Prevalence of Antibiotic Resistance Genes in Multidrug-Resistant Enterobacteriaceae on Portuguese Livestock Manure
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sampling
2.3. Microbiological Analysis and Phenotypic Characterisation
2.4. Multiplex PCR (Polymerase Chain Reaction) for Genes Detection
3. Results and Discussion
3.1. Microbiological Parameters
3.2. Antibiotic Resistance Profile
3.3. Screening of AB Resistance and Integrons Genes
3.3.1. Chloramphenicol
3.3.2. Quinolones
3.3.3. Tetracycline
3.3.4. Sulphonamide
3.3.5. Trimethoprim
3.3.6. Integrons
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- DGAV. Relatório Nacional de Monitorização do Consumo de Antimicrobianos Ano 2013—Portugal; Direcção-Geral de Alimentação e Veterinária/Direcção de Serviços de Medicamentos e Produtos de Uso Veterinário. Ministério da Agricultura, Mar, Ambiente e Ordenamento do Território; Governo de Portugal: Lisbon, Portugal, 2014; pp. 1–13.
- Jury, K.L.; Vancov, T.; Stuetz, R.M.; Khan, S.J. Antibiotic resistance dissemination and sewage treatment plants. In Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology; Méndez-Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2010; pp. 509–519. ISBN 978-84-614-6194-3. [Google Scholar]
- Faldynova, M.; Videnska, P.; Havlickova, H.; Sisak, F.; Juricova, H.; Babak, V.; Steinhauser, L.; Rychlik, I. Prevalence of antibiotic resistance genes in faecal samples from cattle, pigs and poultry. Vet. Med. 2013, 58, 298–304. [Google Scholar] [CrossRef]
- Fluit, A.C.; Schmitz, F.J. Class 1 integrons, gene cassettes, mobility and epidemiology. Eur. J. Clin. Microbiol. Infect. Dis. 1999, 18, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, M.; Kumar, S.; Kapoor, R.K.; Virdi, J.S.; Gulati, P. Integrons in Enterobacteriaceae: Diversity, distribution and epidemiology. Int. J. Antimicrob. Agents 2017, 51, 167–176. [Google Scholar] [CrossRef]
- Martinez-Carballo, E.; Gonzalez-Barreiro, C.; Scharf, S.; Gans, O. Environmental monitoring study of selected veterinary antibiotics in animal manure and soils in Austria. Environ. Pollut. 2007, 148, 570–579. [Google Scholar] [CrossRef]
- Ji, X.L.; Shen, Q.H.; Liu, F.; Ma, J.; Xu, G.; Wang, Y.L.; Wu, M.H. Antibiotic resistance gene abundances associated with antibiotics and heavy metals in animal manures and agricultural soils adjacent to feedlots in Shanghai; China. J. Hazard. Mater. 2012, 235, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, H.; Guo, Y.; Tian, T. Influence of Chicken Manure Fertilization on Antibiotic-Resistant Bacteria in Soil and the Endophytic Bacteria of Pakchoi. Int. J. Environ. Res. Public Health 2016, 13, 662. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Tan, Y.; Zhang, X.; Hu, J.; Miao, Z.; Wei, L.; Chai, T. Emissions of Escherichia coli carrying extended-spectrum β-lactamase resistance from pig farms to the surrounding environment. Int. J. Environ. Res. Public Health 2015, 12, 4203–4213. [Google Scholar] [CrossRef]
- Songe, M.M.; Hang’ombe, B.M.; Knight-Jones, T.J.; Grace, D. Antimicrobial resistant enteropathogenic Escherichia coli and Salmonella spp. in houseflies infesting fish in food markets in Zambia. Int. J. Environ. Res. Public Health 2016, 14, 21. [Google Scholar] [CrossRef]
- Massé, D.I.; Saady, N.M.; Gilbert, Y. Potential of biological processes to eliminate antibiotics in livestock manure: An overview. Animals 2014, 4, 146–163. [Google Scholar] [CrossRef]
- WHO. Global Action Plan on Antimicrobial Resistance; World Health Organization: Geneva, Switzerland, 2015; p. 28. ISBN 978-92-4-150976-3. [Google Scholar]
- WHO. Antimicrobial Resistance: A Manual for Developing National Action Plans; Published by the WHO and the Food and Agriculture Organization of the United Nations and the World Organization for Animal Health; World Health Organization: Geneva, Switzerland, 2016; p. 26. ISBN 978-92-4-154953-0. [Google Scholar]
- King, L.J.; Anderson, L.R.; Blackmore, C.G.; Blackwell, M.J.; Lautner, E.A.; Marcus, L.C.; Meyer, T.E.; Monath, T.P.; Nave, J.E.; Ohle, J.; et al. Executive summary of the AVMA one health initiative task force report. J. Am. Vet. Med. Assoc. 2008, 233, 259–261. [Google Scholar] [CrossRef]
- Rousham, E.K.; Unicomb, L.; Islam, M.A. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: Integrating behavioural, epidemiological and One Health approaches. Proc. R. Soc. Lond. B 2018, 285, 20180332. [Google Scholar] [CrossRef]
- Amador, P.; Fernandes, R.; Prudêncio, C.; Barreto, M.; Duarte, I. Antibiotic resistance in wastewater: Occurrence and fate of Enterobacteriaceae producers of Class A and Class C β-lactamases. J. Environ. Sci. Health A 2015, 50, 26–39. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; CLSI Supplement M100S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Magiorakos, A.P.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; Paterson, D.L.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Amador, P.; Fernandes, R.; Brito, L.; Prudêncio, C. Antibiotic resistance in Enterobacteriaceae isolated from Portuguese deli meats. J. Food Saf. 2011, 31, 1–20. [Google Scholar] [CrossRef]
- Šeputienė, V.; Povilonis, J.; Ružauskas, M.; Pavilonis, A.; Sužiedėlienė, E. Prevalence of trimethoprim resistance genes in Escherichia coli isolates of human and animal origin in Lithuania. J. Med. Microbiol. 2010, 59, 315–322. [Google Scholar] [CrossRef]
- Szmolka, A.; Nagy, B. Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front. Microbiol. 2013, 4, 258. [Google Scholar] [CrossRef]
- De Jong, A.; Smet, A.; Ludwig, C.; Stephan, B.; De Graef, E.; Vanrobaeys, M.; Haesebrouck, F. Antimicrobial susceptibility of Salmonella isolates from healthy pigs and chickens (2008–2011). Vet. Microbiol. 2014, 171, 298–306. [Google Scholar] [CrossRef]
- Moawad, A.A.; Hotzel, H.; Awad, O.; Tomaso, H.; Neubauer, H.; Hafez, H.M.; El Adawy, H. Occurrence of Salmonella enterica and Escherichia coli in raw chicken and beef meat in northern Egypt and dissemination of their antibiotic resistance markers. Gut Pathog. 2017, 9, 57. [Google Scholar] [CrossRef]
- EC. Community procedure on measures to monitor certain substances and residues thereof in live animals and animal products and repealing. Directives 85/358/EEC and 86/469/EEC and Decisions 89/187/EEC and 91/664/EEC. Council Directive 96/23/EC of 29 April 1996. Offic. J. Eur. Communities 1996, 125, 10. [Google Scholar]
- Rasmussen, M.M.; Opintan, J.A.; Frimodt-Møller, N.; Styrishave, B. Beta-lactamase producing Escherichia coli isolates in imported and locally produced chicken meat from Ghana. PLoS ONE 2015, 10, e0139706. [Google Scholar] [CrossRef]
- Gay, N.; Leclaire, A.; Laval, M.; Miltgen, G.; Jégo, M.; Stéphane, R.; Jaubert, J.; Belmonte, O.; Cardinale, E. Risk factors of extended-spectrum β-lactamase producing Enterobacteriaceae occurrence in farms in Reunion, Madagascar and Mayotte Islands, 2016–2017. Vet. Sci. 2018, 5, 22. [Google Scholar] [CrossRef]
- Wu, H.; Wang, M.; Liu, Y.; Wang, X.; Wang, Y.; Lu, J.; Xu, H. Characterization of antimicrobial resistance in Klebsiella species isolated from chicken broilers. Int. J. Food Microbiol. 2016, 232, 95–102. [Google Scholar] [CrossRef]
- Mendonça, N.; Figueiredo, R.; Mendes, C.; Card, R.M.; Anjum, M.F.; Silva, G.J. Microarray evaluation of antimicrobial resistance and virulence of Escherichia coli isolates from Portuguese poultry. Antibiotics 2016, 5, 4. [Google Scholar] [CrossRef]
- Agunos, A.; Léger, D.; Avery, B.P.; Parmley, E.J.; Deckert, A.; Carson, C.A.; Dutil, L. Ciprofloxacin-resistant Campylobacter spp. in retail chicken, western Canada. Emerg. Infect. Dis. 2013, 19, 1121–1124. [Google Scholar] [CrossRef]
- Xie, R.; Huo, S.; Li, Y.; Chen, L.; Zhang, F.; Wu, X. Molecular epidemiological survey on quinolone resistance genotype and phenotype of Escherichia coli in septicemic broilers in Hebei, China. Poult. Sci. 2014, 93, 335–339. [Google Scholar] [CrossRef]
- Gouvêa, R.; dos Santos, F.F.; de Aquino, M.H.C. Fluoroquinolones in industrial poultry production, bacterial resistance and food residues: A review. Rev. Bras. Cienc. Avic. 2015, 17, 1–10. [Google Scholar] [CrossRef]
- Bhushan, C.; Khurana, A.; Sinha, R.; Nagaraju, M. Antibiotic Resistance in Poultry Environment: Spread of Resistance from Poultry Farm to Agricultural Field; Centre for Science and Environment: New Delhi, India, 2017; 36p, Available online: www.cseindia.org (accessed on 14 July 2018).
- Franco, A.; Leekitcharoenphon, P.; Feltrin, F.; Alba, P.; Cordaro, G.; Iurescia, M.; Tolli, R.; D’Incau, M.; Staffolani, M.; Di Giannatale, E.; et al. Emergence of a clonal lineage of multidrug-resistant ESBL-producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS ONE 2015, 10, e0144802. [Google Scholar] [CrossRef]
- Gao, L.; Hu, J.; Zhang, X.; Wei, L.; Li, S.; Miao, Z.; Chai, T. Application of swine manure on agricultural fields contributes to extended-spectrum β-lactamase-producing Escherichia coli spread in Tai’an, China. Front. Microbiol. 2015, 6, 313. [Google Scholar] [CrossRef]
- Li, L.; Ye, L.; Yu, L.; Zhou, C.; Meng, H. Characterization of extended spectrum β-lactamase producing Enterobacteria and methicillin-resistant Staphylococcus aureus isolated from raw pork and cooked pork products in South China. J. Food Sci. 2016, 81, M1773–M1777. [Google Scholar] [CrossRef]
- Jiang, H.X.; Lü, D.H.; Chen, Z.L.; Wang, X.M.; Chen, J.R.; Liu, Y.H.; Liao, X.P.; Liu, J.H.; Zeng, Z.L. High prevalence and widespread distribution of multi-resistant Escherichia coli isolates in pigs and poultry in China. Vet. J. 2011, 187, 99–103. [Google Scholar] [CrossRef]
- Koovapra, S.; Bandyopadhyay, S.; Das, G.; Bhattacharyya, D.; Banerjee, J.; Mahanti, A.; Samanta, I.; Nanda, P.K.; Kumar, A.; Mukherjee, R.; et al. Molecular signature of extended spectrum β-lactamase producing Klebsiella pneumoniae isolated from bovine milk in eastern and north-eastern India. Infect. Genet. Evol. 2016, 44, 395–402. [Google Scholar] [CrossRef]
- Clemente, L.; Manageiro, V.; Jones-Dias, J.; Correia, C.; Patricia Themudo, P.; Albuquerque, T.; Geraldes, M.; Matos, F.; Almendra, C.; Ferreira, E.; et al. Antimicrobial susceptibility and oxymino-β-lactam resistance mechanisms in Salmonella enterica and Escherichia coli isolates from different animal sources. Res. Microbiol. 2015, 166, 574–583. [Google Scholar] [CrossRef]
- Marchant, M.; Vinué, L.; Torres, C.; Moreno, M.A. Change of integrons over time in Escherichia coli isolates recovered from healthy pigs and chickens. Vet. Microbiol. 2013, 163, 124–132. [Google Scholar] [CrossRef]
- Clemente, L.; Manageiro, V.; Ferreira, E.; Jones-Dias, D.; Correia, I.; Themudo, P.; Albuquerque, T.; Caniça, M. Occurrence of extended-spectrum β-lactamases among isolates of Salmonella enterica subsp. enterica from food-producing animals and food products, in Portugal. Int. J. Food Microbiol. 2013, 167, 221–228. [Google Scholar] [CrossRef]
- Jones-Dias, D.; Manageiro, V.; Martins, A.P.; Ferreira, E.; Caniça, M. New class 2 integron in 2-4 among IncI1-positive Escherichia coli isolates carrying ESBL and PMAβ genes from food animals in Portugal. Foodborne Pathog. Dis. 2015, 13, 36–39. [Google Scholar] [CrossRef]
- Karczmarczyk, M.; Walsh, C.; Slowey, R.; Leonard, N.; Fanning, S. Molecular characterization of multidrug-resistant Escherichia coli isolates from Irish cattle farms. Appl. Environ. Microbiol. 2011, 77, 7121–7127. [Google Scholar] [CrossRef]
- Frye, J.G.; Jackson, C.R. Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli and Enteroccocus spp. isolated from U.S. food animals. Front. Microbiol. 2013, 4, 135. [Google Scholar] [CrossRef]
- Shaw, W.V. Chloramphenicol acetyltransferase: Enzymology and molecular biology. Crit. Rev. Biochem. 1983, 14, 1–46. [Google Scholar] [CrossRef]
- Yoo, M.H.; Huh, M.D.; Kim, E.H.; Lee, H.H.; Do Jeong, H. Characterization of chloramphenicol acetyltransferase gene by multiplex polymerase chain reaction in multidrug-resistant strains isolated from aquatic environments. Aquaculture 2003, 217, 11–21. [Google Scholar] [CrossRef]
- Niero, G.; Bortolaia, V.; Vanni, M.; Intorre, L.; Guardabassi, L.; Piccirillo, A. High diversity of genes and plasmids encoding resistance to third-generation cephalosporins and quinolones in clinical Escherichia coli from commercial poultry flocks in Italy. Vet. Microbiol. 2018, 216, 93–98. [Google Scholar] [CrossRef]
- Röderova, M.; Halova, D.; Papousek, I.; Dolejska, M.; Masarikova, M.; Hanulik, V.; Pudova, V.; Broz, P.; Htoutou-Sedlakova, M.; Sauer, P.; et al. Characteristics of quinolone resistance in Escherichia coli isolates from humans, animals and the environment in the Czech Republic. Front. Microbiol. 2017, 7, 2147. [Google Scholar] [CrossRef]
- Akinbami, O.R.; Olofinsae, S.; Ayeni, F.A. Prevalence of extended spectrum beta lactamase and plasmid mediated quinolone resistant genes in strains of Klebsiella pneumonia, Morganella morganii, Leclercia adecarboxylata and Citrobacter freundii isolated from poultry in South Western Nigeria. PeerJ 2018, 6, e5053. [Google Scholar] [CrossRef]
- Li, J.; Wang, T.; Shao, B.; Shen, J.; Wang, S.; Wu, Y. Plasmid-mediated quinolone resistance genes and antibiotic residues in wastewater and soil adjacent to swine feedlots: Potential transfer to agricultural lands. Environ. Health Perspect. 2012, 120, 1144–1149. [Google Scholar] [CrossRef]
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Dönhöfer, A.; Wilson, D.N. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014, 395, 559–575. [Google Scholar] [CrossRef]
- Zhang, S.; Gu, J.; Wang, C.; Wang, P.; Jiao, S.; He, Z.; Han, B. Characterization of antibiotics and antibiotic resistance genes on an ecological farm system. J. Chem. 2015, 2015, 526143. [Google Scholar] [CrossRef]
- Ghosh, S.; LaPara, T.M. The effects of subtherapeutic antibiotic use in farm animals on the proliferation and persistence of antibiotic resistance among soil bacteria. ISME J. 2007, 1, 191–203. [Google Scholar] [CrossRef]
- Koo, H.J.; Woo, G.J. Distribution and transferability of tetracycline resistance determinants in Escherichia coli isolated from meat and meat products. Int. J. Food Microbiol. 2011, 145, 407–413. [Google Scholar] [CrossRef]
- McNeece, G.; Naughton, V.; Woodward, M.J.; Dooley, J.S.G.; Naughton, P.J. Array based detection of antibiotic resistance genes in Gram negative bacteria isolated from retail poultry meat in the UK and Ireland. Int. J. Food Microbiol. 2014, 179, 24–32. [Google Scholar] [CrossRef]
- Chee-Sanford, J.; Maxwell, S.; Tsau, K.; Merrick, K.; Aminov, R. Antibiotic resistance in swine-manure-impacted environments. In Antimicrobial Resistance in the Environment; Keen, P.L., Mark, H.M.M., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012; pp. 203–223. ISBN 9780470905425. [Google Scholar]
- Hammerum, A.M.; Sandvang, D.; Andersen, S.R.; Seyfarth, A.M.; Porsbo, L.J.; Frimodt-Møller, N.; Heuer, O.E. Detection of sul1, sul2 and sul3 in sulphonamide resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. Int. J. Food Microbiol. 2006, 106, 235–237. [Google Scholar] [CrossRef]
- Wu, S.; Dalsgaard, A.; Hammerum, A.M.; Porsbo, L.J.; Jensen, L.B. Prevalence and characterization of plasmids carrying sulfonamide resistance genes among Escherichia coli from pigs, pig carcasses and human. Acta Vet. Scand. 2010, 52, 47. [Google Scholar] [CrossRef]
- Vuthy, Y.; Lay, K.S.; Seiha, H.; Kerleguer, A.; Aidara-Kane, A. Antibiotic susceptibility and molecular characterization of resistance genes among Escherichia coli and among Salmonella subsp. in chicken food chains. Asian Pac. J. Trop. Biomed. 2017, 7, 670–674. [Google Scholar] [CrossRef]
- Tacão, M.; Moura, A.; Correia, A.; Henriques, I. Co-resistance to different classes of antibiotics among ESBL-producers from aquatic systems. Water Res. 2014, 48, 100–107. [Google Scholar] [CrossRef]
- EFSA—European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). The European Union Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2010. EFSA J. 2012, 10, 2598. [Google Scholar] [CrossRef]
- Dillon, B.; Thomas, L.; Mohmand, G.; Zelynski, A.; Iredell, J. Multiplex PCR for screening of integrons in bacterial lysates. J. Microbiol. Methods 2005, 62, 221–232. [Google Scholar] [CrossRef]
- Goldstein, C.; Lee, M.D.; Sanchez, S.; Hudson, C.; Phillips, B.; Register, B.; Grady, M.; Liebert, C.; Summers, A.O.; White, D.G.; et al. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals and exotics. Antimicrob. Agents Chemother. 2001, 45, 723–726. [Google Scholar] [CrossRef]
- Akya, A.; Azam Elahi, A.; Chegenelorestani, R.; Rezaee, M. Dissemination of multidrug-resistant, class I and II integrons and molecular typing of CTX-M-producing Klebsiella pneumoniae. Int. J. Appl. Basic Med. Res. 2018, 8, 100–105. [Google Scholar] [CrossRef]
- Koczura, R.; Mokracka, J.; Jabłońska, L.; Gozdecka, E.; Kubek, M.; Kaznowski, A. Antimicrobial resistance of integron-harboring Escherichia coli isolates from clinical samples, wastewater treatment plant and river water. Sci. Total Environ. 2012, 414, 680–685. [Google Scholar] [CrossRef]
- Ravi, A.; Avershina, E.; Ludvigsen, J.; L’Abée-Lund, T.M.; Rudi, K. Integrons in the intestinal microbiota as reservoirs for transmission of antibiotic resistance genes. Pathogens 2014, 3, 238–248. [Google Scholar] [CrossRef]
 dairy cattle farm;
dairy cattle farm;  poultry farm;
poultry farm;  slaughterhouse;
slaughterhouse;  pig farm.
pig farm.
 dairy cattle farm;
dairy cattle farm;  poultry farm;
poultry farm;  slaughterhouse;
slaughterhouse;  pig farm.
pig farm.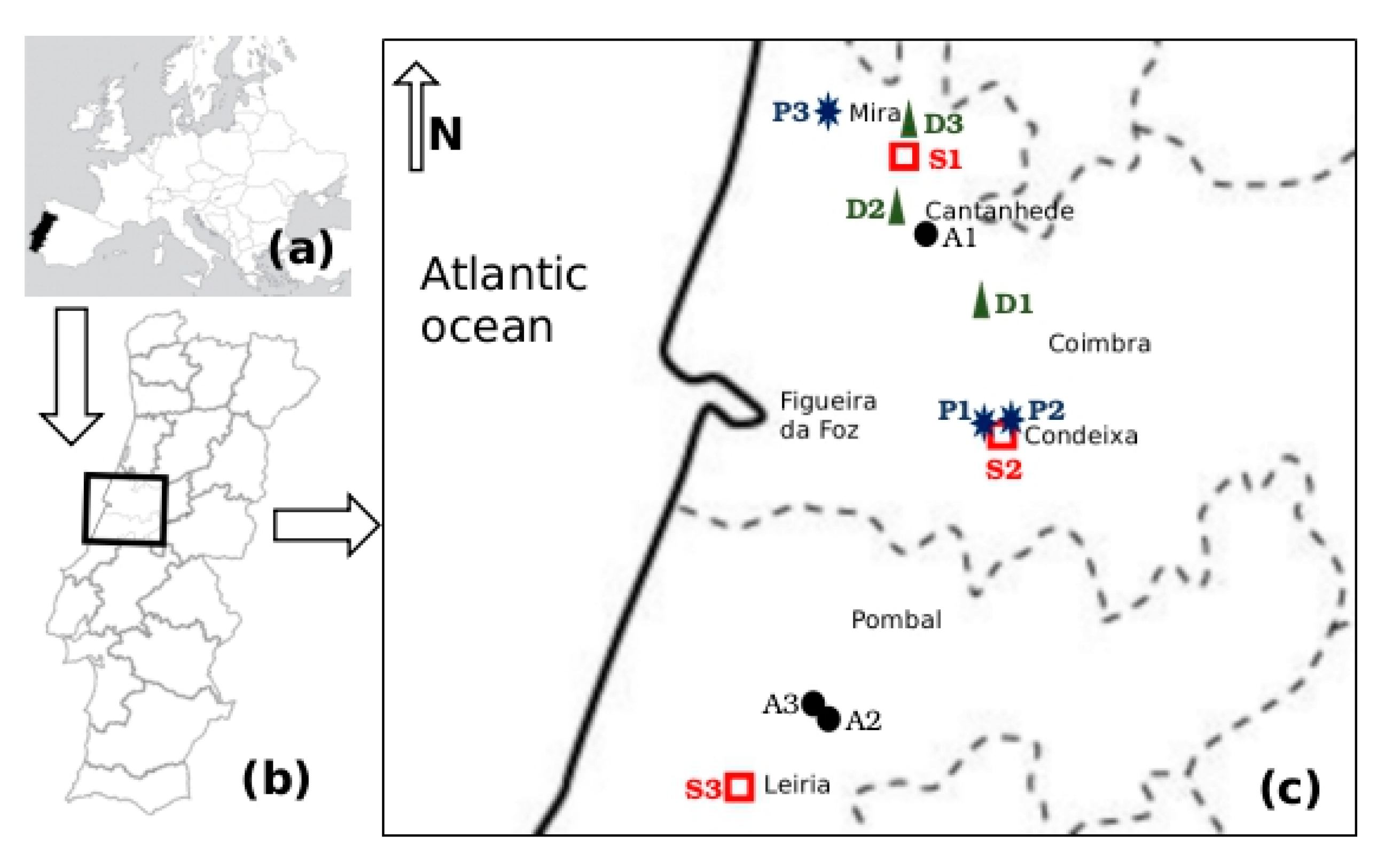
 poultry,
poultry,  pig,
pig,  dairy,
dairy,  slaughterhouse.
slaughterhouse.
 poultry,
poultry,  pig,
pig,  dairy,
dairy,  slaughterhouse.
slaughterhouse.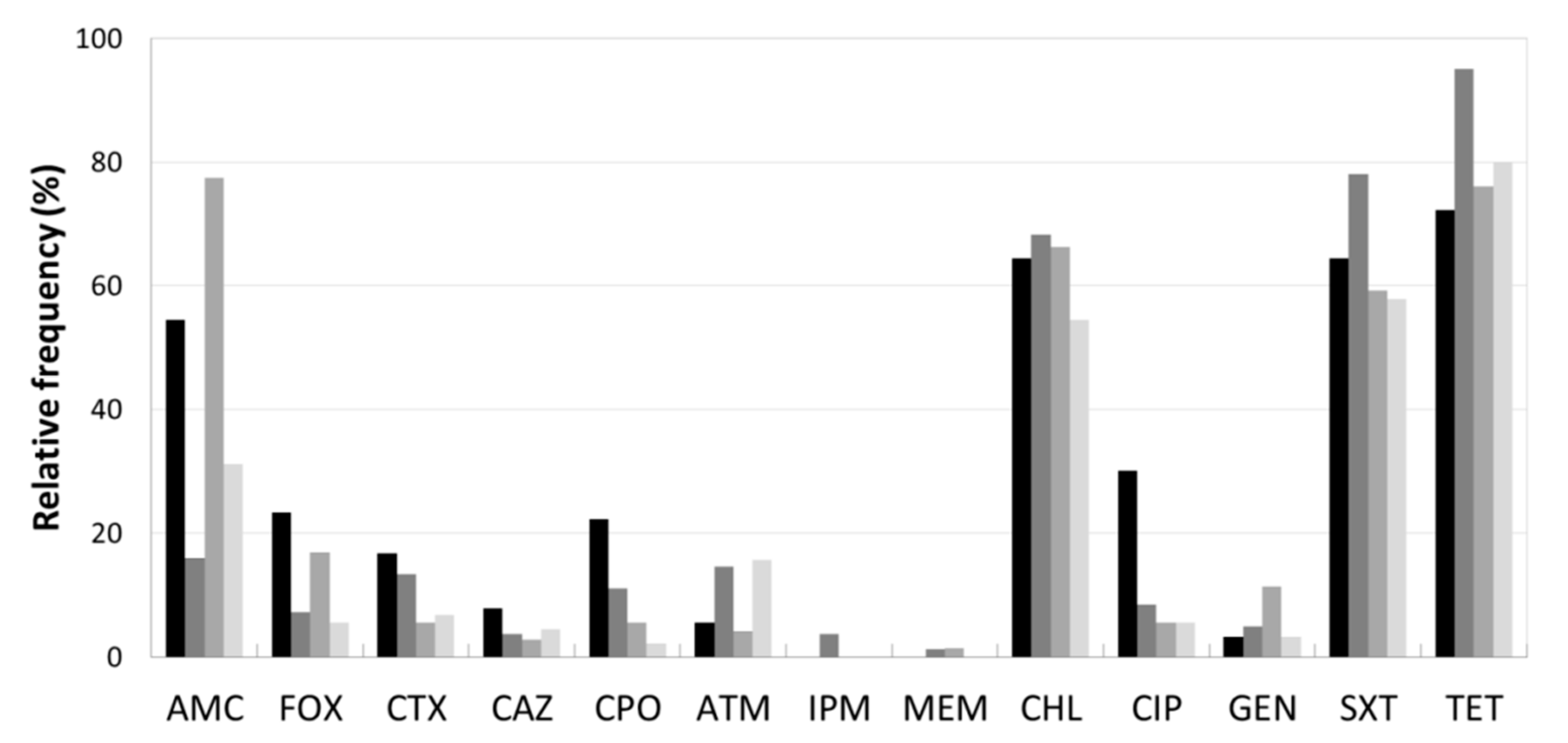
 poultry,
poultry,  pig,
pig,  dairy,
dairy,  slaughterhouse.
slaughterhouse.
 poultry,
poultry,  pig,
pig,  dairy,
dairy,  slaughterhouse.
slaughterhouse.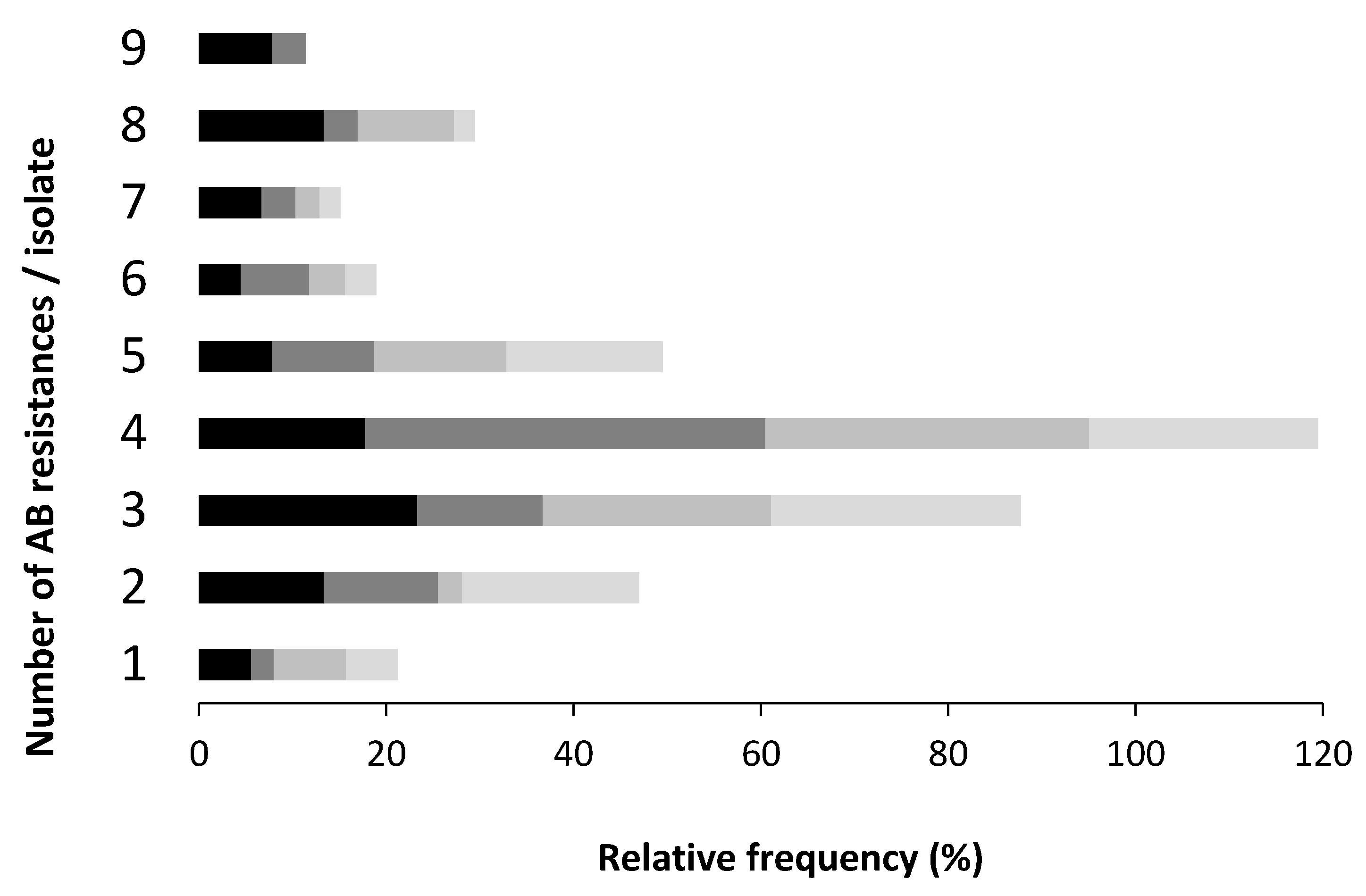
| Characterisation of Sampling Sites | Characterisation of Samples | ||||
|---|---|---|---|---|---|
| Site Code | Enterprise Type | Animal Heads | Enterobacteriaceae (cfu/mL) | Temperature (°C) | pH |
| A1 | Poultry | 10,000 chicks Legorne/pavilion | 1.4 × 107 | 10.2 | 6.80 |
| A2 | Poultry | 72,000 caged laying hens Legorne | 6.9 × 109 | 24.1 | 7.24 |
| A3 | Poultry | 28,000 soil, cage-free laying hens Legorne | 7.6 × 109 | 24.0 | 7.06 |
| D1 | Dairy cattle | 100 Holstein Friesian | 1.7 × 105 | 9.4 | 8.81 |
| D2 | Dairy cattle | 30 Holstein Friesian | 5.7 × 105 | 14.6 | 7.46 |
| D3 | Dairy cattle | 54 Holstein Friesian | 2.4 × 105 | 16.9 | 8.47 |
| P1 | Pig | 48 breeding sow Large Write | 3.4 × 105 | 14.7 | 7.71 |
| P2 | Pig | 700 breeding sow, 5300 piglets Large Write | 5.0 × 105 | 14.0 | 7.16 |
| P3 | Pig | 1640 fattening pigs Large Write | 1.7 × 106 | 13.8 | 7.33 |
| S1 | Slaughterhouse | 505 piglets, 62 cattle (daily abattoir) | 1.0 × 106 | 14.1 | 6.52 |
| S2 | Slaughterhouse | 381 hogs, 740 cattle (daily abattoir) | 1.7 × 106 | 16.0 | 4.92 |
| S3 | Slaughterhouse | 190 hogs, 40 cattle (daily abattoir) | 1.1 × 109 | 23.8 | 7.31 |
| Sample (a) | Species | Resistance Phenotype (b) | AB Resistance Genes | intI | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMC | FOX | CTX | CAZ | CPO | ATM | IPM | MEM | CHL | CIP | GEN | SXT | TET | cat | qnr/oqx | tet | sul | dfr | |||
| Poultry | S. enterica | x | x | x | x | x | x | x | x | - | - | A, M, L, K | 1, 2, 3 | Ia, IIb | - | |||||
| Enterobacter cloacae | x | x | x | x | x | x | x | x | x | I | D, S | A, E, B, K, O | 1, 3 | Ia | - | |||||
| E. hermannii | x | x | x | x | x | x | x | x | x | - | - | C | 1, 2 | Ic | - | |||||
| C. freundii | x | x | x | x | x | x | x | x | I | C, S | A | 1, 2 | Ia, IIb, IIIc | - | ||||||
| C. freundii | x | x | x | x | x | x | x | x | I | S | A, L, K | 1, 2 | Ia, Ic, IIIa, IIIc | - | ||||||
| R. ornithinolytica | x | x | x | x | - | nt | - | 3 | Ia | 1 | ||||||||||
| E. coli | x | x | x | x | x | x | x | x | x | - | nt | A, C | 1, 2, 3 | Ia, Ib, IIIa | - | |||||
| E. coli | x | x | x | x | x | - | nt | A, E, C, M, L | 2, 3 | Ia, IIa | - | |||||||||
| E. coli | x | x | x | x | x | x | x | - | nt | A | 1, 3 | Ia, Ib | - | |||||||
| E. coli | x | x | x | x | x | x | x | x | - | nt | A | 1, 3 | Ia, Ib, Ic, IIIa | - | ||||||
| E. coli | x | x | x | x | x | - | nt | A | 2 | Ia, Ic, IIb | - | |||||||||
| E. coli | x | x | x | x | x | x | x | - | nt | A | 3 | Ia, Ib | - | |||||||
| nt | x | x | x | x | x | x | x | x | I | S | A, M | 1, 2, 3 | Ia, IIb | - | ||||||
| nt | x | x | x | x | x | x | x | x | x | I | S | A, A(P) | 1, 3 | Ia | - | |||||
| nt | x | x | x | x | x | x | x | - | C, S | A | 1, 3 | Ia | 1 | |||||||
| nt | x | x | x | x | x | x | x | x | I | C, S | A | 1, 2, 3 | Ia | 1 | ||||||
| nt | x | x | x | x | x | x | I | B, S | A | 1, 3 | - | I | ||||||||
| nt | x | x | x | x | x | x | I | B, C, D, S | - | 1 | Ia | 1 | ||||||||
| nt | x | x | x | x | - | nt | A, A(P), M | - | IIb | - | ||||||||||
| nt | x | x | x | x | x | x | - | nt | A | - | - | - | ||||||||
| nt | x | x | x | x | x | - | B, S | A, L, K | 3 | - | - | |||||||||
| nt | x | x | x | x | x | - | B, S | A, L | 1, 3 | Ia, Ic, IIb | - | |||||||||
| Pig | S. enterica | x | x | x | x | x | - | nt | A | - | Ia, Ic | 2 | ||||||||
| K. oxytoca | x | x | x | x | x | - | oqxB | A | 1, 2, 3 | Ic, IIa | - | |||||||||
| R. ornithinolytica | x | x | x | x | x | x | x | I | B | M | 2, 3 | - | - | |||||||
| Kluyvera spp. | x | x | x | x | x | x | x | x | x | - | nt | A | 2 | Ia | 2 | |||||
| E. coli | x | x | x | x | - | nt | A, B | - | Ic | - | ||||||||||
| E. coli | x | x | x | x | x | x | x | I | - | A | 1, 2, 3 | Ia, Ic | 1 | |||||||
| E. coli | x | x | x | x | x | x | - | B | A | 1, 2, 3 | Ia | 2 | ||||||||
| E. coli | x | x | x | x | x | x | - | - | - | - | Ia, IIIb | - | ||||||||
| E. coli | x | x | x | x | x | x | x | x | I | - | B, M | 3 | Ia, IIa | 2 | ||||||
| E. coli | x | x | x | x | x | x | x | - | B | A | - | Ia | - | |||||||
| E. coli | x | x | x | x | x | x | x | x | x | - | B | A | - | Ia, IIIb | 1 | |||||
| E. coli | x | x | x | x | x | x | x | x | x | - | B | A | 1, 2 | Ia, Ic | - | |||||
| nt | x | x | x | x | - | oqxB | A, M | 1, 2, 3 | - | 1 | ||||||||||
| nt | x | x | x | x | x | x | - | nt | - | - | - | 2 | ||||||||
| nt | x | x | x | x | x | x | - | B, S | A | 2 | IIIb | - | ||||||||
| nt | x | x | x | x | x | x | - | nt | A, M, K | 2, 3 | - | 2 | ||||||||
| nt | x | x | x | x | x | x | - | S | A, M | 3 | - | 1 | ||||||||
| nt | x | x | x | x | x | x | x | - | nt | A | 2 | IIIb | - | |||||||
| nt | x | x | x | x | x | x | x | - | nt | A | 2, 3 | - | 1 | |||||||
| Dairy | M. morganii | x | x | x | x | x | II | nt | A, O | 1, 2 | - | - | ||||||||
| M. morganii | x | x | x | x | x | x | x | II | nt | A, K | 1, 2 | - | - | |||||||
| C. freundii | x | x | x | x | x | x | x | I | S | A | 1, 2 | Ia | - | |||||||
| C. freundii | x | x | x | x | x | x | I | C, S | - | 1, 3 | Ia | 1 | ||||||||
| C. freundii | x | x | x | x | x | x | x | I | C, S | A | 1, 3 | Ia | 1 | |||||||
| C. braakii | x | x | x | x | x | x | x | - | nt | A | - | Ib | - | |||||||
| C. koseri | x | x | x | x | x | x | x | x | x | - | nt | A, E | 1, 3 | Ib, IIIb | - | |||||
| R. ornithinolytica | x | x | x | x | - | nt | A, E | - | Ia, Ib | - | ||||||||||
| C. koseri | x | x | x | x | - | nt | - | - | IIIc | - | ||||||||||
| E. coli | x | x | x | x | - | nt | A | 2 | Ia, IIa | - | ||||||||||
| E. coli | x | x | x | x | x | x | - | nt | A, E | 1 | IIc | - | ||||||||
| E. coli | x | x | x | x | x | - | nt | - | - | Ia | - | |||||||||
| nt | x | x | x | x | x | x | - | nt | M | 3 | IIIb, IIIc | - | ||||||||
| Slaughterhouse | E. coli | x | x | x | - | nt | A(P), L | - | Ia, Ic | 2 | ||||||||||
| E. coli | x | x | x | x | x | - | nt | O, M | 3 | Ia | - | |||||||||
| E. coli | x | x | x | x | x | x | - | nt | A, M | - | Ic | - | ||||||||
| E. coli | x | x | x | x | x | x | - | - | A, B, O, M | 3 | Ic | - | ||||||||
| E. coli | x | x | x | x | x | x | x | x | x | - | nt | K | 1, 3 | Ia | 1 | |||||
| E. coli | x | x | x | x | x | x | x | x | - | nt | A | 2, 3 | IIb | 1 | ||||||
| E. coli | x | x | x | x | x | x | x | x | I | - | A, M | 2, 3 | Ic, IIb | 1 | ||||||
| S. enterica | x | x | x | x | - | nt | - | 3 | Ia, IIIb | 2 | ||||||||||
| E. vulneris | x | x | x | x | x | - | - | B, M | 1, 2, 3 | Ic, IIb | - | |||||||||
| Enterobacter cloacae | x | x | x | x | x | - | nt | - | - | - | - | |||||||||
| C. freundii | x | x | x | x | x | x | x | - | nt | - | 3 | IIIb | - | |||||||
| nt | x | x | x | - | nt | A(P) | - | - | 1 | |||||||||||
| nt | x | x | x | x | - | B, S | A, M | 3 | - | - | ||||||||||
| nt | x | x | x | x | x | - | B, S | B | 3 | - | - | |||||||||
| nt | x | x | x | x | x | - | nt | A, A(P), M | 3 | IIIb | 2 | |||||||||
| nt | x | x | x | x | x | - | nt | A | 1 | - | - | |||||||||
| Target Gene/Group | Enterprise Type | Total * | ||||
|---|---|---|---|---|---|---|
| Poultry * | Pig * | Dairy * | Slaughterhouse * | |||
| cat | I | 8 (36.4) | 3 (15.7) | 3 (23.1) | 1 (6.3) | 15 (21.4) |
| II | 2 (15.4) | 2 (2.9) | ||||
| III, IV | 0 (0.0) | |||||
| qnr | A | 0 (0.0) | ||||
| B | 4 (30.8) | 6 (50.0) | 2 (40.0) | 12 (36.4) | ||
| C | 4 (30.8) | 2 (66.7) | 6 (18.1) | |||
| D | 2 (15.4) | 2 (6.1) | ||||
| S | 11 (84.6) | 2 (16.7) | 3 (100.0) | 2 (40.0) | 18 (54.6) | |
| oqx | A | 0 (0.0) | ||||
| B | 2 (16.7) | 2 (6.1) | ||||
| aac(6’)-Ib | 0 (0.0) | |||||
| qep | A | 0 (0.0) | ||||
| tet | A | 19 (86.4) | 15 (78.9) | 9 (69.2) | 7 (43.8) | 50 (71.4) |
| B | 1 (4.6) | 2 (10.5) | 3 (18.8) | 6 (8.6) | ||
| C | 3 (13.6) | 3 (4.3) | ||||
| E | 2 (9.1) | 3 (23.1) | 5 (7.1) | |||
| K | 4 (18.2) | 1 (5.3) | 1 (7.7) | 1 (6.3) | 7 (10.0) | |
| L | 5 (22.7) | 1 (6.3) | 6 (8.6) | |||
| M | 4 (18.2) | 5 (26.3) | 1 (7.7) | 7 (43.8) | 17 (24.3) | |
| O | 1 (4.6) | 1 (7.7) | 2 (12.5) | 4 (5.7) | ||
| A(P) | 2 (9.1) | 3 (18.8) | 5 (7.1) | |||
| D, G, S, Q, X | 0 (0.0) | |||||
| sul | 1 | 15 (68.2) | 5 (26.3) | 7 (53.8) | 3 (18.8) | 30 (42.9) |
| 2 | 9 (40.9) | 11 (57.9) | 4 (30.8) | 3 (18.8) | 27 (38.6) | |
| 3 | 15 (68.2) | 9 (47.4) | 4 (30.8) | 11 (68.8) | 40 (57.1) | |
| dfr | Ia | 17 (77.2) | 9 (47.4) | 6 (46.2) | 4 (25.0) | 36 (51.4) |
| Ib | 4 (18.2) | 3 (23.1) | 7 (10.0) | |||
| Ic | 5 (22.7) | 5 (26.3) | 5 (31.3) | 15 (21.4) | ||
| IIa | 1 (4.6) | 2 (10.5) | 1 (7.7) | 4 (5.7) | ||
| IIb | 6 (27.3) | 3 (18.8) | 9 (12.9) | |||
| IIc | 1 (7.7) | 1 (1.4) | ||||
| IIIa | 3 (13.6) | 3 (4.3) | ||||
| IIIb | 4 (21.1) | 2 (15.4) | 3 (18.8) | 9 (12.9) | ||
| IIIc | 2 (9.1) | 2 (15.4) | 4 (5.7) | |||
| IVa, IVb, IVc | 0 (0.0) | |||||
| Va, Vb, Vc, Vd | 0 (0.0) | |||||
| intI | 1 | 5 (22.7) | 5 (26.3) | 2 (15.4) | 4 (25.0) | 16 (22.9) |
| 2 | 6 (31.6) | 3 (18.8) | 9 (12.9) | |||
| 3 | 0 (0.0) | |||||
| AB | Resistant Isolates (N) | Resistant Isolates with Integrons, N (%) | Resistant Isolates without Integrons, N (%) | |
|---|---|---|---|---|
| intI1 | intI2 | |||
| CHL | 58 | 14 (24.1%) | 8 (13.8%) | 36 (62.1%) |
| CIP | 29 | 10 (34.5%) | 1 (3.4%) | 18 (62.1%) |
| GEN | 7 | 1 (14.3%) | 0 (0.0%) | 6 (85.7%) |
| SXT | 56 | 15 (26.8%) | 8 (14.3%) | 33 (58.9%) |
| TET | 63 | 13 (20.6%) | 8 (12.7%) | 42 (66.7%) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amador, P.; Fernandes, R.; Prudêncio, C.; Duarte, I. Prevalence of Antibiotic Resistance Genes in Multidrug-Resistant Enterobacteriaceae on Portuguese Livestock Manure. Antibiotics 2019, 8, 23. https://doi.org/10.3390/antibiotics8010023
Amador P, Fernandes R, Prudêncio C, Duarte I. Prevalence of Antibiotic Resistance Genes in Multidrug-Resistant Enterobacteriaceae on Portuguese Livestock Manure. Antibiotics. 2019; 8(1):23. https://doi.org/10.3390/antibiotics8010023
Chicago/Turabian StyleAmador, Paula, Ruben Fernandes, Cristina Prudêncio, and Isabel Duarte. 2019. "Prevalence of Antibiotic Resistance Genes in Multidrug-Resistant Enterobacteriaceae on Portuguese Livestock Manure" Antibiotics 8, no. 1: 23. https://doi.org/10.3390/antibiotics8010023
APA StyleAmador, P., Fernandes, R., Prudêncio, C., & Duarte, I. (2019). Prevalence of Antibiotic Resistance Genes in Multidrug-Resistant Enterobacteriaceae on Portuguese Livestock Manure. Antibiotics, 8(1), 23. https://doi.org/10.3390/antibiotics8010023






