Antimicrobial Activity of Chemokine CXCL10 for Dermal and Oral Microorganisms
Abstract
:1. Introduction
2. Results
2.1. Activity of CXCL10 on Microorganisms Commonly Found on the Skin
| Microorganism | CXCL10 µg/mL MIC (Standard Error) | CXCL10 µg/mL MBC (Standard Error) | SMAP28 µg/mL MIC (Standard Error) | SMAP28 µg/mL MBC (Standard Error) |
|---|---|---|---|---|
| S. aureus | >50.00 | >50.00 | 4.17 (1.04) | 6.25 (0.00) |
| E. coli | >50.00 | >50.00 | 3.13 (0.00) | 3.13 (0.00) |
| C. bovis | >50.00 | >50.00 | 6.25 (0.00) | 6.25 (0.00) |
| C. striatum | 5.21 (1.04) | 25.00 (0.00) | 0.07 (0.01) | 0.07 (0.01) |
| C. jeikeium | >50.00 | >50.00 | 0.31 (0.00) | 0.31 (0.00) |
| Microorganism | CXCL10 µg/mL MIC (Standard Error) | SMAP28 µg/mL MIC (Standard Error) |
|---|---|---|
| S. aureus | 125.89 (0.00) | 10.49 (1.66) |
| E. coli | 11.62 (4.23) | 3.69 (0.75) |
| C. bovis | >200.00 | 17.24 (0.43) |
| C. striatum | 3.42 (1.55) | 8.00 (1.62) |
| C. jeikeium | 21.52 (0.23) | 4.44 (0.26) |
2.2. Activity of CXCL10 on Microorganisms Commonly Found in the Oral Cavity
| Microorganism | CXCL10 µg/mL MIC (Standard Error) | SMAP28 µg/mL MIC (Standard Error) |
|---|---|---|
| S. mutans | >200.00 | 40.51 (2.70) |
| S. mitis | >200.00 | 125.89 (0.00) |
| S. sanguinis | >200.00 | 61.55 (2.80) |
| F. nucleatum | >1000.00 | 39.47 (5.62) |
| P. gingivalis 381 | >1000.00 | 69.70 (22.74) |
| P. gingivalis ATCC 33277 | >1000.00 | 74.42 (11.32) |
| A. actinomycetemcomitans | >1000.00 | 26.18 (3.81) |
2.3. Activity of CXCL10 on C. albicans Commonly Found on the Skin and in the Oral Cavity
| Microorganism | CXCL10 µg/mL MIC (Standard Error) | CXCL10 µg/mL MBC (Standard Error) | SMAP28 µg/mL MIC (Standard Error) | SMAP28 µg/mL MBC (Standard Error) |
|---|---|---|---|---|
| Broth microdilution assay | ||||
| C. albicans ATCC 64124 | >50.00 | >50.00 | 12.50 (0.00) | 12.50 (0.00) |
| C. albicans HMV4C | >50.00 | >50.00 | 12.50 (0.00) | 16.67 (4.17) |
| Radial diffusion assay | ||||
| C. albicans ATCC 64124 | >1,000.00 | n/a | 39.73 (18.60) | n/a |
| C. albicans HMV4C | 23.90 (10.38) | n/a | 18.90 (2.13) | n/a |
2.4. Proposed Structure of CXCL10
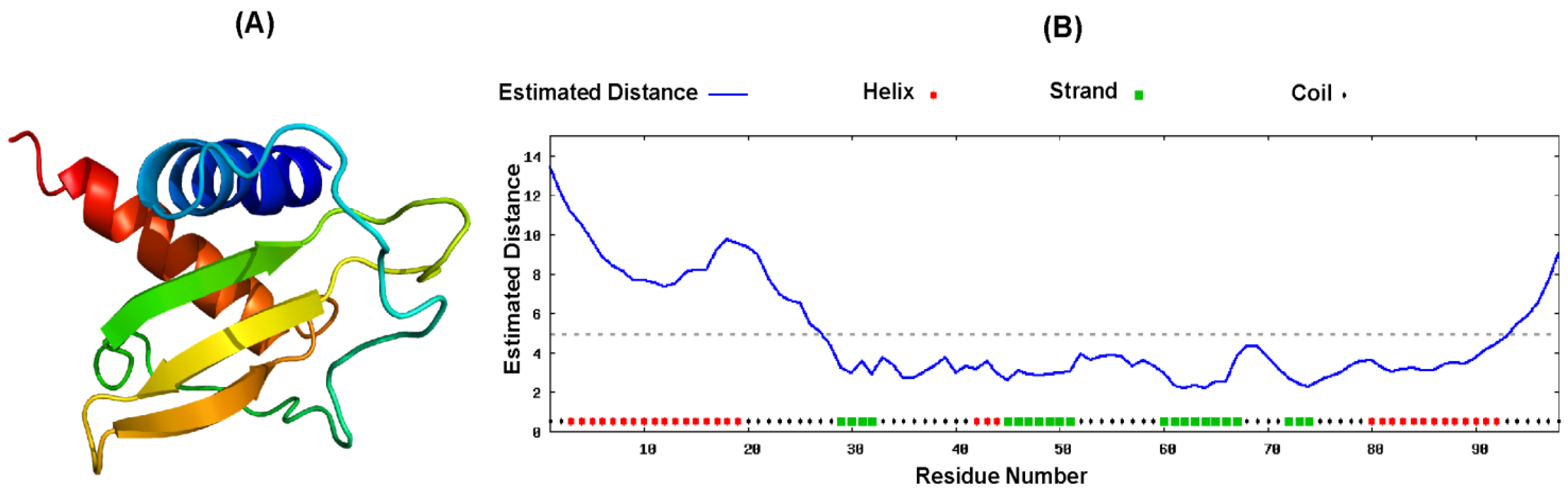
3. Discussion
4. Experimental Section
4.1. Microorganisms and Culture Conditions
4.2. Chemokine and Antimicrobial Peptide
4.3. Broth Microdilution Assay
4.4. Radial Diffusion Assay
4.5. Viable Plate Count Assay
4.6. Modeling the Structure of CXCL10
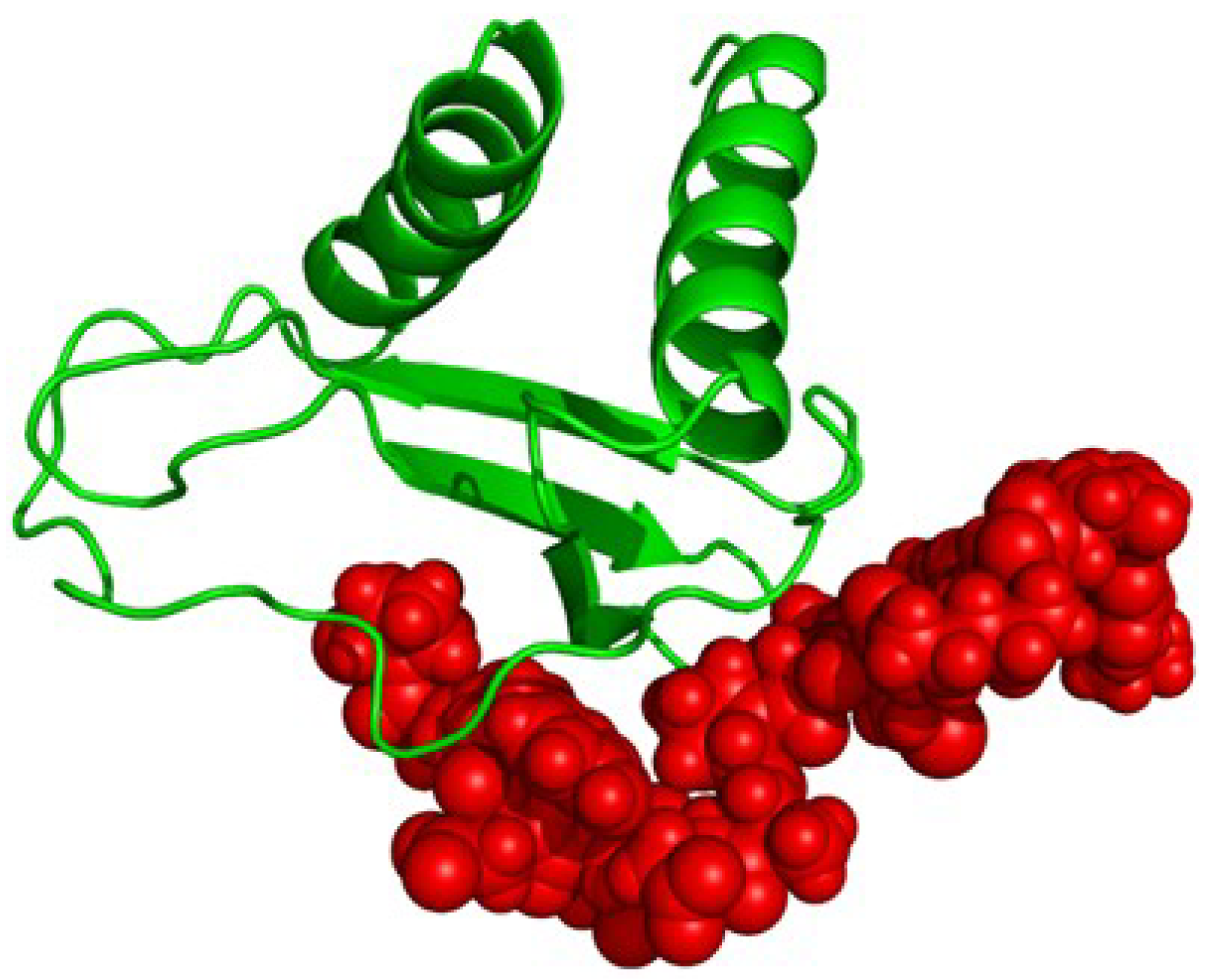
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zlotnik, A.; Yoshie, O. Chemokines: A new classification system and their role in immunity. Immunity 2000, 12, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Chen, Q.; Hoover, D.M.; Staley, P.; Tucker, K.D.; Lubkowski, J.; Oppenheim, J.J. Many chemokines including CCL20/MIP-3alpha display antimicrobial activity. J. Leukoc. Biol. 2003, 74, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Luster, A.D.; Unkeless, J.C.; Ravetch, J.V. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature 1985, 315, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Dyer, K.D.; Percopo, C.M.; Fischer, E.R.; Gabryszewski, S.J.; Rosenberg, H.F. Pneumoviruses infect eosinophils and elicit MyD88-dependent release of chemoattractant cytokines and interleukin-6. Blood 2009, 114, 2649–2656. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Guo, S.; Stiles, J.K. The emerging role of CXCL10 in cancer (Review). Oncol. Lett. 2011, 2, 583–589. [Google Scholar] [PubMed]
- Bonecchi, R.; Bianchi, G.; Bordignon, P.P.; D’Ambrosio, D.; Lang, R.; Borsatti, A.; Sozzani, S.; Allavena, P.; Gray, P.A.; Mantovani, A.; et al. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J. Exp. Med. 1998, 187, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Guo, S.; Hibbert, J.M.; Jain, V.; Singh, N.; Wilson, N.O.; Stiles, J.K. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. 2011, 22, 121–130. [Google Scholar] [PubMed]
- Goebeler, M.; Toksoy, A.; Spandau, U.; Engelhardt, E.; Brocker, E.B.; Gillitzer, R. The C-X-C chemokine Mig is highly expressed in the papillae of psoriatic lesions. J. Pathol. 1998, 184, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Lee, Z.H.; Song, Y.W. CXCL10 and autoimmune diseases. Autoimmun. Rev. 2009, 8, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Flier, J.; Boorsma, D.M.; van Beek, P.J.; Nieboer, C.; Stoof, T.J.; Willemze, R.; Tensen, C.P. Differential expression of CXCR3 targeting chemokines CXCL10, CXCL9, and CXCL11 in different types of skin inflammation. J. Pathol. 2001, 194, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Belperio, J.A.; Keane, M.P.; Arenberg, D.A.; Addison, C.L.; Ehlert, J.E.; Burdick, M.D.; Strieter, R.M. CXC chemokines in angiogenesis. J. Leukoc. Biol. 2000, 68, 1–8. [Google Scholar] [PubMed]
- Cole, A.M.; Ganz, T.; Liese, A.M.; Burdick, M.D.; Liu, L.; Strieter, R.M. Cutting edge: IFN-inducible ELR-CXC chemokines display defensin-like antimicrobial activity. J. Immunol. 2001, 167, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Egesten, A.; Eliasson, M.; Johansson, H.M.; Olin, A.I.; Morgelin, M.; Mueller, A.; Pease, J.E.; Frick, I.M.; Bjorck, L. The CXC chemokine MIG/CXCL9 is important in innate immunity against Streptococcus pyogenes. J. Infect. Dis. 2007, 195, 684–693. [Google Scholar] [CrossRef] [PubMed]
- Crawford, M.A.; Lowe, D.E.; Fisher, D.J.; Stibitz, S.; Plaut, R.D.; Beaber, J.W.; Zemansky, J.; Mehrad, B.; Glomski, I.J.; Strieter, R.M.; et al. Identification of the bacterial protein FtsX as a unique target of chemokine-mediated antimicrobial activity against Bacillus anthracis. Proc. Natl. Acad. Sci. USA 2011, 108, 17159–17164. [Google Scholar] [CrossRef] [PubMed]
- Crawford, M.A.; Burdick, M.D.; Glomski, I.J.; Boyer, A.E.; Barr, J.R.; Mehrad, B.; Strieter, R.M.; Hughes, M.A. Interferon-inducible CXC chemokines directly contribute to host defense against inhalational anthrax in a murine model of infection. PLoS Pathog. 2010, 6, e1001199. [Google Scholar] [CrossRef] [PubMed]
- Tymkiw, K.D.; Thunell, D.H.; Johnson, G.K.; Joly, S.; Burnell, K.K.; Cavanaugh, J.E.; Brogden, K.A.; Guthmiller, J.M. Influence of smoking on gingival crevicular fluid cytokines in severe chronic periodontitis. J. Clin. Periodontol. 2011, 38, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Thunell, D.H.; Tymkiw, K.D.; Johnson, G.K.; Joly, S.; Burnell, K.K.; Cavanaugh, J.E.; Brogden, K.A.; Guthmiller, J.M. A multiplex immunoassay demonstrates reductions in gingival crevicular fluid cytokines following initial periodontal therapy. J. Periodontal. Res. 2010, 45, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Holdren, G.O.; Rosenthal, D.J.; Yang, J.; Bates, A.M.; Fischer, D.L.; Zhang, Y.; Brogden, N.K.; Brogden, K.A.; Dows Institute for Dental Research, College of Dentistry, The University of Iowa, Iowa City, IA, USA. Unpublished data set 1. 2014.
- Holdren, G.O.; Rosenthal, D.J.; Yang, J.; Bates, A.M.; Fischer, D.L.; Zhang, Y.; Brogden, N.K.; Brogden, K.A.; Dows Institute for Dental Research, College of Dentistry, The University of Iowa, Iowa City, IA, USA. Unpublished data set 2. 2014.
- Holdren, G.O.; Rosenthal, D.J.; Yang, J.; Bates, A.M.; Fischer, D.L.; Zhang, Y.; Brogden, N.K.; Brogden, K.A.; Dows Institute for Dental Research, College of Dentistry, The University of Iowa, Iowa City, IA, USA. Unpublished data set 3. 2014.
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Brogden, N.K.; Mehalick, L.; Fischer, C.L.; Wertz, P.W.; Brogden, K.A. The emerging role of peptides and lipids as antimicrobial epidermal barriers and modulators of local inflammation. Skin Pharmacol. Physiol. 2012, 25, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Gorr, S.U. Antimicrobial peptides of the oral cavity. Periodontol. 2000 2009, 51, 152–180. [Google Scholar] [CrossRef] [PubMed]
- Gorr, S.U. Antimicrobial peptides in periodontal innate defense. Front. Oral Biol. 2012, 15, 84–98. [Google Scholar] [PubMed]
- Gorr, S.U.; Abdolhosseini, M. Antimicrobial peptides and periodontal disease. J. Clin. Periodontol. 2011, 38, S126–S141. [Google Scholar] [CrossRef]
- Holdren, G.O.; Fischer, D.L.; Brogden, K.A.; Brogden, N.K.; Division of Pharmaceutics and Translational Therapeutics, Department of Pharmaceutical Sciences and Experimental Therapeutics, College of Pharmacy, The University of Iowa: Iowa City, IA, USA. Unpublished data set 4. 2014.
- Khan, A. Detection and quantitation of forty eight cytokines, chemokines, growth factors and nine acute phase proteins in healthy human plasma, saliva and urine. J. Proteomics 2012, 75, 4802–4819. [Google Scholar] [CrossRef] [PubMed]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef] [PubMed]
- Paster, B.J.; Boches, S.K.; Galvin, J.L.; Ericson, R.E.; Lau, C.N.; Levanos, V.A.; Sahasrabudhe, A.; Dewhirst, F.E. Bacterial diversity in human subgingival plaque. J. Bacteriol. 2001, 183, 3770–3783. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Schloss, P.D. Dynamics and associations of microbial community types across the human body. Nature 2014, 509, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Jorth, P.; Turner, K.H.; Gumus, P.; Nizam, N.; Buduneli, N.; Whiteley, M. Metatranscriptomics of the human oral microbiome during health and disease. MBio 2014, 5, e01012–e01014. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.L.; Drake, D.R.; Dawson, D.V.; Blanchette, D.R.; Brogden, K.A.; Wertz, P.W. Antibacterial activity of sphingoid bases and fatty acids against Gram-positive and Gram-negative bacteria. Antimicrob. Agents Chemother. 2012, 56, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Fischer, C.L.; Walters, K.S.; Drake, D.R.; Dawson, D.V.; Blanchette, D.R.; Brogden, K.A.; Wertz, P.W. Oral mucosal lipids are antibacterial against Porphyromonas gingivalis, induce ultrastructural damage, and alter bacterial lipid and protein compositions. Int. J. Oral. Sci. 2013, 5, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A.; Nordholm, G.; Ackermann, M. Antimicrobial activity of cathelicidins BMAP28, SMAP28, SMAP29, and PMAP23 against Pasteurella multocida is more broad-spectrum than host species specific. Vet. Microbiol. 2007, 119, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Joly, S.; Maze, C.; McCray, P.B., Jr.; Guthmiller, J.M. Human beta-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J. Clin. Microbiol. 2004, 42, 1024–1029. [Google Scholar] [CrossRef] [PubMed]
- Brogden, N.K.; Brogden, K.A. Will new generations of modified antimicrobial peptides improve their potential as pharmaceuticals? Int. J. Antimicrob. Agents 2011, 38, 217–225. [Google Scholar]
- Brogden, K.A.; Kalfa, V.C.; Ackermann, M.R.; Palmquist, D.E.; McCray, P.B., Jr.; Tack, B.F. The ovine cathelicidin SMAP29 kills ovine respiratory pathogens in vitro and in an ovine model of pulmonary infection. Antimicrob. Agents Chemother. 2001, 45, 331–334. [Google Scholar] [CrossRef] [PubMed]
- Repp, K.K.; Menor, S.A.; Pettit, R.K. Microplate Alamar blue assay for susceptibility testing of Candida albicans biofilms. Med. Mycol. 2007, 45, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Borgwardt, D.S.; Martin, A.; van Hemert, J.R.; Yang, J.; Fischer, C.L.; Recker, E.N.; Nair, P.R.; Vidva, R.; Chandrashekaraiah, S.; Progulske-Fox, A.; et al. Histatin 5 binds to Porphyromonas gingivalis hemagglutinin B (HagB) and alters HagB-induced chemokine responses. Sci. Rep. 2014. [Google Scholar] [CrossRef]
- Yang, J.; Roy, A.; Zhang, Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 2013, 29, 2588–2595. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Roy, A.; Zhang, Y. BioLiP: A semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic Acids Res. 2013, 41, D1096–D1103. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Holdren, G.O.; Rosenthal, D.J.; Yang, J.; Bates, A.M.; Fischer, C.L.; Zhang, Y.; Brogden, N.K.; Brogden, K.A. Antimicrobial Activity of Chemokine CXCL10 for Dermal and Oral Microorganisms. Antibiotics 2014, 3, 527-539. https://doi.org/10.3390/antibiotics3040527
Holdren GO, Rosenthal DJ, Yang J, Bates AM, Fischer CL, Zhang Y, Brogden NK, Brogden KA. Antimicrobial Activity of Chemokine CXCL10 for Dermal and Oral Microorganisms. Antibiotics. 2014; 3(4):527-539. https://doi.org/10.3390/antibiotics3040527
Chicago/Turabian StyleHoldren, Grant O., David J. Rosenthal, Jianyi Yang, Amber M. Bates, Carol L. Fischer, Yang Zhang, Nicole K. Brogden, and Kim A. Brogden. 2014. "Antimicrobial Activity of Chemokine CXCL10 for Dermal and Oral Microorganisms" Antibiotics 3, no. 4: 527-539. https://doi.org/10.3390/antibiotics3040527
APA StyleHoldren, G. O., Rosenthal, D. J., Yang, J., Bates, A. M., Fischer, C. L., Zhang, Y., Brogden, N. K., & Brogden, K. A. (2014). Antimicrobial Activity of Chemokine CXCL10 for Dermal and Oral Microorganisms. Antibiotics, 3(4), 527-539. https://doi.org/10.3390/antibiotics3040527





