Comparative Analysis of Biofilm Formation and Antibiotic Resistance in Five ESKAPE Pathogen Species from a Tertiary Hospital in Bangladesh
Abstract
1. Introduction
2. Results
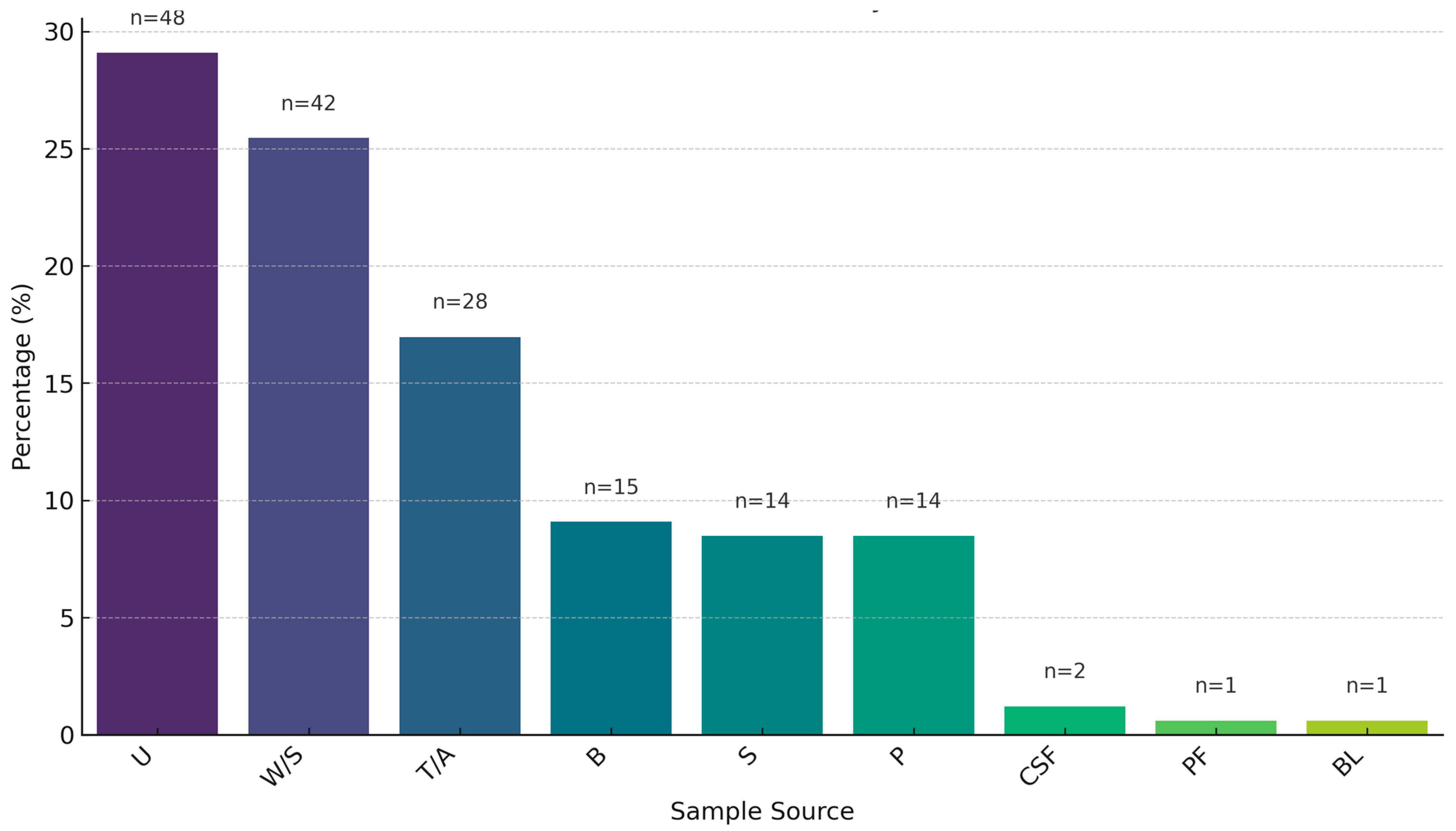
2.1. Sample Prevalence
2.2. Antimicrobial Resistance Pattern of Gram-Positive Isolates of ESKAPE Pathogens
2.3. Antimicrobial Resistance Pattern of Gram-Negative Isolates of ESKAPE Pathogens
2.4. Biofilm Formation
2.5. Correlation Between Antibiotic Resistance and Biofilm Formation
2.6. Detection of Carbapenemase and Metallo-β-Lactamase Production
2.7. Screening of MRSA
2.8. Screening of VRE and VRSA
2.9. Prevalence of Biofilm-Related Genes Compared to Pan-Genomes
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Detection
4.2. Antimicrobial Susceptibility Testing (AST)
4.3. Determination of Multiple Antibiotic Resistance Index
4.4. Determination of Colistin Resistance by MIC
4.5. Detection of Methicillin-Resistant Staphylococcus aureus (MRSA)
4.6. Screening of VRE and VRSA
4.7. Detection of Carbapenemase and Metallo-β-Lactamase Production
4.8. Phenotypic Biofilm Formation Assay
4.9. Detection and Comparison of Biofilm-Forming Genes
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Antimicrobial Resistance; WHO: Geneva, Switzerland, 2023; Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 28 May 2025).
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Navidinia, M. The clinical importance of emerging ESKAPE pathogens in nosocomial infections. J. Med. Microb. Diagn. 2016, 7, 43–57. [Google Scholar]
- Codjoe, F.S.; Donkor, E.S. Carbapenem resistance: A review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef]
- Bales, P.M.; Renke, E.M.; May, S.L.; Shen, Y.; Nelson, D.C. Purification and characterization of biofilm-associated EPS exopolysaccharides from ESKAPE organisms and other pathogens. PLoS ONE 2013, 8, e67950. [Google Scholar] [CrossRef]
- Alshaikh, S.A.; El-Banna, T.; Sonbol, F.; Farghali, M.H. Correlation between antimicrobial resistance, biofilm formation, and virulence determinants in uropathogenic Escherichia coli from Egyptian hospitals. Ann. Clin. Microbiol. Antimicrob. 2024, 23, 20. [Google Scholar] [CrossRef]
- Mirghani, R.; Almagboul, B.; Mohammed, A.A.; Gashash, A.A. Biofilms: Formation, drug resistance, and alternatives to conventional approaches. AIMS Microbiol. 2022, 8, 239–277. [Google Scholar] [CrossRef]
- The Role of Bacterial Biofilms in Antimicrobial Resistance | ASM.org [Internet]. ASM.org. 2023. Available online: https://asm.org/articles/2023/march/the-role-of-bacterial-biofilms-in-antimicrobial-re (accessed on 28 May 2025).
- Liu, H.Y.; Prentice, E.L.; Webber, M.A. Mechanisms of antimicrobial resistance in biofilms. NPJ Antimicrob. Resist. 2024, 2, 27. Available online: https://www.nature.com/articles/s44259-024-00046-3 (accessed on 28 May 2025). [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Zhen, X.; Stålsby Lundborg, C.; Sun, X.; Hu, X.; Dong, H. Economic burden of antibiotic resistance in ESKAPE organisms: A systematic review. Antimicrob. Resist. Infect. Control 2019, 8, 137. [Google Scholar] [CrossRef]
- Ramsamy, Y.; Essack, S.Y.; Sartorius, B.; Patel, M.; Mlisana, K.P. Antibiotic resistance trends of ESKAPE pathogens in KwaZulu-Natal, South Africa: A five-year retrospective analysis. Afr. J. Lab. Med. 2018, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Ayobami, O.; Brinkwirth, S.; Eckmanns, T.; Markwart, R. Antibiotic resistance in hospital-acquired ESKAPE-E infections in low- and lower-middle-income countries: A systematic review and meta-analysis. Emerg. Microbes Infect. 2022, 11, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.Z.; Sturm-Ramirez, K.; Rahman, M.Z.; Hossain, K.; Aleem, M.A.; Bhuiyan, M.U.; Islam, M.M.; Rahman, M.; Gurley, E.S. Contamination of hospital surfaces with respiratory pathogens in Bangladesh. PLoS ONE 2019, 14, e0220857. [Google Scholar] [CrossRef] [PubMed]
- Noor, R.; Munna, M.S. Emerging diseases in Bangladesh: Current microbiological research perspective. Tzu Chi Med. J. 2015, 27, 49–53. [Google Scholar] [CrossRef]
- Hammour, K.A.; Abu-Farha, R.; Itani, R.; Karout, S.; Allan, A.; Manaseer, Q.; Hammour, W.A. The prevalence of carbapenem-resistant Gram-negative pathogens in a tertiary teaching hospital in Jordan. BMC Infect. Dis. 2023, 23, 634. [Google Scholar] [CrossRef]
- Farzana, R.; Swedberg, G.; Giske, C.G.; Hasan, B. Molecular and genetic characterization of emerging carbapenemase-producing Acinetobacter baumannii strains from patients and hospital environments in Bangladesh. Infect. Prev. Pract. 2022, 4, 100215. [Google Scholar] [CrossRef]
- Mahmud, Z.H.; Uddin, S.Z.; Moniruzzaman, M.; Ali, S.; Hossain, M.; Islam, T.; Costa, D.T.D.; Islam, M.R.; Islam, S.; Hassan, Z.; et al. Healthcare facilities as potential reservoirs of antimicrobial-resistant Klebsiella pneumoniae: An emerging concern to public health in Bangladesh. Pharmaceuticals 2022, 15, 1116. [Google Scholar] [CrossRef]
- Aurilio, C.; Sansone, P.; Barbarisi, M.; Pota, V.; Giaccari, L.G.; Coppolino, F.; Barbarisi, A.; Passavanti, M.B.; Pace, M.C. Mechanisms of action of carbapenem resistance. Antibiotics 2022, 11, 421. [Google Scholar] [CrossRef]
- Bialvaei, A.Z.; Kafil, H.S.; Javan, A.O.; Ebrahimzadeh, S.; Asgharzadeh, M. Modified CIM test as a useful tool to detect carbapenemase activity among extensively drug-resistant Klebsiella pneumoniae, Escherichia coli, and Acinetobacter baumannii. Ann. Microbiol. 2021, 71, 23. [Google Scholar]
- Guignard, B.; Entenza, J.M.; Moreillon, P. β-lactams against methicillin-resistant Staphylococcus aureus. Curr. Opin. Pharmacol. 2005, 5, 479–489. [Google Scholar] [CrossRef]
- Islam, T.; Kubra, K.; Mehadi, M. Prevalence of methicillin-resistant Staphylococcus aureus in hospitals in Chittagong, Bangladesh: A threat of nosocomial infection. J. Microsc. Ultrastruct. 2018, 6, 188–191. [Google Scholar] [PubMed]
- Haq, J.A.; Rahman, M.M.; Asna, S.Z.; Hossain, M.A.; Iftekhar Ahmed, I.A.; Tahniyah Haq, T.H.; Morshed, M.G. Methicillin-resistant Staphylococcus aureus in Bangladesh—A multicentre study. Int. J. Antimicrob. Agents 2005, 25, 276–277. [Google Scholar] [CrossRef] [PubMed]
- Parvez, M.A.; Ferdous, R.N.; Rahman, M.S.; Islam, S. Healthcare-associated (HA) and community-associated (CA) methicillin-resistant Staphylococcus aureus (MRSA) in Bangladesh—Source, diagnosis, and treatment. J. Genet. Eng. Biotechnol. 2018, 16, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Palchak, M.; Pappas, P.G. Vancomycin-Resistant Enterococcus. U.S. Pharmacist—The Leading Journal in Pharmacy. 2014. Available online: https://www.uspharmacist.com/article/vancomycinresistant-enterococcus (accessed on 4 March 2025).
- Pajohesh, R.; Tajbakhsh, E.; Momtaz, H.; Rahimi, E. Relationship between biofilm formation and antibiotic resistance and adherence genes in Staphylococcus aureus strains isolated from raw cow milk in Shahrekord, Iran. Int. J. Microbiol. 2022, 2022, 6435774. [Google Scholar] [CrossRef]
- Qian, W.; Sun, Z.; Bai, Y.; Zeng, A.P.; Liu, Z. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Escherichia coli isolates from Ningbo, China. Infect. Drug Resist. 2022, 15, 2865–2878. [Google Scholar] [CrossRef]
- Ong, C.Y.; Gilligan, P.H.; Beveridge, T.J.; Mattick, J.S. Molecular analysis of type 3 fimbrial genes from Escherichia coli, Klebsiella, and Citrobacter species. BMC Microbiol. 2010, 10, 183. [Google Scholar] [CrossRef]
- Li, Y.; Ni, M. Regulation of biofilm formation in Klebsiella pneumoniae. Front. Microbiol. 2023, 14, 1238482. [Google Scholar] [CrossRef]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, invasion, and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2013, 12, 49–62. [Google Scholar] [CrossRef]
- Speziale, P.; Pietrocola, G. The multivalent role of fibronectin-binding proteins A and B (FnBPA and FnBPB) of Staphylococcus aureus in host infections. Front. Microbiol. 2020, 11, 2054. [Google Scholar] [CrossRef]
- Su, Y.A.; Sulavik, M.C.; He, P.I.; Makinen, K.K.; Makinen, P.L.; Fiedler, S.; Wirth, R.; Clewell, D.B. Nucleotide sequence of the gelatinase gene (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect. Immun. 1991, 59, 415–420. [Google Scholar] [CrossRef]
- Francois, P.; Pittet, D.; Bento, M.; Pepey, B.; Vaudaux, P.; Lew, D.; Schrenzel, J. Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay. J. Clin. Microbiol. 2003, 41, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Turton, J.F.; Woodford, N.; Glover, J.; Yarde, S.; Kaufmann, M.E.; Pitt, T.L. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 2006, 44, 2974–2976. [Google Scholar] [CrossRef] [PubMed]
- Arabestani, M.R.; Nasaj, M.; Mousavi, S.M. Correlation between infective factors and antibiotic resistance in Enterococci clinical isolates in the west of Iran. Chonnam Med. J. 2017, 53, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Taee, S.R.; Nezhad, B.K.; Abtahi, H.; Najafimosleh, M.; Ghaznavi-Rad, E. Detection of algD, oprL, and exoA genes by new specific primers as an efficient, rapid, and accurate procedure for direct diagnosis of Pseudomonas aeruginosa strains in clinical samples. Jundishapur J. Microbiol. 2014, 7, e13583. [Google Scholar]
- Liu, Y.; Liu, C.; Zheng, W.; Zhang, X.; Yu, J.; Gao, Q.; Hou, Y.; Huang, X. PCR detection of Klebsiella pneumoniae in infant formula based on 16S–23S internal transcribed spacer. Int. J. Food Microbiol. 2008, 125, 230–235. [Google Scholar] [CrossRef]
- Hudzicki, J. Kirby-Bauer disk diffusion susceptibility test protocol. Am. Soc. Microbiol. 2009, 15, 1–23. Available online: https://asm.org/getattachment/2594ce26-bd44-47f6-8287-0657aa9185ad/Kirby-Bauer-Disk-Diffusion-Susceptibility-Test-Protocol-pdf.pdf (accessed on 15 June 2024).
- CLSI M100; Performance Standards for Antimicrobial Susceptibility Testing. CLSI: Wayne, PA, USA, 2024.
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant, and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Becker, K.; Roth, R.; Peters, G.; Von Eiff, C. Evaluation of a modular multiplex-PCR methicillin-resistant Staphylococcus aureus detection assay adapted for mecC detection. J. Clin. Microbiol. 2013, 51, 1917–1919. [Google Scholar] [CrossRef]
- George, S.K.; Selvaraj, A.; Pratheepa, V.; Ghosh, S.; Vasudevan, A. Molecular determination of van genes among clinical isolates of enterococci at a hospital setting. Saudi J. Biol. Sci. 2021, 28, 2895–2899. [Google Scholar] [CrossRef]
- Pierce, V.M.; Simner, P.J.; Lonsway, D.R.; Roe-Carpenter, D.E.; Johnson, J.K.; Brasso, W.B.; Bobenchik, A.M.; Lockett, Z.C.; Charnot-Katsikas, A.; Ferraro, M.J.; et al. Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among Enterobacteriaceae. J. Clin. Microbiol. 2017, 55, 2321–2323. [Google Scholar] [CrossRef] [PubMed]
- Sfeir, M.M.; Hayden, J.A.; Fauntleroy, K.A.; Mazur, C.; Johnson, J.K.; Simner, P.J.; Das, S.; Satlin, M.J.; Jenkins, S.G.; Westblade, L.F. EDTA-modified carbapenem inactivation method: A phenotypic method for detecting metallo-β-lactamase-producing Enterobacteriaceae. J. Clin. Microbiol. 2019, 57, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Coffey, B.M.; Anderson, G.G. Biofilm formation in the 96-well microtiter plate. Methods Mol. Biol. 2014, 1149, 631–641. [Google Scholar] [PubMed]
- Yang, C.H.; Su, P.W.; Moi, S.H.; Chuang, L.Y. Biofilm formation in Acinetobacter baumannii: Genotype-phenotype correlation. Molecules 2019, 24, 1849. [Google Scholar] [CrossRef]
- Hou, Z.; Xu, B.; Liu, L.; Yan, R.; Zhang, J.; Yin, J.; Li, P.; Wei, J. Prevalence, drug resistance, molecular typing, and comparative genomics analysis of MRSA strains from a tertiary hospital in Shanxi Province, China. Front. Microbiol. 2023, 14, 1273397. [Google Scholar] [CrossRef]
- Tristan, A.; Ying, L.; Bes, M.; Etienne, J.; Vandenesch, F. Use of multiplex PCR to identify Staphylococcus aureus adhesins involved in human hematogenous infections. J. Clin. Microbiol. 2003, 41, 4465–4467. [Google Scholar] [CrossRef]
- Verkaik, N.J.; Boelens, H.A.; de Vogel, C.P.; Tavakol, M.; Bode, L.G.; Verbrugh, H.A.; van Belkum, A.; van Wamel, W.J. Heterogeneity of the humoral immune response following Staphylococcus aureus bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 509–518. [Google Scholar] [CrossRef]
- Waryah, C.B.; Ulett, G.C.; Benjamin, W.H., Jr.; Sheikh, M.A. Diversity of virulence factors associated with West Australian methicillin-sensitive Staphylococcus aureus isolates of human origin. BioMed Res. Int. 2016, 2016, 8651918. [Google Scholar] [CrossRef]
- Sabat, A.; Melles, D.C.; Martirosian, G.; Grundmann, H.; van Belkum, A.; Hryniewicz, W. Distribution of the serine-aspartate repeat protein-encoding sdr genes among nasal-carriage and invasive Staphylococcus aureus strains. J. Clin. Microbiol. 2006, 44, 1135–1138. [Google Scholar] [CrossRef]
- Vankerckhoven, V.; Van Autgaerden, T.; Vael, C.; Lammens, C.; Chapelle, S.; Rossi, R.; Jabes, D.; Goossens, H. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J. Clin. Microbiol. 2004, 42, 4473–4479. [Google Scholar] [CrossRef]
- Nallapareddy, S.R.; Weinstock, G.M.; Murray, B.E. Clinical isolates of Enterococcus faecium exhibit strain-specific collagen binding mediated by Acm, a new member of the MSCRAMM family. Mol. Microbiol. 2003, 47, 1733–1747. [Google Scholar] [CrossRef] [PubMed]
- Eaton, T.J.; Gasson, M.J. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 2001, 67, 1628–1635. [Google Scholar] [CrossRef] [PubMed]
- Khoshnood, S.; Savari, M.; Montazeri, E.A.; Sheikh, A.F. Survey on genetic diversity, biofilm formation, and detection of colistin resistance genes in clinical isolates of Acinetobacter baumannii. Infect. Drug Resist. 2020, 13, 1547–1758. [Google Scholar] [CrossRef] [PubMed]
- Khanna, N.; Girija, A.S.S.; Priyadharsini, J.V. Detection of the early putative biofilm marker pgaB among the MDR strains of A. baumannii. Heliyon 2024, 10, e27020. [Google Scholar] [CrossRef]
- Ansari, H.; Doosti, A.; Kargar, M.; Bijanzadeh, M.; Jaafarinia, M. Cloning of ompA gene from Acinetobacter baumannii into the eukaryotic expression vector pBudCE4.1 as DNA vaccine. Indian J. Microbiol. 2018, 58, 174–181. [Google Scholar] [CrossRef]
- Azizi, O.; Shakibaie, M.R.; Modarresi, F.; Shahcheraghi, F.; Motamedifar, M.; Mansouri, S. Molecular analysis and expression of bap gene in biofilm-forming multi-drug-resistant Acinetobacter baumannii. Rep. Biochem. Mol. Biol. 2016, 5, 62–72. [Google Scholar]
- Bahador, A.; Bazargani, A.; Taheri, M.; Hashemizadeh, Z.; Khaledi, A.; Rostami, H.; Esmaili, D. Clonal lineages and virulence factors among Acinetobacter baumannii isolated from Southwest Iran. J. Pure Appl. Microbiol. 2013, 7, 1959–1966. [Google Scholar]
- Qaralleh, H.; Saghir, S.A.; Al-Limoun, M.O.; Dmor, S.M.; Khleifat, K.; Al-Ahmad, B.E.; Al-Omari, L.; Tabana, Y.; Mothana, R.A.; Al-Yousef, H.M.; et al. Effect of Matricaria aurea essential oils on biofilm development, virulence factors and quorum sensing-dependent genes of Pseudomonas aeruginosa. Pharmaceuticals 2024, 17, 386. [Google Scholar] [CrossRef]
- Bakhtiari, R.; Javadi, A.; Aminzadeh, M.; Molaee-Aghaee, E.; Shaffaghat, Z. Association between presence of rmpA, mrkA and mrkD genes and antibiotic resistance in clinical Klebsiella pneumoniae isolates from hospitals in Tehran, Iran. Iran J. Public Health 2021, 50, 1009–1016. [Google Scholar] [CrossRef]
- Shadkam, S.; Goli, H.R.; Mirzaei, B.; Gholami, M.; Ahanjan, M. Correlation between antimicrobial resistance and biofilm formation capability among Klebsiella pneumoniae strains isolated from hospitalized patients in Iran. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 13. [Google Scholar] [CrossRef]
- Ding, W.; Baumdicker, F.; Neher, R.A. panX: Pan-genome analysis and exploration. Nucleic Acids Res. 2018, 46, e5. [Google Scholar] [CrossRef]








| Antibiotics Classes | Antibiotics | Gram-Negative | Gram-Positive | |||
|---|---|---|---|---|---|---|
| A. baumannii | P. aeruginosa | K. pneumoniae | S. aureus | E. faecium | ||
| Aminoglycosides | Gentamicin (10 μg) | Yes | Yes | Yes | Yes | Yes |
| Amikacin (30 μg) | Yes | Yes | Yes | No | No | |
| Fluoroquinolones | Ciprofloxacin (5 μg) | Yes | Yes | Yes | Yes | Yes |
| Levofloxacin (5 μg) | Yes | Yes | Yes | Yes | Yes | |
| Cephalosporins | Ceftazidime (30 μg) | Yes | Yes | Yes | No | No |
| Cefepime (30 μg) | Yes | Yes | Yes | No | No | |
| Carbapenems | Imipenem (10 μg) | Yes | Yes | Yes | No | No |
| Meropenem (10 μg) | Yes | Yes | Yes | No | No | |
| Penicillins + β-lactamase inhibitors | Piperacillin/tazobactam (100/10 μg) | Yes | Yes | Yes | No | No |
| Polymyxins | Colistin (MIC) | Yes | Yes | Yes | No | No |
| Tetracyclines | Tetracycline (30 μg) | Yes | No | Yes | Yes | Yes |
| Doxycycline (30 μg) | Yes | No | Yes | Yes | Yes | |
| Monobactams | Aztreonam (30 μg) | No | Yes | Yes | No | No |
| Folate pathway inhibitors | Trimethoprim/ Sulfamethoxazole (25 μg) | Yes | No | Yes | Yes | No |
| Oxazolidinones | Linezolid (30 μg) | No | No | No | Yes | Yes |
| Glycopeptides | Vancomycin (MIC) | No | No | No | Yes | Yes |
| Phenicols | Chloramphenicol (30 μg) | No | No | No | Yes | Yes |
| Nitrofurans | Nitrofurantoin (300 μg) | No | No | No | Yes | Yes |
| Anti-staphylococcal β-lactams | Cefoxitin (30 μg) | No | No | No | Yes | No |
| Penicillins | Ampicillin (10 μg) | No | No | No | No | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anik, T.A.; Uzzaman, R.; Rahman, K.T.; Hossain, A.; Islam, F.; Tasnim, M.N.; Begum, S.A.; Akhter, H.; Begum, A. Comparative Analysis of Biofilm Formation and Antibiotic Resistance in Five ESKAPE Pathogen Species from a Tertiary Hospital in Bangladesh. Antibiotics 2025, 14, 842. https://doi.org/10.3390/antibiotics14080842
Anik TA, Uzzaman R, Rahman KT, Hossain A, Islam F, Tasnim MN, Begum SA, Akhter H, Begum A. Comparative Analysis of Biofilm Formation and Antibiotic Resistance in Five ESKAPE Pathogen Species from a Tertiary Hospital in Bangladesh. Antibiotics. 2025; 14(8):842. https://doi.org/10.3390/antibiotics14080842
Chicago/Turabian StyleAnik, Tasnimul Arabi, Rahat Uzzaman, Khandaker Toyabur Rahman, Abir Hossain, Faruk Islam, Mosammod Nowshin Tasnim, Shahin Ara Begum, Humaira Akhter, and Anowara Begum. 2025. "Comparative Analysis of Biofilm Formation and Antibiotic Resistance in Five ESKAPE Pathogen Species from a Tertiary Hospital in Bangladesh" Antibiotics 14, no. 8: 842. https://doi.org/10.3390/antibiotics14080842
APA StyleAnik, T. A., Uzzaman, R., Rahman, K. T., Hossain, A., Islam, F., Tasnim, M. N., Begum, S. A., Akhter, H., & Begum, A. (2025). Comparative Analysis of Biofilm Formation and Antibiotic Resistance in Five ESKAPE Pathogen Species from a Tertiary Hospital in Bangladesh. Antibiotics, 14(8), 842. https://doi.org/10.3390/antibiotics14080842






