassim, K. prevalence and role of isaba1 associated with blaoxa-51 and blaoxa-23 genes in carbapenem-resistant acinetobacter baumannii. Eur. J. Pharm. Med Res. 2016, 3, 49–53. [Google Scholar]
- Turton, J.F.; Woodford, N.; Glover, J.; Yarde, S.; Kaufmann, M.E.; Pitt, T.L. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 2006, 44, 2974–2976. [Google Scholar] [CrossRef]
- Figueiredo, S.; Poirel, L.; Papa, A.; Koulourida, V.; Nordmann, P. Overexpression of the Naturally Occurring blaOXA-51 Gene in Acinetobacter baumannii Mediated by Novel Insertion Sequence ISAba9. Antimicrob. Agents Chemother. 2011, 53, 4045–4047. [Google Scholar] [CrossRef]
- Alsultan, A.; Evans, B.; Elsayed, E.; Al-Thawadi, S.; Al-Taher, A.; Amyes, S.; Al-Dughaym, A.M.; Hamouda, A. High frequency of carbapenem-resistant Acinetobacter baumannii in patients with diabetes mellitus in Saudi Arabia. J. Med Microbiol. 2013, 62, 885–888. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-R.; Lee, J.H.; Park, M.; Park, K.S.; Bae, I.K.; Kim, Y.B.; Cha, C.-J.; Jeong, B.C.; Lee, S.H. Biology of Acinetobacter baumannii: Pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell. Infect. Microbiol. 2017, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Sarada, V.; Rao, R.; Mani, R.; Ramana, K. Colistin, polymyxin B and tigecycline suspectibility to metallo betalactamase producing Acinetobacter baumannii isolated from tertiary health care hospital. Am. J. Microbiol. Res. 2014, 2, 60–62. [Google Scholar] [CrossRef][Green Version]
- Viehman, J.A.; Nguyen, M.H.; Doi, Y. Treatment Options for Carbapenem-Resistant and Extensively Drug-Resistant Acinetobacter baumannii Infections. Drugs 2014, 74, 1315–1333. [Google Scholar] [CrossRef] [PubMed]
- Nepka, M.; Perivolioti, E.; Kraniotaki, E.; Politi, L.; Tsakris, A.; Pournaras, S. In Vitro Bactericidal Activity of Trimethoprim-Sulfamethoxazole alone and in Combination with Colistin against Carbapenem-Resistant Acinetobacter baumannii Clinical Isolates. Antimicrob. Agents Chemother. 2016, 60, 6903–6906. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saeed, A.; Bosch, A.; Bettiol, M.; Nossa, G.D.; Erben, M.; Lamberti, Y. Novel Guanidine Compound against Multidrug-Resistant Cystic Fibrosis-Associated Bacterial Species. Molecules 2018, 23, 1158. [Google Scholar] [CrossRef]
- Figueiredo, J.; Serrano, J.L.; Soares, M.; Ferreira, S.; Domingues, F.C.; Almeida, P.; Silvestre, S. 5-Hydrazinylethylidenepyrimidines effective against multidrug-resistant Acinetobacter baumannii: Synthesis and in vitro biological evaluation of antibacterial, radical scavenging and cytotoxic activities. Eur. J. Pharm. Sci. 2019, 137, 104964. [Google Scholar] [CrossRef]
- Surendran-Nair, M.; Lau, P.; Liu, Y.; Venkitanarayanan, K. Efficacy of selenium in controlling Acinetobacter baumannii associated wound infections. Wound Med. 2019, 26, 100165. [Google Scholar] [CrossRef]
- Beigverdi, R.; Sattari-Maraji, A.; Emaneini, M.; Jabalameli, F. Status of carbapenem-resistant Acinetobacter baumannii harboring carbapenemase: First systematic review and meta-analysis from Iran. Infect. Genet. Evol. 2019, 73, 433–443. [Google Scholar] [CrossRef]

| Sequence (5′-3′) | Target Genes | Product Size (bp) | Origin | Volume of Reaction (nM) | Reference |
|---|---|---|---|---|---|
| Primers for Real-time PCR | |||||
| F: CACTAGGAGAAGCCATGAAGC R: CAGCATTACCGAAACCAATACG P: 5′-Cy5-TTGCGCGACGTATCGGTCTTGATC-BHQ2-3′ | blaOXA–23 | 114 | intrinsically disordered protein (IDP) | 200 | [23,30] |
| 100 | |||||
| F: GAAGTGAAGCGTGTTGGTTATG R: GCCTCTTGCTGAGGAGTAAT P: 5′-FAM-CGACTTGGGTACCGATATCTGCATTGC-BHQ1-3′ | blaOXA–51 | 148 | IDP | 200 | |
| 100 | |||||
| F: ATATTTAAGTGGGATGGAAAGCC R: CGTGCCAATTCTTGATATACAGG P: 5′-Texas Red-TTTACTTTGGGCGAAGCCATGCAAG-BHQ2-3′ | blaOXA–58 | 110 | IDP | 200 | |
| 100 | |||||
| F: CCAGTGACAAACTGGAGGAAG R: GCTGTGTAGCAACCCTTTGTA P: 5′-HEX-ACGTCAAGTCATCATGGCCCTTACG-BHQ1-3′ | 16S rRNA | 199 | IDP | 400 | |
| 200 | |||||
| Primers for PCR | |||||
| F: CGATTGGCCAGCAAATGGAAACTG R: CATACCGCCCATCTTGTCCTGATG | blaNDM-1 | 287 | IDP | 20 | [26] |
| F: TGCATTCGATACTGGTGAGC R: CTAGTATGTCAAGGCCAGGTAAG | 16S rRNA | 370 | IDP | 20 |
| Level of Resistance | Clinical Wards | Total (n = 97) n (%) | P * | |||
|---|---|---|---|---|---|---|
| ICU (n = 71) n (%) | Dept of Internal Medicine (n = 24) n (%) | Dept of Urology (n = 1) n (%) | Dept of Cardiovascular (n = 1) | |||
| Not multidrug-resistant (MDR) | 1 (10) | 10 (41.7) | 0 | 0 | 11 (11.3) | <0.0001 |
| MDR | 70 (98.6) | 14 (58.3) | 1 (100) | 1 (100) | 86 (88.7) | |
| not extensively drug-resistant (XDR) | 3 (4.2) | 0 | 0 | 0 | 3 (3.1) | <0.0001 |
| XDR | 67 (94.4) | 14 (58.3) | 1 (100) | 1 (100) | 83 (85.6) | |
| Total | 71 (85.5) | 24 (24.7) | 1(1) | 1(1) | 97 (100) | |
| Antibiotics | blaOXA-23-like | blaOXA-58-like | blaNDM-1 | blaOXA-23-like | blaOXA-58-like | blaNDM-1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (−) | (+) | (−) | (+) | (−) | (+) | (−) | (+) | (− | (+) | (−) | (+) | |||
| Amikacin | S | 16 | 7 | 22 | 1 | 22 | 1 | LVX | 5 | 25 | 11 | 0 | 11 | 0 |
| NS | 5 | 69 | 65 | 9 | 69 | 5 | 16 | 51 | 50 | 6 | 50 | 6 | ||
| P | <0.0001 | >0.1 | >0.1 | <0.0001 | >0.1 | >0.1 | ||||||||
| Gentamicin | S | 10 | 0 | 10 | 0 | 10 | 0 | IMP | 14 | 0 | 14 | 0 | 14 | 0 |
| NS | 11 | 76 | 77 | 10 | 81 | 6 | 7 | 76 | 73 | 10 | 77 | 6 | ||
| P | <0.0001 | >0.1 | >0.1 | <0.0001 | >0.1 | >0.1 | ||||||||
| Ceftazidime | S | 9 | 0 | 9 | 0 | 9 | 0 | MEM | 13 | 0 | 13 | 0 | 13 | 0 |
| NS | 12 | 76 | 78 | 10 | 82 | 6 | 8 | 76 | 74 | 10 | 78 | 6 | ||
| P | <0.0001 | >0.1 | >0.1 | <0.0001 | >0.1 | >0.1 | ||||||||
| Ceftriaxone | S | 2 | 0 | 2 | 0 | 2 | 0 | TZP | 15 | 0 | 15 | 0 | 15 | 0 |
| NS | 5 | 26 | 27 | 4 | 31 | 0 | 6 | 76 | 72 | 10 | 76 | 6 | ||
| P | <0.002 | >0.1 | >0.1 | <0.0001 | >0.1 | >0.1 | ||||||||
| Cefepime | S | 11 | 0 | 11 | 0 | 11 | 0 | SAM | 10 | 0 | 10 | 0 | 10 | 0 |
| NS | 10 | 76 | 76 | 10 | 80 | 6 | 6 | 52 | 51 | 7 | 52 | 6 | ||
| P | <0.0001 | >0.1 | >0.1 | <0.0001 | >0.1 | >0.1 | ||||||||
| Ciprofloxacin | S | 11 | 0 | 11 | 0 | 11 | 0 | SXT | 13 | 33 | 41 | 5 | 44 | 2 |
| NS | 10 | 76 | 76 | 10 | 80 | 6 | 8 | 43 | 46 | 5 | 47 | 4 | ||
| P | <0.0001 | >0.1 | >0.1 | >0.1 | >0.1 | >0.1 | ||||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoang Quoc, C.; Nguyen Thi Phuong, T.; Nguyen Duc, H.; Tran Le, T.; Tran Thi Thu, H.; Nguyen Tuan, S.; Phan Trong, L. Carbapenemase Genes and Multidrug Resistance of Acinetobacter Baumannii: A Cross Sectional Study of Patients with Pneumonia in Southern Vietnam. Antibiotics 2019, 8, 148. https://doi.org/10.3390/antibiotics8030148
Hoang Quoc C, Nguyen Thi Phuong T, Nguyen Duc H, Tran Le T, Tran Thi Thu H, Nguyen Tuan S, Phan Trong L. Carbapenemase Genes and Multidrug Resistance of Acinetobacter Baumannii: A Cross Sectional Study of Patients with Pneumonia in Southern Vietnam. Antibiotics. 2019; 8(3):148. https://doi.org/10.3390/antibiotics8030148
Chicago/Turabian StyleHoang Quoc, Cuong, Thao Nguyen Thi Phuong, Hai Nguyen Duc, Trung Tran Le, Hang Tran Thi Thu, Si Nguyen Tuan, and Lan Phan Trong. 2019. "Carbapenemase Genes and Multidrug Resistance of Acinetobacter Baumannii: A Cross Sectional Study of Patients with Pneumonia in Southern Vietnam" Antibiotics 8, no. 3: 148. https://doi.org/10.3390/antibiotics8030148
APA StyleHoang Quoc, C., Nguyen Thi Phuong, T., Nguyen Duc, H., Tran Le, T., Tran Thi Thu, H., Nguyen Tuan, S., & Phan Trong, L. (2019). Carbapenemase Genes and Multidrug Resistance of Acinetobacter Baumannii: A Cross Sectional Study of Patients with Pneumonia in Southern Vietnam. Antibiotics, 8(3), 148. https://doi.org/10.3390/antibiotics8030148





