Phage Resistance Modulates Escherichia coli B Response to Metal-Based Antimicrobials
Abstract
1. Introduction
2. Results
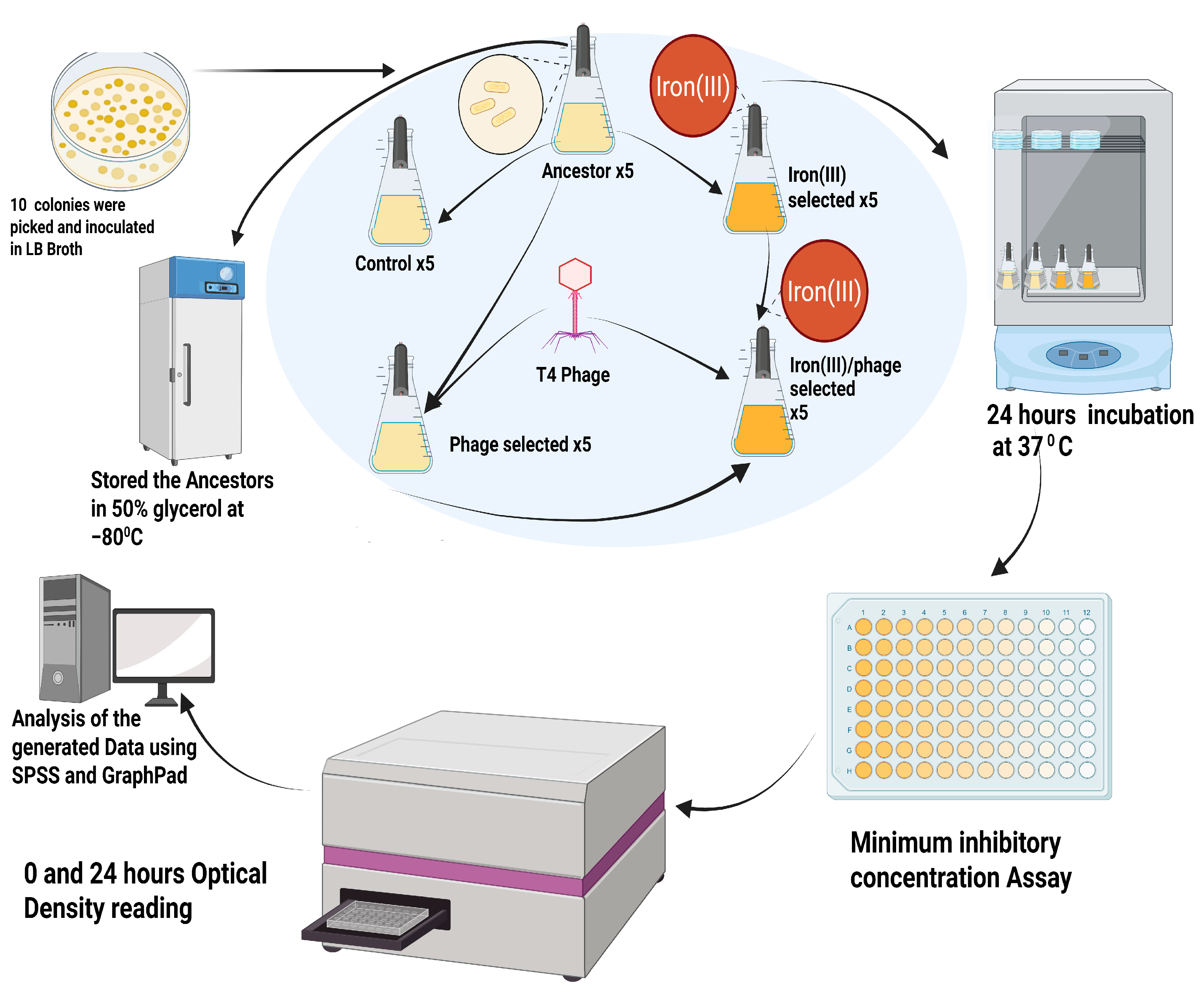
2.1. Experimental Evolution Design
- Control: Serially passaged in LB broth;
- Phage-selected: Exposed to T4 phage in LB broth daily;
- Iron (III)-selected: Cultured in LB broth with 1500 mg/L iron (III) sulfate;
- Phage/iron (III)-selected: First evolved for phage resistance in LB with T4 phage, then transferred to LB broth with 1500 mg/L iron (III) sulfate. All populations were transferred daily at a 1:100 dilution. Populations were archived and analyzed via phenotypic assays and whole-genome sequencing after 35 days of serial transfer.
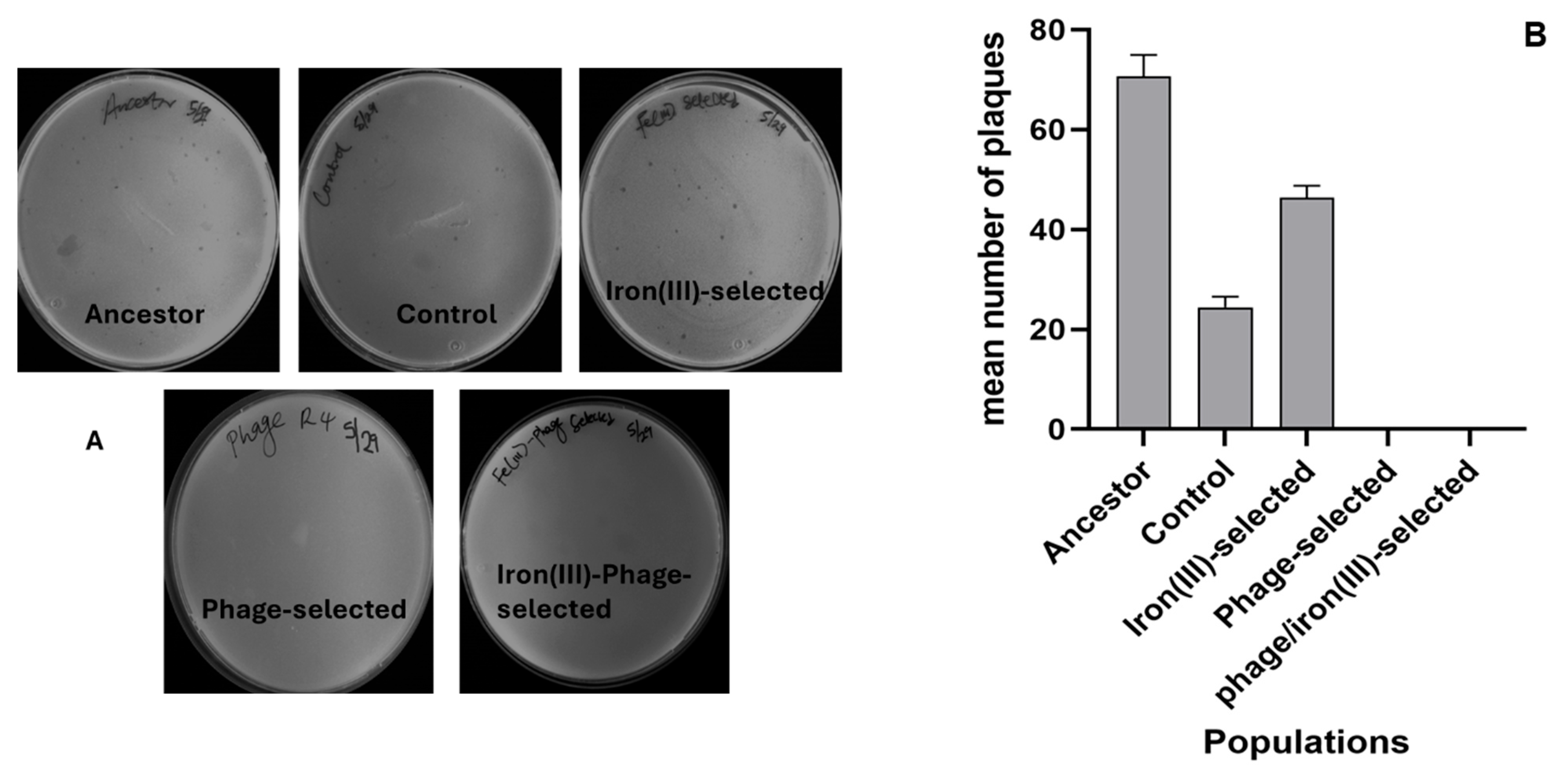
2.2. T4 Phage Resistance
2.3. Dual Resistance to Iron (III) Sulfate and T4 Phage
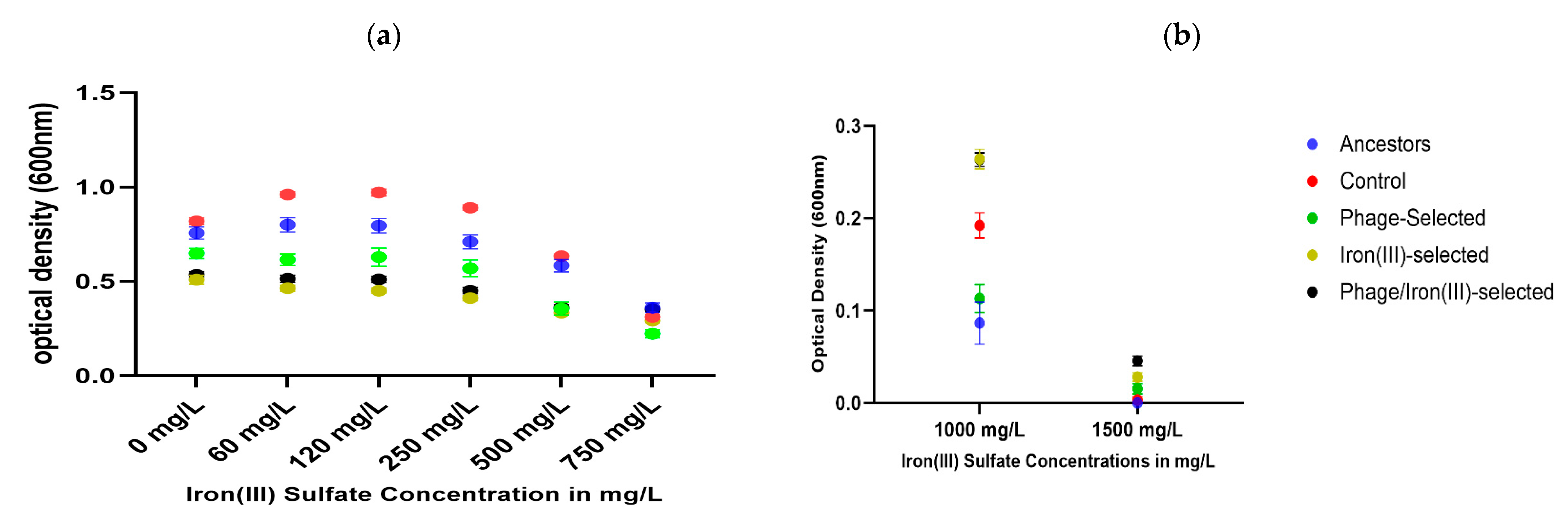
2.4. Iron (III) Sulfate Tolerance
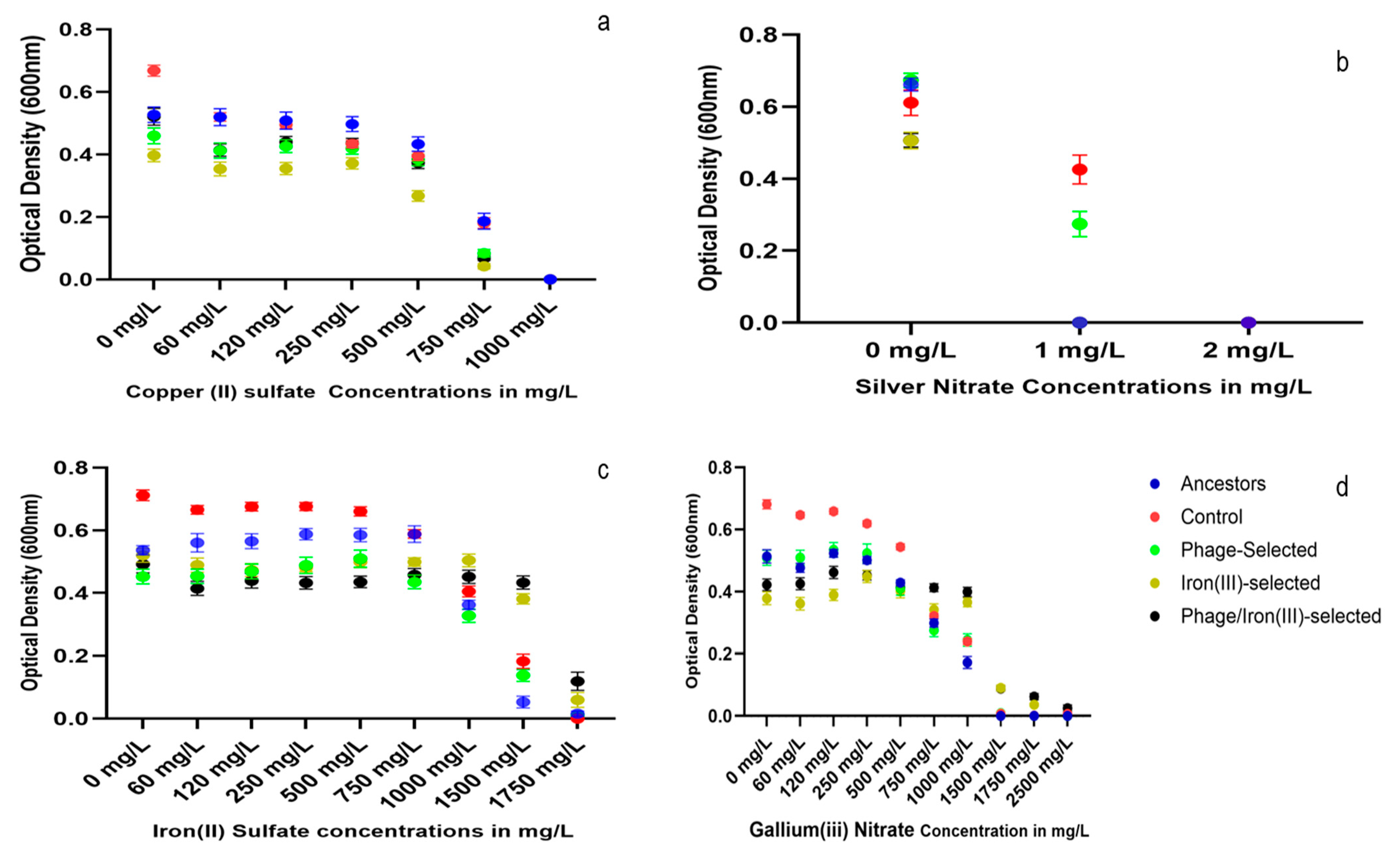
2.5. Heavy Metal Resistance
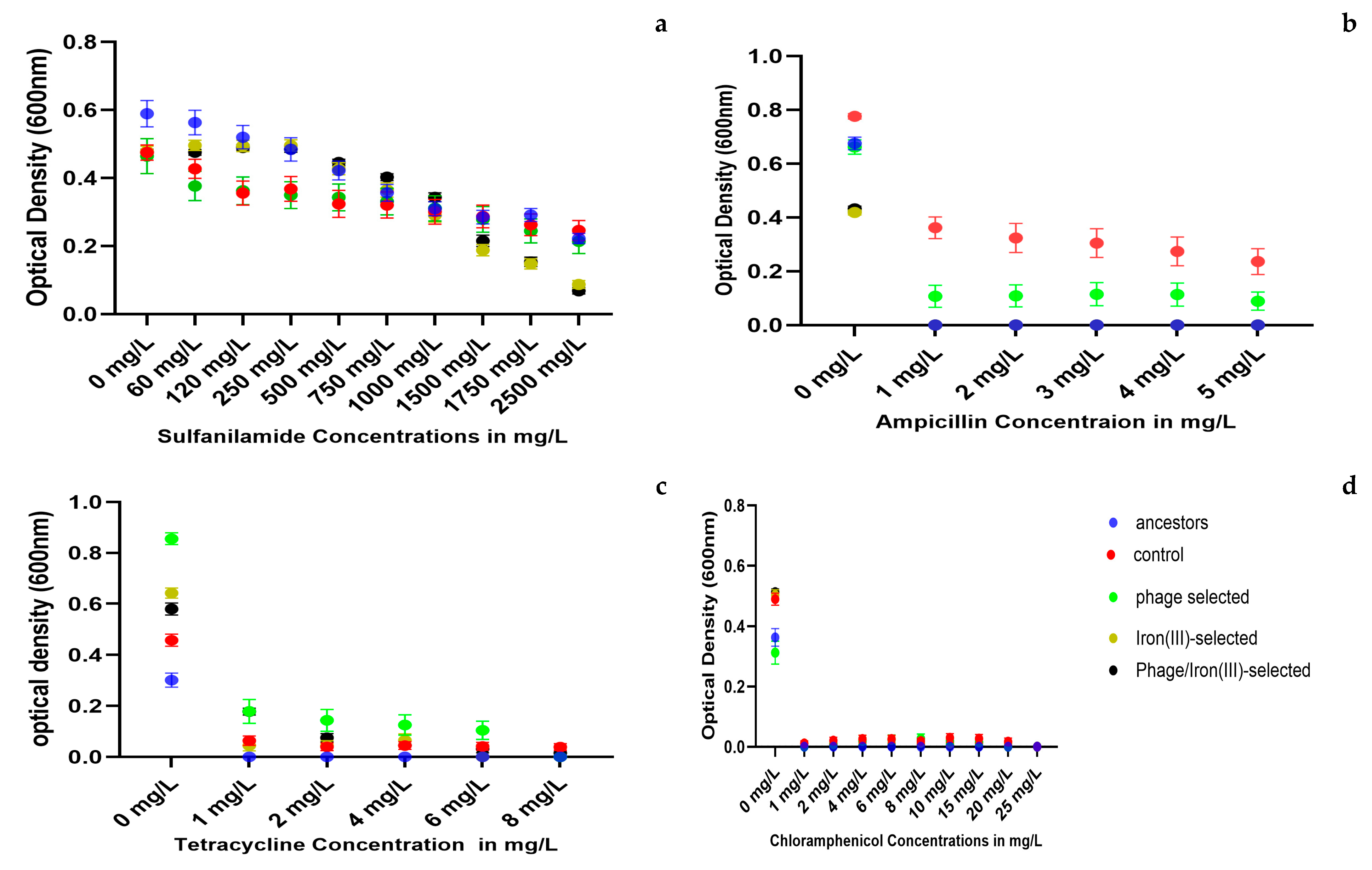
2.6. Antibiotic Resistance
2.7. Statistical Analysis of Growth Responses to Metals and Antibiotics
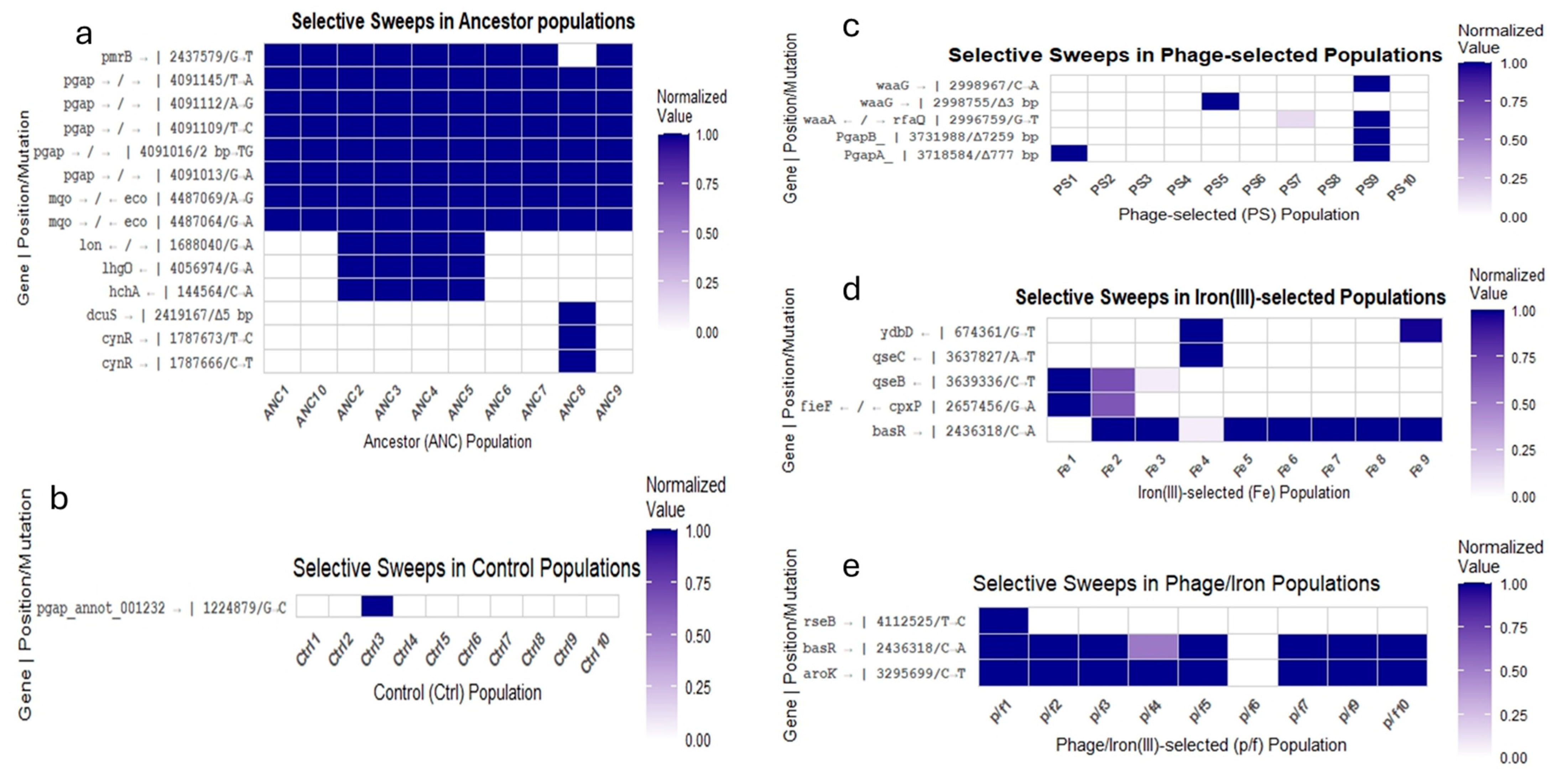
2.8. Genomic Analysis
3. Discussion
3.1. Phage Resistance and Sequence of Selection
3.2. Iron (III) Tolerance and Growth Trade-Offs
3.3. Cross-Resistance and Pleiotropy in Metals and Antibiotics
3.4. Genomic Signatures of Adaptation
3.5. Fitness Epistasis and Evolutionary Trajectories
4. Materials and Methods
4.1. Media Conditions and Experimental Layout
Experimental Populations
4.2. Determination of Minimal Inhibitory Concentration (MIC) of Iron (III) Sulfate
4.3. Assessing T4-Phage Resistance E. coli B
4.4. Dual Resistance Assay: T4-Phage and Excess Iron (III)
4.5. Phenotypic Analysis
4.6. Genomic Sequencing
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial Resistance |
| p/f | Phage/iron |
| PS | Phage-selected |
| Ctrl | Control |
| ACN | Ancestor |
| MIC | Minimum Inhibitory Concentration |
| Fe III | Iron (III) |
| Fe II | Iron (II) |
| Ga III | Gallium (III) |
| Cu II | Copper (II) |
| Ag I | Silver Nitrate |
References
- Silver, S. Bacterial silver resistance: Molecular biology and uses and misuses of silver compounds. FEMS Microbiol. Rev. 2003, 27, 341. [Google Scholar] [CrossRef]
- Guerinot, M.L. Microbial iron transport. Annu. Rev. Microbiol. 1994, 48, 743–772. [Google Scholar] [CrossRef]
- Levin, B.R.; Stewart, F.M.; Chao, L. Resource-limited growth, competition, and predation: A model and experimental studies with bacteria and bacteriophage. Am. Nat. 1977, 111, 3–24. [Google Scholar] [CrossRef]
- Lenski, R.E.; Levin, B.R. Constraints on the Coevolution of Bacteria and Virulent Phage: A Model, Some Experiments, and Predictions for Natural Communities. Am. Nat. 1985, 125, 585–602. [Google Scholar] [CrossRef]
- Graves, J.L.; Ewunkem, A.J.; Ward, J.; Staley, C.; Thomas, M.D.; Rhinehardt, K.L.; Han, J.; Harrison, S.H. Experimental evolution of gallium resistance in Escherichia coli. Evol. Med. Public Health 2019, 2019, 169. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.D.; Ewunkem, A.J.; Boyd, S.; Williams, D.K.; Moore, A.; Rhinehardt, K.L.; Van Beveren, E.; Yang, B.; Tapia, A.; Han, J.; et al. Too much of a good thing. Evol. Med. Public Health 2021, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Jeje, O.; Ewunkem, A.J.; Jeffers-Francis, L.K.; Graves, J.L. Serving Two Masters: Effect of Escherichia coli Dual Resistance on Antibiotic Susceptibility. Antibiotics 2023, 12, 603. [Google Scholar] [CrossRef] [PubMed]
- Ewunkem, A.J.; Rodgers, L.; Campbell, D.; Staley, C.; Subedi, K.; Boyd, S.; Graves, J.L. Experimental Evolution of Magnetite Nanoparticle Resistance in Escherichia coli. Nanomaterials 2021, 11, 790. [Google Scholar] [CrossRef]
- Frenoy, A.; Bonhoeffer, S. Death and population dynamics affect mutation rate estimates and evolvability under stress in bacteria. PLoS Biol. 2018, 16, e2005056. [Google Scholar] [CrossRef]
- Braun, V.; Ijmm, I.J. Iron uptake mechanisms and their regulation in pathogenic bacteria. Int. J. Med. Microbiol. 2001, 291, 67–79. [Google Scholar] [CrossRef]
- Lacroix, R.A.; Sandberg, T.E.; O’brien, E.J.; Utrilla, J.; Ebrahim, A.; Guzman, G.I.; Szubin, R.; Palsson, B.O.; Feist, A.M. Use of Adaptive Laboratory Evolution to Discover Key Mutations Enabling Rapid Growth of Escherichia coli K-12 MG1655 on Glucose Minimal Medium. Appl. Environ. Microbiol. 2015, 81, 17. [Google Scholar] [CrossRef] [PubMed]
- Kutter, E.; Sulakvelidze, A. (Eds.) Bacteriophages: Biology and Applications; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Campbell, E.A.; Korzheva, N.; Mustaev, A.; Murakami, K.; Nair, S.; Goldfarb, A.; Darst, S.A. Structural Mechanism for Rifampicin Inhibition of Bacterial RNA Polymerase. Cell 2001, 104, 901–912. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163. [Google Scholar] [CrossRef]
- Deatherage, D.E.; Barrick, J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014, 1151, 165–188. [Google Scholar]
- Miller, E.S.; Kutter, E.; Mosig, G.; Arisaka, F.; Kunisawa, T.; Rüger, W. Bacteriophage T4 Genome. Microbiol. Mol. Biol. Rev. 2003, 67, 86. [Google Scholar] [CrossRef]
- Perry, E.B.; Barrick, J.E.; Bohannan, B.J.M. The Molecular and Genetic Basis of Repeatable Coevolution between Escherichia coli and Bacteriophage T3 in a Laboratory Microcosm. PLoS ONE 2015, 10, e0130639. [Google Scholar] [CrossRef]
- Soundararajan, M.; Von Bünau, R.; Oelschlaeger, T.A. K5 Capsule and Lipopolysaccharide Are Important in Resistance to T4 Phage Attack in Probiotic E. coli Strain Nissle 1917. Front. Microbiol. 2019, 10, 2783. [Google Scholar] [CrossRef]
- Bryant, J.A.; Sellars, L.E.; Busby, S.J.W.; Lee, D.J. Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res. 2014, 42, 11383. [Google Scholar] [CrossRef]
- Breland, E.J.; Zhang, E.W.; Bermudez, T.; Martinez, C.R.; Hadjifrangiskou, M. The Histidine Residue of QseC Is Required for Canonical Signaling between QseB and PmrB in Uropathogenic Escherichia coli. J. Bacteriol. 2017, 199, e00060-17. [Google Scholar] [CrossRef] [PubMed]
- Wollmann, P.; Zeth, K. The Structure of RseB: A Sensor in Periplasmic Stress Response of E. coli. J. Mol. Biol. 2007, 372, 927. [Google Scholar] [CrossRef] [PubMed]
- Grass, G.; Otto, M.; Fricke, B.; Haney, C.J.; Rensing, C.; Nies, D.H.; Munkelt, D. FieF (YiiP) from Escherichia coli mediates decreased cellular accumulation of iron and relieves iron stress. Arch. Microbiol. 2004, 183, 9. [Google Scholar] [CrossRef] [PubMed]
- Masi, M.; Pinet, E.; Pagès, J. Complex Response of the CpxAR Two-Component System to β-Lactams on Antibiotic Resistance and Envelope Homeostasis in Enterobacteriaceae. Antimicrob. Agents Chemother. 2020, 64, e00291-20. [Google Scholar] [CrossRef]
- Lenski, R.E.; Rose, M.R.; Simpson, S.C.; Tadler, S.C. Long Term Experimental Evolution-1991. Available online: https://www.jstor.org/stable/2462549?seq=1 (accessed on 3 April 2025).
- Studier, F.W.; Daegelen, P.; Lenski, R.E.; Maslov, S.; Kim, J.F. Understanding the Differences between Genome Sequences of Escherichia coli B Strains REL606 and BL21(DE3) and Comparison of the E. coli B and K-12 Genomes. J. Mol. Biol. 2009, 394, 653. [Google Scholar] [CrossRef] [PubMed]
- Cooper, R.A. Metabolism of methylglyoxal in microorganisms. Annu. Rev. Microbiol. 1984, 38, 49–68. [Google Scholar] [CrossRef]
- Wan, Y.; Ye, L.; Zheng, J.; Tang, Y.; Chan, E.W.; Chen, S. Starvation-induced mutagenesis in rhsC and ybfD genes extends bacterial tolerance to various stresses by boosting efflux function. Microbiol. Res. 2025, 295, 128106. [Google Scholar] [CrossRef]
- Magnusson, L.U.; Farewell, A.; Nyström, T. ppGpp: A global regulator in Escherichia coli. Trends Microbiol. 2005, 13, 236. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Garushyants, S.K.; Van Den Hurk, A.; Aparicio-Maldonado, C.; Kushwaha, S.K.; King, C.M.; Ou, Y.; Todeschini, T.C.; Clokie, M.R.J.; Millard, A.D.; et al. Bacterial defense systems exhibit synergistic anti-phage activity. Cell Host Microbe 2025, 32, 557. [Google Scholar] [CrossRef]
- Mcphee, J.B.; Bains, M.; Winsor, G.; Lewenza, S.; Kwasnicka, A.; Brazas, M.D.; Brinkman, F.S.L.; Hancock, R.E.W. Contribution of the PhoP-PhoQ and PmrA-PmrB Two-Component Regulatory Systems to Mg2+-Induced Gene Regulation in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 3995. [Google Scholar] [CrossRef]
- Kwon, O.; Kim, S.; Hahm, D.; Lee, S.Y.; Kim, P. Potential Application of the Recombinant Escherichia coli-Synthesized Heme as a Bioavailable Iron Source. J. Microbiol. Biotechnol. 2009, 19, 604–609. [Google Scholar] [CrossRef]
- Conrad, T.M.; Frazier, M.; Joyce, A.R.; Cho, B.; Knight, E.M.; Lewis, N.E.; Landick, R.; Palsson, B.Ø. RNA polymerase mutants found through adaptive evolution reprogram Escherichia coli for optimal growth in minimal media. Proc. Natl. Acad. Sci. USA 2010, 107, 20500–20505. [Google Scholar] [CrossRef]
- Cooke, K.; Browning, D.F.; Lee, D.J.; Blair, J.M.A.; Mcneill, H.E.; Huber, D.; Busby, S.J.W.; Bryant, J.A. Position effects on promoter activity in Escherichia colis and their consequences for antibiotic-resistance determinants. Biochem. Soc. Trans. 2019, 47, 839. [Google Scholar] [CrossRef] [PubMed]


| Population | Iron (III) | T4-Phage |
|---|---|---|
| Phage/iron (III)-selected | 2 | 1 |
| Phage | 3 | 2 |
| Iron (III)-selected | 1 | 3 |
| Control | 5 | 4 |
| Ancestor | 4 | 5 |
| Dilution Factor | Ancestor (CFU/mL) | Control (CFU/mL) | Phage-Selected (CFU/mL) | Iron (III)-Selected (CFU/mL) | Phage/Iron (III)-Selected (CFU/mL) |
|---|---|---|---|---|---|
| 10−1 | 0 | 0 | 0 | TMC | 1.77 × 104 ± 3.51 |
| 10−2 | 0 | 0 | 0 | 6.53 × 105 ± 15.37 | 2 × 104 ± 1 |
| 10−3 | 0 | 0 | 0 | 5.67 × 105 ± 1.53 | 0 |
| Fe(III) Sulfate Bonferroni’s Multiple Comparisons | 95% Confidence Interval | |||||
|---|---|---|---|---|---|---|
| (I) Population | (J) Population | Mean Difference (I-J) | Std. Error | Sig. | Lower Bound | Upper Bound |
| Ancestor | Control | −0.0864 * | 0.01145 | 0.000 | −0.1186 | −0.0542 |
| Phage | 0.1154 * | 0.01145 | 0.000 | 0.0832 | 0.1476 | |
| Fe(III)-selected | 0.1739 * | 0.01145 | 0.000 | 0.1417 | 0.2061 | |
| Phage/Iron (III)-selected | 0.1406 * | 0.01145 | 0.000 | 0.1084 | 0.1728 | |
| Control | Phage | 0.2018 * | 0.01145 | 0.000 | 0.1696 | 0.2340 |
| Fe(III)-selected | 0.2603 * | 0.01145 | 0.000 | 0.2281 | 0.2925 | |
| Phage/Iron (III)-selected | 0.2270 * | 0.01145 | 0.000 | 0.1948 | 0.2592 | |
| Phage-selected | Fe(III)-selected | 0.0585 * | 0.01145 | 0.000 | 0.0263 | 0.0907 |
| Phage/Iron (III)-selected | 0.0252 | 0.01145 | 0.276 | −0.0069 | 0.0574 | |
| Fe(III)-selected | Phage/Iron (III)-selected | 0.0332 * | 0.01145 | 0.038 | 0.0010 | 0.0654 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ezeanowai, F.C.; Ewunkem, A.J.; Morikwe, U.C.; Kiki, L.C.; McGee, L.W.; Graves, J.L., Jr.; Jeffers-Francis, L.K. Phage Resistance Modulates Escherichia coli B Response to Metal-Based Antimicrobials. Antibiotics 2025, 14, 942. https://doi.org/10.3390/antibiotics14090942
Ezeanowai FC, Ewunkem AJ, Morikwe UC, Kiki LC, McGee LW, Graves JL Jr., Jeffers-Francis LK. Phage Resistance Modulates Escherichia coli B Response to Metal-Based Antimicrobials. Antibiotics. 2025; 14(9):942. https://doi.org/10.3390/antibiotics14090942
Chicago/Turabian StyleEzeanowai, Franklin C., Akamu J. Ewunkem, Ugonna C. Morikwe, Larisa C. Kiki, Lindsey W. McGee, Joseph L. Graves, Jr., and Liesl K. Jeffers-Francis. 2025. "Phage Resistance Modulates Escherichia coli B Response to Metal-Based Antimicrobials" Antibiotics 14, no. 9: 942. https://doi.org/10.3390/antibiotics14090942
APA StyleEzeanowai, F. C., Ewunkem, A. J., Morikwe, U. C., Kiki, L. C., McGee, L. W., Graves, J. L., Jr., & Jeffers-Francis, L. K. (2025). Phage Resistance Modulates Escherichia coli B Response to Metal-Based Antimicrobials. Antibiotics, 14(9), 942. https://doi.org/10.3390/antibiotics14090942






