Resistance to Antimicrobial Peptides in Vibrios
Abstract
:1. Introduction
2. Antimicrobial Peptides in Host-Vibrio Interactions
2.1. Vibrios Colonizing Epithelial Surfaces
| Species or strain | Host | Tissues | References |
|---|---|---|---|
| V. cholerae | human | intestine | [20] |
| V. vulnificus | human | skin, wounds | [27] |
| V. parahemolyticus | human | intestine | [19] |
| V. anguillarum | fish | skin, intestine | [25] |
| V. shiloi | coral | oral ectoderm | [26] |
| V. coralliilyticus | coral | oral ectoderm | [28] |
| V. fisheri | squid | light organ | [24] |
2.2. AMPs and Epithelial Defenses
| Species | AMP family | Examples | Epithelial Tissues | References |
|---|---|---|---|---|
| Human | α-defensins | HD-5, HD-6 | Small intestine, female genital tract | [37,57] |
| β-defensins | hBD-1/-2/-3 | Respiratory tract, large intestine, urogenital epithelium, oral cavity, skin | [58,59,60,61,62] | |
| Cathelicidins | LL-37(hCAP-18) | Skin, gastrointestinal tract, epididymis, lungs, oral cavity, ocular surface | [31,63,64], for review see [65] | |
| Bactericidal-permeability increasing proteins | BPI | Esophagus, respiratory tract, large intestine | For review see [66] | |
| C-type lectins | HIP/PAP | Small intestine | [36] | |
| Fish | β-defensins | omDB-1/-2/-3/-4 | Skin, gills, intestine | [45,67] |
| Cathelicidins | rtCATH_1/-2A-2B, asCATH-1/-2 HFIAP-1/-2/-3 | Skin, gills, intestine | [47,68] | |
| Liver-expressed antimicrobial peptides (LEAPs) | Hepcidin (LEAP-1), LEAP-2 Sal-1 Sal-2 | Skin, intestine | [69], for review see [44] | |
| α-helical peptides | Pleurocidin, Piscidins Chrysophsins Moronecidin | Skin, gills | [70,71,72] | |
| Bactericidal-permeability increasing proteins | BPI | Intestine, gills | [48,49] | |
| Histone-derived AMPs | Parasin-1 Hipposin Oncorhyncin | Skin mucus | [73,74] [44,50] | |
| Squid | LPS-binding/ Bactericidal-permeability increasing proteins | Es-LBP1 | Light organ | [51] |
| Oyster | CS-αβ defensins | Cg-Defm | Mantle tissue | [53] |
| Bactericidal-permeability increasing proteins | Cg-BPI | Gills, mantle, labial palps, gastrointestinal tract | [52] | |
| Histone-derived AMPs | cvH2B-1/-2/-3/-4 | Gills | [55] | |
| Coral | Cysteine Rich peptides | Damicornin Mytimacin-like | Oral ectoderm | [28] |
| LPS-binding/ Bactericidal-permeability increasing proteins | LBP–BPI | Oral ectoderm | [28] |
2.3. Vibrios Adapted to Intracellular Life in Phagocytes
2.4. AMPs of Phagocytes
| Species | AMP | Examples | Phagocytes | References |
|---|---|---|---|---|
| Human | α-defensins | HNP-1/-2/-3/-4 | Neutrophils | [80] |
| β-defensins | hBD-1/-2 | Macrophages, Dendritic cells | [81,82] | |
| Cathelicidins | LL-37 | Neutrophils | [89,90] | |
| Liver-expressed antimicrobial peptides (LEAPs) | Hepcidin | Granulocytes Macrophages | [91,92] | |
| Bactericidal-permeability increasing proteins | BPI | Neutrophils, (Eosinophils/to a lesser extent) | [30,93,94] | |
| Fish | α-helical peptides | Piscidins | Granulocytes | [85] |
| LPS-Binding/Bactericidal-permeability increasing proteins | LBP/BPI | Head–kidney leukocytes | [49] | |
| Oyster | CS-αβ defensins | Cg-Defh-1/h2 | Hemocytes | [1] |
| Big-defensins | Cg-big-defensin-1/-2/-3 | Hemocytes | [87] | |
| Proline-rich peptides | Cg-Prp | Hemocytes | [14] | |
| Bactericidal-permeability increasing protein | Cg-BPI | Hemocytes | [14,52] | |
| Histone-derived AMPs | H1- and H5-like histones | Hemocytes | [56] |
3. Known Mechanisms of Resistance/Evasion to AMPs in Vibrios
3.1. Outer Membrane Remodeling
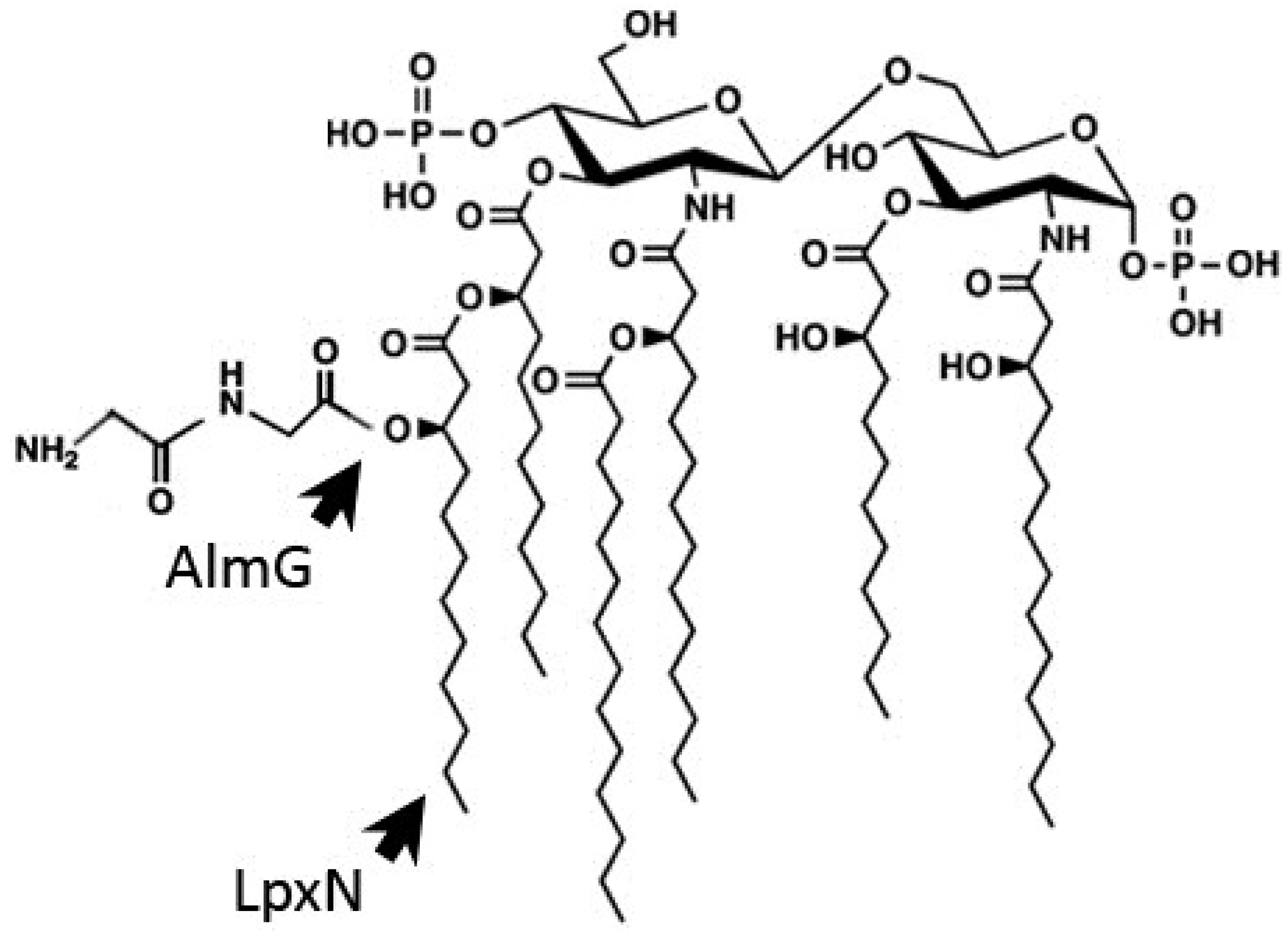
3.2. Induction of the Envelope Stress Response
3.3. AMP Titration by Outer Membrane Vesicles
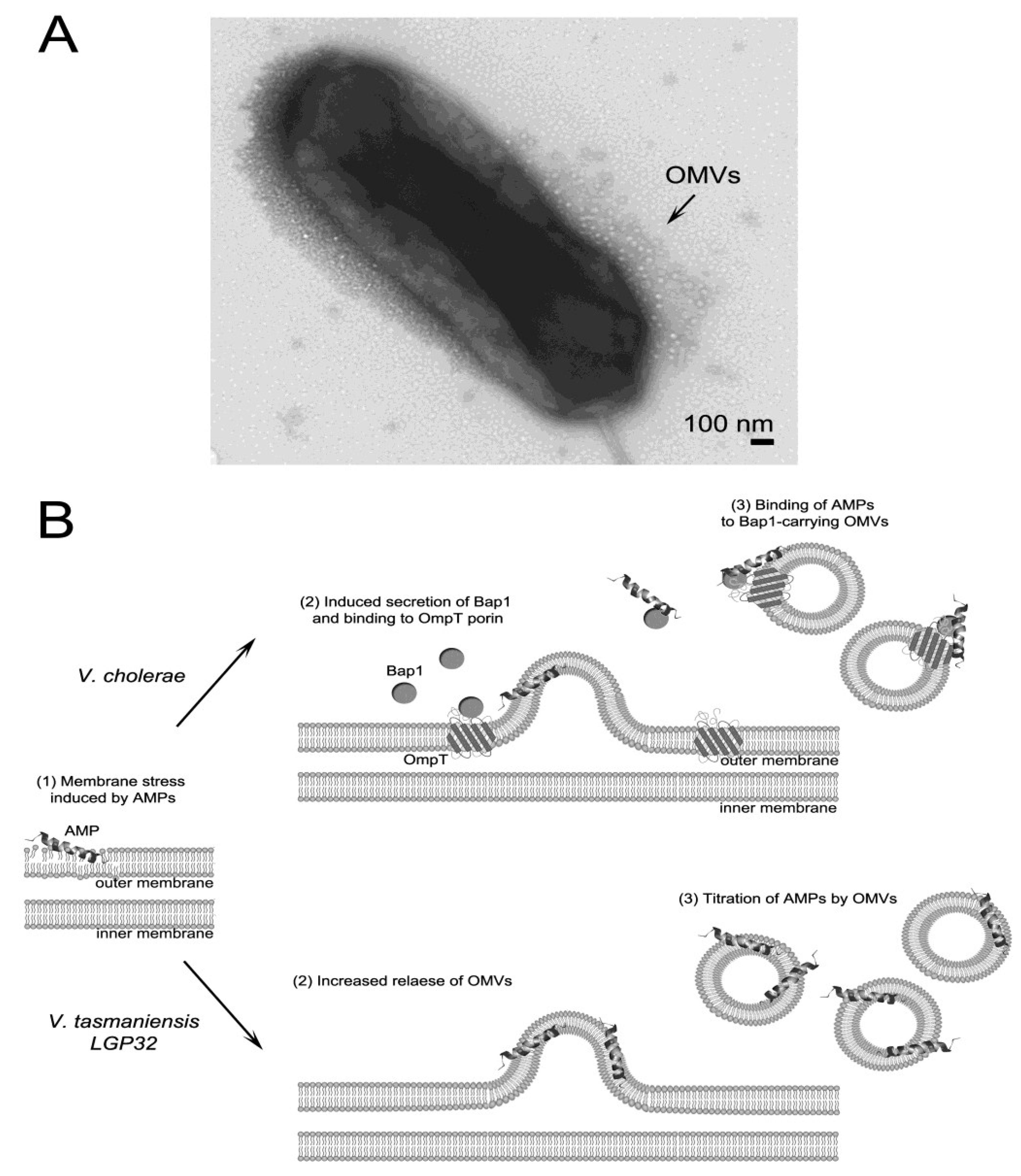
3.4. Efflux of AMPs
3.5. Suppression of AMP Expression
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schmitt, P.; Rosa, R.D.; Duperthuy, M.; de Lorgeril, J.; Bachere, E.; Destoumieux-Garzon, D. The antimicrobial defense of the pacific oyster, Crassostrea gigas. How diversity may compensate for scarcity in the regulation of resident/pathogenic microflora. Front. Microbiol. 2012, 3, e160. [Google Scholar] [CrossRef]
- Jutla, A.; Whitcombe, E.; Hasan, N.; Haley, B.; Akanda, A.; Huq, A.; Alam, M.; Sack, R.B.; Colwell, R. Environmental factors influencing epidemic cholera. Am. J. Trop. Med. Hyg. 2013, 89, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 2003, 3, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Gallo, R.L.; Hooper, L.V. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 2012, 12, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.T.; McAuley, S.; Pukatzki, S.; Mekalanos, J.J. Translocation of a Vibrio cholerae type vi secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 2009, 5, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Duperthuy, M.; Schmitt, P.; Garzon, E.; Caro, A.; Rosa, R.D.; Le Roux, F.; Lautredou-Audouy, N.; Got, P.; Romestand, B.; de Lorgeril, J.; et al. Use of ompu porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc. Natl. Acad. Sci. USA 2011, 108, 2993–2998. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar]
- Kragol, G.; Lovas, S.; Varadi, G.; Condie, B.A.; Hoffmann, R.; Otvos, L., Jr. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 2001, 40, 3016–3026. [Google Scholar] [CrossRef] [PubMed]
- Brotz, H.; Josten, M.; Wiedemann, I.; Schneider, U.; Gotz, F.; Bierbaum, G.; Sahl, H.G. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 1998, 30, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Park, C.B.; Kim, H.S.; Kim, S.C. Mechanism of action of the antimicrobial peptide buforin II: Buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 1998, 244, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Patrzykat, A.; Friedrich, C.L.; Zhang, L.; Mendoza, V.; Hancock, R.E. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 2002, 46, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Beema Shafreen, R.M.; Nithyanand, P.; Manisankar, P.; Pandian, S.K. Synthesis and in vitro antimicrobial evaluation of novel fluoroquinolone derivatives. Eur. J. Med. Chem. 2010, 45, 6101–6105. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, M.; Cammue, B.P.; Sahl, H.G.; Thevissen, K. Antibiotic activities of host defense peptides: More to it than lipid bilayer perturbation. Nat. Prod. Rep. 2011, 28, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, P.; de Lorgeril, J.; Gueguen, Y.; Destoumieux-Garzon, D.; Bachere, E. Expression, tissue localization and synergy of antimicrobial peptides and proteins in the immune response of the oyster Crassostrea gigas. Dev. Comp. Immunol. 2012, 37, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Hilchie, A.L.; Wuerth, K.; Hancock, R.E. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat. Chem. Biol. 2013, 9, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Duquesne, S.; Destoumieux-Garzon, D.; Peduzzi, J.; Rebuffat, S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat. Prod. Rep. 2007, 24, 708–734. [Google Scholar] [CrossRef] [PubMed]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar]
- Martinez, J.L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef] [PubMed]
- Qadri, F.; Alam, M.S.; Nishibuchi, M.; Rahman, T.; Alam, N.H.; Chisti, J.; Kondo, S.; Sugiyama, J.; Bhuiyan, N.A.; Mathan, M.M.; et al. Adaptive and inflammatory immune responses in patients infected with strains of Vibrio parahaemolyticus. J. Infect. Dis. 2003, 187, 1085–1096. [Google Scholar] [CrossRef] [PubMed]
- Qadri, F.; Bhuiyan, T.R.; Dutta, K.K.; Raqib, R.; Alam, M.S.; Alam, N.H.; Svennerholm, A.M.; Mathan, M.M. Acute dehydrating disease caused by Vibrio cholerae serogroups o1 and o139 induce increases in innate cells and inflammatory mediators at the mucosal surface of the gut. Gut 2004, 53, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Vanden Broeck, D.; Horvath, C.; de Wolf, M.J. Vibrio cholerae: Cholera toxin. Int. J. Biochem. Cell. Biol. 2007, 39, 1771–1775. [Google Scholar] [CrossRef] [PubMed]
- Park, K.S.; Ono, T.; Rokuda, M.; Jang, M.H.; Okada, K.; Iida, T.; Honda, T. Functional characterization of two type III secretion systems of vibrio parahaemolyticus. Infect. Immun. 2004, 72, 6659–6665. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Gewurz, B.E.; Ritchie, J.M.; Takasaki, K.; Greenfeld, H.; Kieff, E.; Davis, B.M.; Waldor, M.K. A Vibrio parahaemolyticus t3ss effector mediates pathogenesis by independently enabling intestinal colonization and inhibiting tak1 activation. Cell. Rep. 2013, 3, 1690–1702. [Google Scholar] [CrossRef] [PubMed]
- McFall-Ngai, M.; Nyholm, S.V.; Castillo, M.G. The role of the immune system in the initiation and persistence of the euprymna scolopes—Vibrio fischeri symbiosis. Semin. Immunol. 2010, 22, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.; Chen, C.; Milton, D.L. Colonization of fish skin is vital for Vibrio anguillarum to cause disease. Environ. Microbiol. Rep. 2010, 2, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.; Falkovitz, L. The Vibrio shiloi / Oculina patagonica model system of coral bleaching. Annu. Rev. Microbiol. 2004, 58, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Daniels, N.A. Vibrio vulnificus oysters: Pearls and perils. Clin. Infect. Dis. 2011, 52, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Dupiol, J.; Ladriere, O.; Destoumieux-Garzon, D.; Sautiere, P.E.; Meistertzheim, A.L.; Tambutte, E.; Tambutte, S.; Duval, D.; Foure, L.; Adjeroud, M.; et al. Innate immune responses of a scleractinian coral to vibriosis. J. Biol. Chem. 2011, 286, 22688–22698. [Google Scholar] [CrossRef] [PubMed]
- Pitman, R.S.; Blumberg, R.S. First line of defense: The role of the intestinal epithelium as an active component of the mucosal immune system. J. Gastroenterol. 2000, 35, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Levy, O.; Canny, G.; Serhan, C.N.; Colgan, S.P. Expression of bpi (bactericidal/permeability-increasing protein) in human mucosal epithelia. Biochem. Soc. Trans. 2003, 31, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Bals, R.; Wang, X.; Zasloff, M.; Wilson, J.M. The peptide antibiotic ll-37/hcap-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc. Natl. Acad. Sci. USA 1998, 95, 9541–9546. [Google Scholar] [CrossRef] [PubMed]
- Frohm Nilsson, M.; Sandstedt, B.; Sorensen, O.; Weber, G.; Borregaard, N.; Stahle-Backdahl, M. The human cationic antimicrobial protein (hcap18), a peptide antibiotic, is widely expressed in human squamous epithelia and colocalizes with interleukin-6. Infect. Immun. 1999, 67, 2561–2566. [Google Scholar] [PubMed]
- Bals, R.; Wilson, J.M. Cathelicidins—A family of multifunctional antimicrobial peptides. Cell. Mol. Life Sci. 2003, 60, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Frohm, M.; Agerberth, B.; Ahangari, G.; Stahle-Backdahl, M.; Liden, S.; Wigzell, H.; Gudmundsson, G.H. The expression of the gene coding for the antibacterial peptide LL-37 is induced in human keratinocytes during inflammatory disorders. J. Biol. Chem. 1997, 272, 15258–15263. [Google Scholar] [CrossRef] [PubMed]
- Scheetz, T.; Bartlett, J.A.; Walters, J.D.; Schutte, B.C.; Casavant, T.L.; McCray, P.B., Jr. Genomics-based approaches to gene discovery in innate immunity. Immunol. Rev. 2002, 190, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Cash, H.L.; Whitham, C.V.; Behrendt, C.L.; Hooper, L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 2006, 313, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Ouellette, A.J. Paneth cell alpha-defensins in enteric innate immunity. Cell. Mol. Life Sci. 2011, 68, 2215–2229. [Google Scholar] [CrossRef] [PubMed]
- Pukatzki, S.; Provenzano, D. Vibrio cholerae as a predator: Lessons from evolutionary principles. Front. Microbiol. 2013, 4, e384. [Google Scholar] [CrossRef]
- Salzman, N.H.; Ghosh, D.; Huttner, K.M.; Paterson, Y.; Bevins, C.L. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 2003, 422, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Pazgier, M.; Jung, G.; Nuccio, S.P.; Castillo, P.A.; de Jong, M.F.; Winter, M.G.; Winter, S.E.; Wehkamp, J.; Shen, B.; et al. Human alpha-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 2012, 337, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Shin, O.S.; Uddin, T.; Citorik, R.; Wang, J.P.; della Pelle, P.; Kradin, R.L.; Bingle, C.D.; Bingle, L.; Camilli, A.; Bhuiyan, T.R.; et al. Lplunc1 modulates innate immune responses to Vibrio cholerae. J. Infect. Dis. 2011, 204, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [PubMed]
- Rakers, S.; Niklasson, L.; Steinhagen, D.; Kruse, C.; Schauber, J.; Sundell, K.; Paus, R. Antimicrobial peptides (amps) from fish epidermis: Perspectives for investigative dermatology. J. Invest. Dermatol. 2013, 133, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Smith, V.J.; Desbois, A.P.; Dyrynda, E.A. Conventional and unconventional antimicrobials from fish, marine invertebrates and micro-algae. Mar. Drugs. 2010, 8, 1213–1262. [Google Scholar] [CrossRef] [PubMed]
- Casadei, E.; Wang, T.; Zou, J.; Gonzalez Vecino, J.L.; Wadsworth, S.; Secombes, C.J. Characterization of three novel beta-defensin antimicrobial peptides in rainbow trout (Oncorhynchus mykiss). Mol. Immunol. 2009, 46, 3358–3366. [Google Scholar] [CrossRef] [PubMed]
- Caipang, C.M.; Lazado, C.C.; Brinchmann, M.F.; Kiron, V. Infection-induced changes in expression of antibacterial and cytokine genes in the gill epithelial cells of Atlantic cod, Gadus morhua during incubation with bacterial pathogens. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010, 156, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.I.; Zhang, Y.A.; Zou, J.; Nie, P.; Secombes, C.J. Two cathelicidin genes are present in both rainbow trout (Oncorhynchus mykiss) and Atlantic salmon (Salmo salar). Antimicrob. Agents Chemother. 2006, 50, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Kono, T.; Sakai, M. Molecular cloning of a novel bactericidal permeability-increasing protein/lipopolysaccharide-binding protein (bpi/lbp) from common carp Cyprinus carpio l. and its expression. Mol. Immunol. 2003, 40, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Stenvik, J.; Solstad, T.; Strand, C.; Leiros, I.; Jorgensen, T.T. Cloning and analyses of a bpi/lbp cDNA of the Atlantic cod (Gadus morhua L.). Dev. Comp. Immunol. 2004, 28, 307–323. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Park, I.Y.; Kim, H.S.; Lee, W.T.; Kim, M.S.; Kim, S.C. Cathepsin D produces antimicrobial peptide parasin I from histone H2A in the skin mucosa of fish. FASEB J. 2002, 16, 429–431. [Google Scholar] [PubMed]
- Krasity, B.C.; Troll, J.V.; Weiss, J.P.; McFall-Ngai, M.J. Lbp/bpi proteins and their relatives: Conservation over evolution and roles in mutualism. Biochem. Soc. Trans. 2011, 39, 1039–1044. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.; Gueguen, Y.; Destoumieux-Garzon, D.; Romestand, B.; Fievet, J.; Pugniere, M.; Roquet, F.; Escoubas, J.M.; Vandenbulcke, F.; Levy, O.; et al. Evidence of a bactericidal permeability increasing protein in an invertebrate, the Crassostrea gigas Cg-BPI. Proc. Natl. Acad. Sci. USA 2007, 104, 17759–17764. [Google Scholar] [CrossRef] [PubMed]
- Gueguen, Y.; Herpin, A.; Aumelas, A.; Garnier, J.; Fievet, J.; Escoubas, J.M.; Bulet, P.; Gonzalez, M.; Lelong, C.; Favrel, P.; et al. Characterization of a defensin from the oyster Crassostrea gigas. Recombinant production, folding, solution structure, antimicrobial activities, and gene expression. J. Biol. Chem. 2006, 281, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Lee, M.J.; Nam, B.H.; Park, N.G. Cgmolluscidin, a novel dibasic residue repeat rich antimicrobial peptide, purified from the gill of the pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 2013, 35, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.K.; Stephenson, J.; Noga, E.J. Multiple antibacterial histone H2B proteins are expressed in tissues of American oyster. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2011, 158, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Poirier, A.C.; Schmitt, P.; Rosa, R.D.; Vanhove, A.S.; Kieffer-Jaquinod, S.; Rubio, T.P.; Charriere, G.M.; Destoumieux-Garzon, D. Antimicrobial histones and DNA traps in invertebrate immunity: Evidences in Crassostrea gigas. J. Biol. Chem. 2014, 289, 24821–24831. [Google Scholar] [CrossRef] [PubMed]
- Quayle, A.J.; Porter, E.M.; Nussbaum, A.A.; Wang, Y.M.; Brabec, C.; Yip, K.P.; Mok, S.C. Gene expression, immunolocalization, and secretion of human defensin-5 in human female reproductive tract. Am. J. Pathol. 1998, 152, 1247–1258. [Google Scholar] [PubMed]
- Singh, P.K.; Jia, H.P.; Wiles, K.; Hesselberth, J.; Liu, L.; Conway, B.A.; Greenberg, E.P.; Valore, E.V.; Welsh, M.J.; Ganz, T.; et al. Production of beta-defensins by human airway epithelia. Proc. Natl. Acad. Sci. USA 1998, 95, 14961–14966. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wang, I.; Lehrer, R.I. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett. 1996, 396, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Mathews, M.; Jia, H.P.; Guthmiller, J.M.; Losh, G.; Graham, S.; Johnson, G.K.; Tack, B.F.; McCray, P.B., Jr. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect. Immun. 1999, 67, 2740–2745. [Google Scholar] [PubMed]
- O’Neil, D.A.; Porter, E.M.; Elewaut, D.; Anderson, G.M.; Eckmann, L.; Ganz, T.; Kagnoff, M.F. Expression and regulation of the human beta-defensins HBD-1 and HBD-2 in intestinal epithelium. J. Immunol. 1999, 163, 6718–6724. [Google Scholar] [PubMed]
- Liu, L.; Wang, L.; Jia, H.P.; Zhao, C.; Heng, H.H.; Schutte, B.C.; McCray, P.B., Jr.; Ganz, T. Structure and mapping of the human beta-defensin HBD-2 gene and its expression at sites of inflammation. Gene 1998, 222, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Chromek, M.; Slamova, Z.; Bergman, P.; Kovacs, L.; Podracka, L.; Ehren, I.; Hokfelt, T.; Gudmundsson, G.H.; Gallo, R.L.; Agerberth, B.; et al. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med. 2006, 12, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Hase, K.; Eckmann, L.; Leopard, J.D.; Varki, N.; Kagnoff, M.F. Cell differentiation is a key determinant of cathelicidin LL-37/human cationic antimicrobial protein 18 expression by human colon epithelium. Infect. Immun. 2002, 70, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Durr, U.H.; Sudheendra, U.S.; Ramamoorthy, A. Ll-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim. Biophys. Acta 2006, 1758, 1408–1425. [Google Scholar] [CrossRef] [PubMed]
- Canny, G.; Levy, O. Bactericidal/permeability-increasing protein (bpi) and bpi homologs at mucosal sites. Trends Immunol. 2008, 29, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Falco, A.; Chico, V.; Marroqui, L.; Perez, L.; Coll, J.M.; Estepa, A. Expression and antiviral activity of a beta-defensin-like peptide identified in the rainbow trout (Oncorhynchus mykiss) EST sequences. Mol. Immunol. 2008, 45, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Uzzell, T.; Stolzenberg, E.D.; Shinnar, A.E.; Zasloff, M. Hagfish intestinal antimicrobial peptides are ancient cathelicidins. Peptides 2003, 24, 1655–1667. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.E.; Gallant, J.W.; Liebscher, R.S.; Dacanay, A.; Tsoi, S.C. Identification and expression analysis of hepcidin-like antimicrobial peptides in bony fish. Dev. Comp. Immunol. 2003, 27, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Cole, A.M.; Weis, P.; Diamond, G. Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biol. Chem. 1997, 272, 12008–12013. [Google Scholar] [CrossRef] [PubMed]
- Noga, E.J.; Silphaduang, U. Piscidins: A novel family of peptide antibiotics from fish. Drug News Perspect. 2003, 16, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Salerno, G.; Parrinello, N.; Roch, P.; Cammarata, M. cDNA sequence and tissue expression of an antimicrobial peptide, dicentracin; a new component of the moronecidin family isolated from head kidney leukocytes of sea bass, Dicentrarchus labrax. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2007, 146, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Park, I.Y.; Park, C.B.; Kim, M.S.; Kim, S.C. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish, Parasilurus asotus. FEBS Lett. 1998, 437, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Birkemo, G.A.; Luders, T.; Andersen, O.; Nes, I.F.; Nissen-Meyer, J. Hipposin, a histone-derived antimicrobial peptide in Atlantic halibut (Hippoglossus hippoglossus L.). Biochim. Biophys. Acta 2003, 1646, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Abd, H.; Saeed, A.; Weintraub, A.; Nair, G.B.; Sandstrom, G. Vibrio cholerae O1 strains are facultative intracellular bacteria, able to survive and multiply symbiotically inside the aquatic free-living amoeba Acanthamoeba castellanii. FEMS Microbiol. Ecol. 2007, 60, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Abd, H.; Valeru, S.P.; Sami, S.M.; Saeed, A.; Raychaudhuri, S.; Sandstrom, G. Interaction between Vibrio mimicus and Acanthamoeba castellanii. Environ. Microbiol. Rep. 2010, 2, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Abd, H.; Weintraub, A.; Sandstrom, G. Intracellular survival and replication of Vibrio cholerae o139 in aquatic free-living amoebae. Environ. Microbiol. 2005, 7, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Ormonde, P.; Horstedt, P.; O’Toole, R.; Milton, D.L. Role of motility in adherence to and invasion of a fish cell line by Vibrio anguillarum. J. Bacteriol. 2000, 182, 2326–2328. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Oon, H.L.; Ho, G.W.; Wong, W.S.; Lim, T.M.; Leung, K.Y. Internalization and cytotoxicity are important virulence mechanisms in vibrio-fish epithelial cell interactions. Microbiology 1998, 144, 2987–3002. [Google Scholar] [CrossRef] [PubMed]
- Ganz, T. Defensins: Antimicrobial peptides of vertebrates. C. R. Biol. 2004, 327, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Duits, L.A.; Ravensbergen, B.; Rademaker, M.; Hiemstra, P.S.; Nibbering, P.H. Expression of beta-defensin 1 and 2 mrna by human monocytes, macrophages and dendritic cells. Immunology 2002, 106, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.T.; Modlin, R.L. Human macrophage host defense against Mycobacterium tuberculosis. Curr. Opin. Immunol. 2008, 20, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.T.; Stenger, S.; Li, H.; Wenzel, L.; Tan, B.H.; Krutzik, S.R.; Ochoa, M.T.; Schauber, J.; Wu, K.; Meinken, C.; et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 2006, 311, 1770–1773. [Google Scholar] [CrossRef] [PubMed]
- Sonawane, A.; Santos, J.C.; Mishra, B.B.; Jena, P.; Progida, C.; Sorensen, O.E.; Gallo, R.; Appelberg, R.; Griffiths, G. Cathelicidin is involved in the intracellular killing of mycobacteria in macrophages. Cell. Microbiol. 2011, 13, 1601–1617. [Google Scholar] [CrossRef] [PubMed]
- Mulero, I.; Noga, E.J.; Meseguer, J.; Garcia-Ayala, A.; Mulero, V. The antimicrobial peptides piscidins are stored in the granules of professional phagocytic granulocytes of fish and are delivered to the bacteria-containing phagosome upon phagocytosis. Dev. Comp. Immunol. 2008, 32, 1531–1538. [Google Scholar] [CrossRef] [PubMed]
- Cuesta, A.; Meseguer, J.; Esteban, M.A. The antimicrobial peptide hepcidin exerts an important role in the innate immunity against bacteria in the bony fish Gilthead seabream. Mol. Immunol. 2008, 45, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Rosa, R.D.; Santini, A.; Fievet, J.; Bulet, P.; Destoumieux-Garzon, D.; Bachere, E. Big defensins, a diverse family of antimicrobial peptides that follows different patterns of expression in hemocytes of the oyster Crassostrea gigas. PLoS One 2011, 6, e25594. [Google Scholar] [CrossRef] [PubMed]
- Leippe, M.; Herbst, R. Ancient weapons for attack and defense: The pore-forming polypeptides of pathogenic enteric and free-living amoeboid protozoa. J. Eukaryot. Microbiol. 2004, 51, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, O.; Arnljots, K.; Cowland, J.B.; Bainton, D.F.; Borregaard, N. The human antibacterial cathelicidin, hCAP-18, is synthesized in myelocytes and metamyelocytes and localized to specific granules in neutrophils. Blood 1997, 90, 2796–2803. [Google Scholar] [PubMed]
- Agerberth, B.; Charo, J.; Werr, J.; Olsson, B.; Idali, F.; Lindbom, L.; Kiessling, R.; Jornvall, H.; Wigzell, H.; Gudmundsson, G.H. The human antimicrobial and chemotactic peptides LL-37 and alpha-defensins are expressed by specific lymphocyte and monocyte populations. Blood 2000, 96, 3086–3093. [Google Scholar] [PubMed]
- Knutson, M.D.; Oukka, M.; Koss, L.M.; Aydemir, F.; Wessling-Resnick, M. Iron release from macrophages after erythrophagocytosis is up-regulated by ferroportin 1 overexpression and down-regulated by hepcidin. Proc. Natl. Acad. Sci. USA 2005, 102, 1324–1328. [Google Scholar] [CrossRef] [PubMed]
- Sow, F.B.; Florence, W.C.; Satoskar, A.R.; Schlesinger, L.S.; Zwilling, B.S.; Lafuse, W.P. Expression and localization of hepcidin in macrophages: A role in host defense against tuberculosis. J. Leukoc. Biol. 2007, 82, 934–945. [Google Scholar] [CrossRef] [PubMed]
- Weiss, J.; Olsson, I. Cellular and subcellular localization of the bactericidal/permeability-increasing protein of neutrophils. Blood 1987, 69, 652–659. [Google Scholar] [PubMed]
- Calafat, J.; Janssen, H.; Tool, A.; Dentener, M.A.; Knol, E.F.; Rosenberg, H.F.; Egesten, A. The bactericidal/permeability-increasing protein (bpi) is present in specific granules of human eosinophils. Blood 1998, 91, 4770–4775. [Google Scholar] [PubMed]
- Peschel, A.; Otto, M.; Jack, R.W.; Kalbacher, H.; Jung, G.; Gotz, F. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 1999, 274, 8405–8410. [Google Scholar] [CrossRef] [PubMed]
- Abi Khattar, Z.; Rejasse, A.; Destoumieux-Garzon, D.; Escoubas, J.M.; Sanchis, V.; Lereclus, D.; Givaudan, A.; Kallassy, M.; Nielsen-Leroux, C.; Gaudriault, S. The dlt operon of Bacillus cereus is required for resistance to cationic antimicrobial peptides and for virulence in insects. J. Bacteriol. 2009, 191, 7063–7073. [Google Scholar] [CrossRef] [PubMed]
- Roy, H. Tuning the properties of the bacterial membrane with aminoacylated phosphatidylglycerol. IUBMB Life 2009, 61, 940–953. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Hankins, J.V.; Madsen, J.A.; Giles, D.K.; Childers, B.M.; Klose, K.E.; Brodbelt, J.S.; Trent, M.S. Elucidation of a novel Vibrio cholerae lipid a secondary hydroxy-acyltransferase and its role in innate immune recognition. Mol. Microbiol. 2011, 81, 1313–1329. [Google Scholar] [CrossRef] [PubMed]
- Phillips, N.J.; Adin, D.M.; Stabb, E.V.; McFall-Ngai, M.J.; Apicella, M.A.; Gibson, B.W. The lipid a from Vibrio fischeri lipopolysaccharide: A unique structure bearing a phosphoglycerol moiety. J. Biol. Chem. 2011, 286, 21203–21219. [Google Scholar] [CrossRef] [PubMed]
- Hankins, J.V.; Madsen, J.A.; Giles, D.K.; Brodbelt, J.S.; Trent, M.S. Amino acid addition to Vibrio cholerae lps establishes a link between surface remodeling in Gram-positive and Gram-negative bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 8722–8727. [Google Scholar] [CrossRef] [PubMed]
- Guilhelmelli, F.; Vilela, N.; Albuquerque, P.; Derengowski Lda, S.; Silva-Pereira, I.; Kyaw, C.M. Antibiotic development challenges: The various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front. Microbiol. 2013, 4, e353. [Google Scholar] [CrossRef]
- Needham, B.D.; Trent, M.S. Fortifying the barrier: The impact of lipid a remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 2013, 11, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Matson, J.S.; Yoo, H.J.; Hakansson, K.; Dirita, V.J. Polymyxin B resistance in El Tor Vibrio cholerae requires lipid acylation catalyzed by msbb. J. Bacteriol. 2010, 192, 2044–2052. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Lim, K.B.; Gunn, J.S.; Bainbridge, B.; Darveau, R.P.; Hackett, M.; Miller, S.I. Regulation of lipid a modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 1997, 276, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Dalebroux, Z.D.; Miller, S.I. Salmonellae phopq regulation of the outer membrane to resist innate immunity. Curr. Opin. Microbiol. 2014, 17, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Bashyam, M.D.; Hasnain, S.E. The extracytoplasmic function sigma factors: Role in bacterial pathogenesis. Infect. Genet. Evol. 2004, 4, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.M.; Waldor, M.K. High-throughput sequencing reveals suppressors of Vibrio cholerae rpoe mutations: One fewer porin is enough. Nucleic Acids Res. 2009, 37, 5757–5767. [Google Scholar] [CrossRef] [PubMed]
- Mathur, J.; Davis, B.M.; Waldor, M.K. Antimicrobial peptides activate the Vibrio cholerae sigmae regulon through an OmpU-dependent signalling pathway. Mol. Microbiol. 2007, 63, 848–858. [Google Scholar] [PubMed]
- Mathur, J.; Waldor, M.K. The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect. Immun. 2004, 72, 3577–3583. [Google Scholar] [CrossRef] [PubMed]
- Duperthuy, M.; Binesse, J.; Le Roux, F.; Romestand, B.; Caro, A.; Got, P.; Givaudan, A.; Mazel, D.; Bachere, E.; Destoumieux-Garzon, D. The major outer membrane protein ompu of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ. Microbiol. 2010, 12, 951–963. [Google Scholar] [CrossRef] [PubMed]
- Beveridge, T.J. Structures of Gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 1999, 181, 4725–4733. [Google Scholar] [PubMed]
- Wai, S.N.; Takade, A.; Amako, K. The release of outer membrane vesicles from the strains of enterotoxigenic Escherichia coli. Microbiol. Immunol. 1995, 39, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Kulp, A.; Kuehn, M.J. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010, 64, 163–184. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.N.; Das, J. Electron microscopic observations on the excretion of cell-wall material by Vibrio cholerae. J. Gen. Microbiol. 1967, 49, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.E.; Kim, D.G.; Park, E.M.; Nam, B.H.; Kim, Y.O.; Kong, I.S. Identification of Vibrio anguillarum outer membrane vesicles related to immunostimulation in the Japanese flounder, Paralichthys olivaceus. Biosci. Biotech. Biochem. 2009, 73, 437–439. [Google Scholar] [CrossRef]
- Kim, Y.R.; Kim, B.U.; Kim, S.Y.; Kim, C.M.; Na, H.S.; Koh, J.T.; Choy, H.E.; Rhee, J.H.; Lee, S.E. Outer membrane vesicles of Vibrio vulnificus deliver cytolysin-hemolysin VvhA into epithelial cells to induce cytotoxicity. Biochem. Biophys. Res. Commun. 2010, 399, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Manning, A.J.; Kuehn, M.J. Functional advantages conferred by extracellular prokaryotic membrane vesicles. J. Mol. Microbiol. Biotechnol. 2013, 23, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Duperthuy, M.; Sjostrom, A.E.; Sabharwal, D.; Damghani, F.; Uhlin, B.E.; Wai, S.N. Role of the Vibrio cholerae matrix protein bap1 in cross-resistance to antimicrobial peptides. PLoS Pathog. 2013, 9, e1003620. [Google Scholar] [CrossRef] [PubMed]
- Vanhove, A.S.; Duperthuy, M.; Charriere, G.M.; Le Roux, F.; Goudenege, D.; Gourbal, B.; Kieffer-Jaquinod, S.; Coute, Y.; Wai, S.N.; Destoumieux-Garzon, D. Outer membrane vesicles are vehicles for the delivery of Vibrio tasmaniensis virulence factors to oyster immune cells. Environ. Microbiol. 2014. [Google Scholar] [CrossRef]
- Song, T.; Mika, F.; Lindmark, B.; Liu, Z.; Schild, S.; Bishop, A.; Zhu, J.; Camilli, A.; Johansson, J.; Vogel, J.; et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 2008, 70, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H.; Pages, J.M. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 2012, 36, 340–363. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J. Multidrug-resistance efflux pumps—Not just for resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Putman, M.; van Veen, H.W.; Konings, W.N. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 2000, 64, 672–693. [Google Scholar] [CrossRef] [PubMed]
- Hinchliffe, P.; Symmons, M.F.; Hughes, C.; Koronakis, V. Structure and operation of bacterial tripartite pumps. Annu. Rev. Microbiol. 2013, 67, 221–242. [Google Scholar] [CrossRef] [PubMed]
- Bina, J.E.; Mekalanos, J.J. Vibrio cholerae tolc is required for bile resistance and colonization. Infect. Immun. 2001, 69, 4681–4685. [Google Scholar] [CrossRef] [PubMed]
- Bina, X.R.; Provenzano, D.; Nguyen, N.; Bina, J.E. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect. Immun. 2008, 76, 3595–3605. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H.; Zgurskaya, H.I. AcrAB and related multidrug efflux pumps of Escherichia coli. J. Mol. Microbiol. Biotechnol. 2001, 3, 215–218. [Google Scholar] [PubMed]
- Buckley, A.M.; Webber, M.A.; Cooles, S.; Randall, L.P.; La Ragione, R.M.; Woodward, M.J.; Piddock, L.J. The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol. 2006, 8, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 2001, 3, 255–264. [Google Scholar] [PubMed]
- Kitaoka, M.; Miyata, S.T.; Unterweger, D.; Pukatzki, S. Antibiotic resistance mechanisms of Vibrio cholerae. J. Med. Microbiol. 2011, 60, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Bina, J.E.; Provenzano, D.; Wang, C.; Bina, X.R.; Mekalanos, J.J. Characterization of the Vibrio cholerae vexab and vexcd efflux systems. Arch. Microbiol. 2006, 186, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.L.; Bina, X.R.; Bina, J.E. Vibrio cholerae vexh encodes a multiple drug efflux pump that contributes to the production of cholera toxin and the toxin co-regulated pilus. PLoS One 2012, 7, e38208. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.J.; Kuo, T.Y.; Lin, C.C.; Chow, L.P.; Chen, W.J. Proteomic identification of membrane proteins regulating antimicrobial peptide resistance in Vibrio parahaemolyticus. J. Appl. Microbiol. 2010, 108, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Chuang, Y.C.; Chang, C.C.; Jeang, C.L.; Chang, M.C. A K+ uptake protein, TrkA, is required for serum, protamine, and polymyxin B resistance in Vibrio vulnificus. Infect. Immun. 2004, 72, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Bossemeyer, D.; Borchard, A.; Dosch, D.C.; Helmer, G.C.; Epstein, W.; Booth, I.R.; Bakker, E.P. K+-transport protein TrkA of Escherichia coli is a peripheral membrane protein that requires other trk gene products for attachment to the cytoplasmic membrane. J. Biol. Chem. 1989, 264, 16403–16410. [Google Scholar] [PubMed]
- Chakraborty, K.; Ghosh, S.; Koley, H.; Mukhopadhyay, A.K.; Ramamurthy, T.; Saha, D.R.; Mukhopadhyay, D.; Roychowdhury, S.; Hamabata, T.; Takeda, Y.; et al. Bacterial exotoxins downregulate cathelicidin (hCAP-18/LL-37) and human beta-defensin 1 (HBD-1) expression in the intestinal epithelial cells. Cell. Microbiol. 2008, 10, 2520–2537. [Google Scholar] [CrossRef] [PubMed]
- Shirin, T.; Rahman, A.; Danielsson, A.; Uddin, T.; Bhuyian, T.R.; Sheikh, A.; Qadri, S.S.; Qadri, F.; Hammarstrom, M.L. Antimicrobial peptides in the duodenum at the acute and convalescent stages in patients with diarrhea due to Vibrio cholerae o1 or enterotoxigenic Escherichia coli infection. Microbes Infect. 2011, 13, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Dupiol, J.; Dheilly, N.M.; Rondon, R.; Grunau, C.; Cosseau, C.; Smith, K.M.; Freitag, M.; Adjeroud, M.; Mitta, G. Thermal stress triggers broad Pocillopora damicornis transcriptomic remodeling, while Vibrio coralliilyticus infection induces a more targeted immuno-suppression response. PLoS One 2014. [Google Scholar] [CrossRef]
- Venier, P.; Varotto, L.; Rosani, U.; Millino, C.; Celegato, B.; Bernante, F.; Lanfranchi, G.; Novoa, B.; Roch, P.; Figueras, A.; et al. Insights into the innate immunity of the mediterranean mussel Mytilus galloprovincialis. BMC Genomics 2011, 12, e69. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Destoumieux-Garzón, D.; Duperthuy, M.; Vanhove, A.S.; Schmitt, P.; Wai, S.N. Resistance to Antimicrobial Peptides in Vibrios. Antibiotics 2014, 3, 540-563. https://doi.org/10.3390/antibiotics3040540
Destoumieux-Garzón D, Duperthuy M, Vanhove AS, Schmitt P, Wai SN. Resistance to Antimicrobial Peptides in Vibrios. Antibiotics. 2014; 3(4):540-563. https://doi.org/10.3390/antibiotics3040540
Chicago/Turabian StyleDestoumieux-Garzón, Delphine, Marylise Duperthuy, Audrey Sophie Vanhove, Paulina Schmitt, and Sun Nyunt Wai. 2014. "Resistance to Antimicrobial Peptides in Vibrios" Antibiotics 3, no. 4: 540-563. https://doi.org/10.3390/antibiotics3040540
APA StyleDestoumieux-Garzón, D., Duperthuy, M., Vanhove, A. S., Schmitt, P., & Wai, S. N. (2014). Resistance to Antimicrobial Peptides in Vibrios. Antibiotics, 3(4), 540-563. https://doi.org/10.3390/antibiotics3040540




