Unique Regulation of Sed-1 β-Lactamase in Citrobacter sedlakii: Insights on Resistance to Third-Generation Cephalosporin
Abstract
1. Introduction
2. Results
2.1. Identification of C. sedlakii NR2807
2.2. Antimicrobial Susceptibility and Resistance Gene
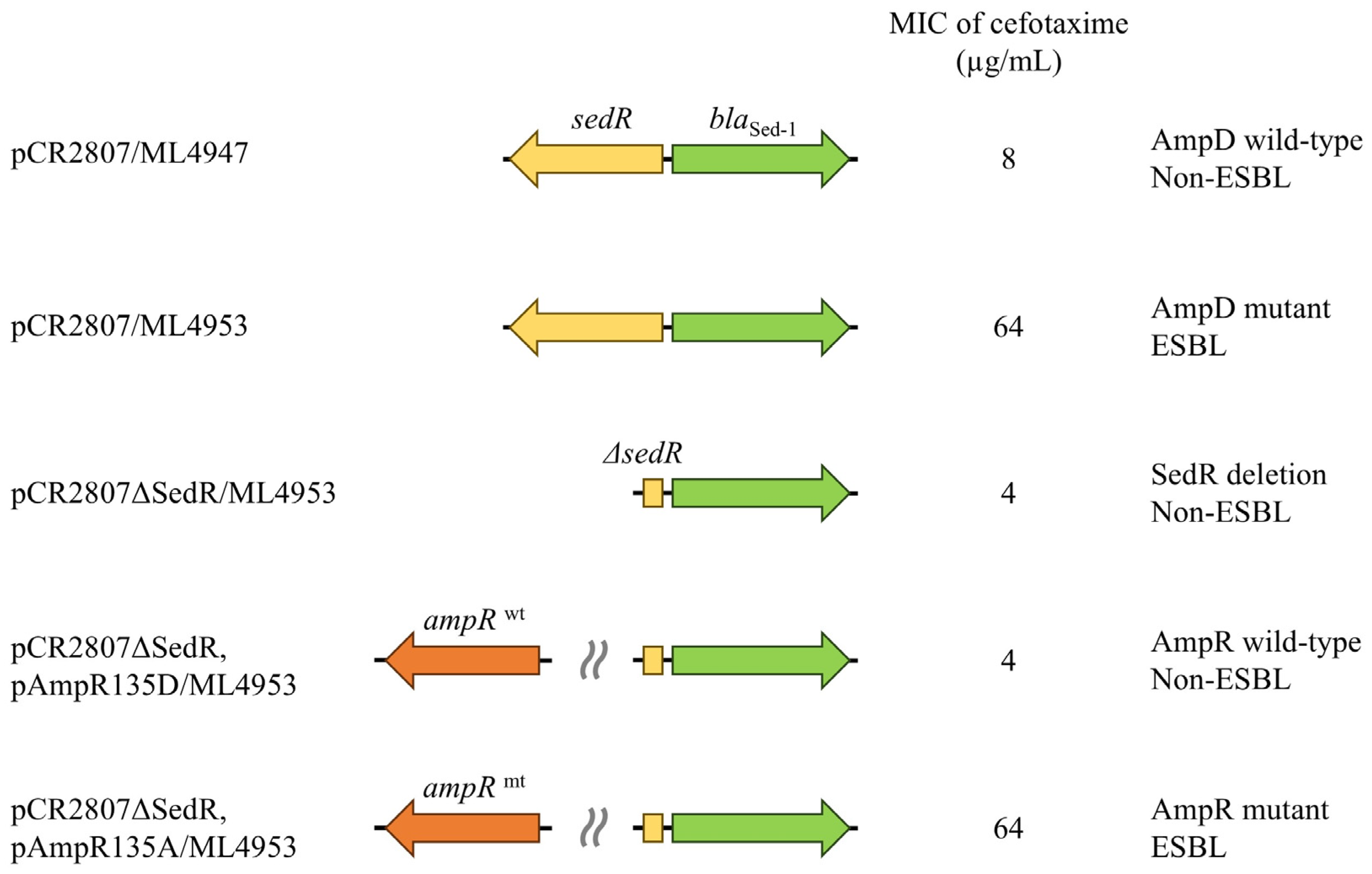
2.3. Effect of Regulator Genes on β-Lactamase Expression
2.4. Characteristics of Various Antibiotic-Resistant Mutants
2.5. Kinetic Parameters of Ceftazidime-Resistant Mutants
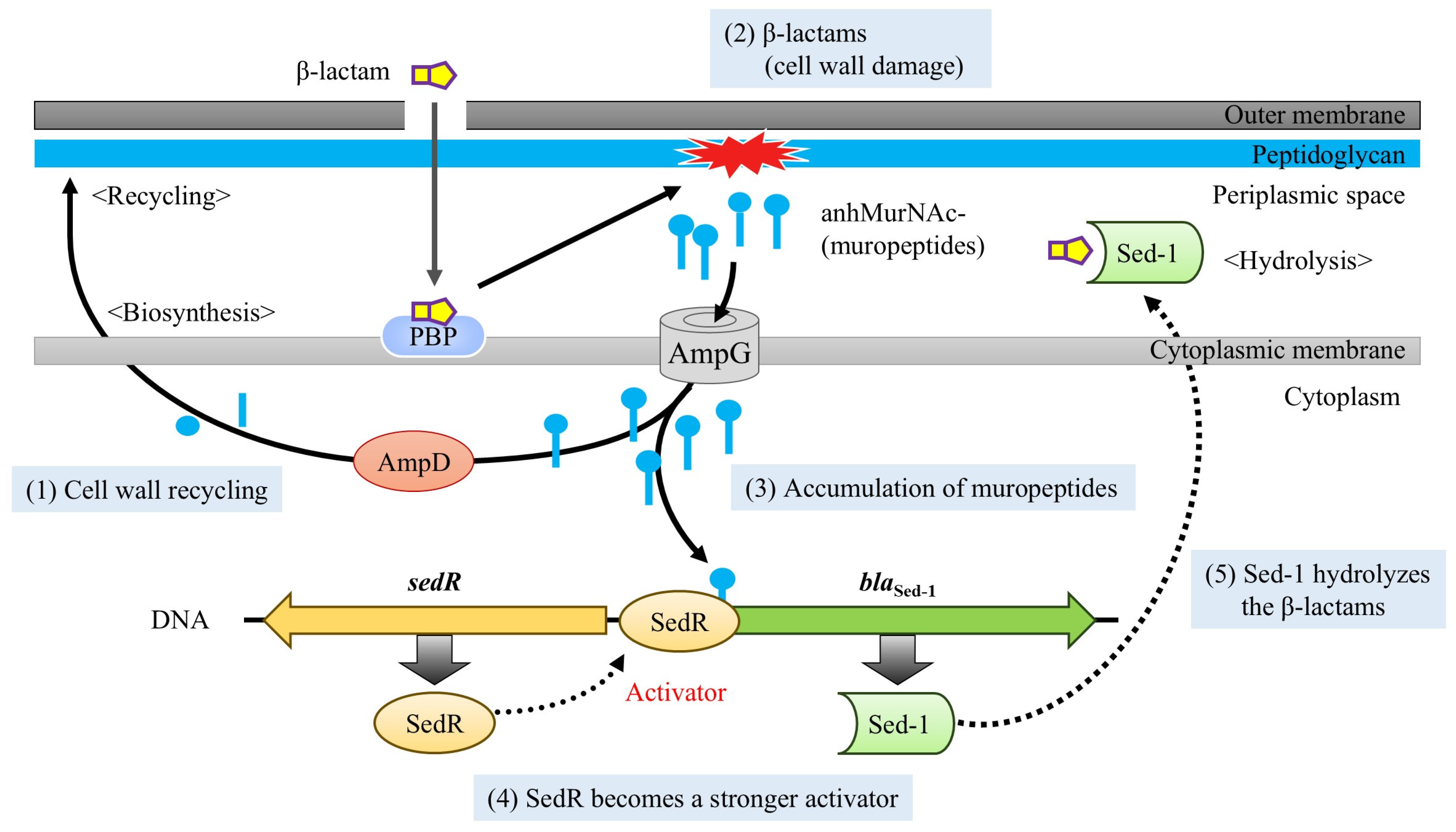
3. Discussion
3.1. Genomic and Clinical Significance of Sed-1 in C. Sedlakii
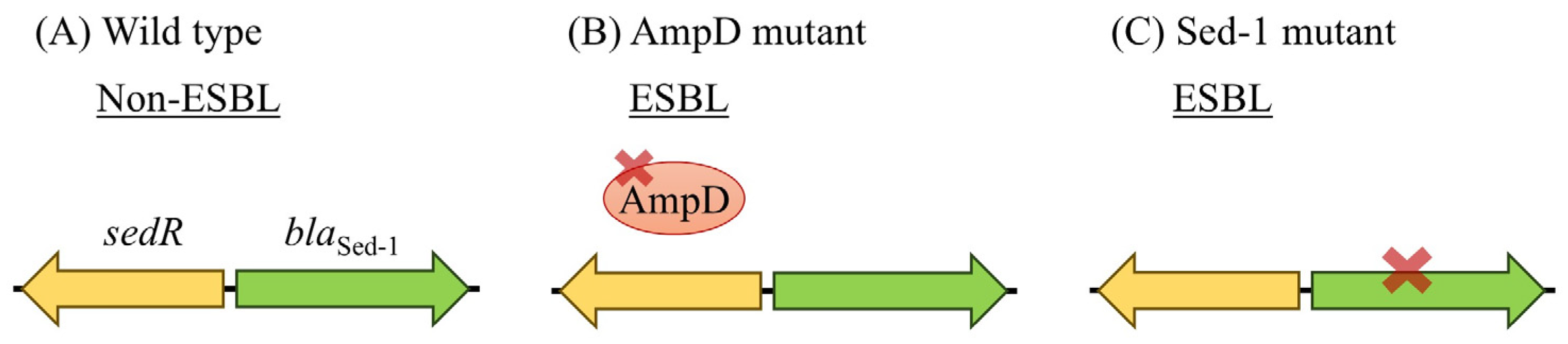
3.2. Functional Impact of AmpD Mutation on Sed-1 Expression and Antibiotic Resistance
3.3. SedR Functions as a Constitutive Activator Similar to Mutant AmpR
3.4. Ceftazidime-Driven Mutations in Sed-1 Expand Substrate Specificity
3.5. ESBL-like Features and Future Risk in Sed-1-Producing C. Sedlakii
3.6. Study Limitations
4. Materials and Methods
4.1. Bacterial Strains and Antimicrobial Susceptibility Testing
4.2. Whole-Genome Sequencing and Analysis
4.3. Measurement of β-Lactamase Activity and Induction Assays
4.4. Selection of Antibiotic-Resistant Strains and Detection of Sequence Changes
4.5. Measurement of Kinetic Parameters in NR2807 and Mutants
4.6. Nucleotide Sequence Accession Numbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3GC | Third-generation cephalosporins |
| CFU | Colony-forming units |
| ESBL | Extended-spectrum β-lactamase |
| LB | Luria–Bertani |
References
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Melchiorri, D.; Rocke, T.; Alm, R.A.; Cameron, A.M.; Gigante, V. Addressing Urgent Priorities in Antibiotic Development: Insights from WHO 2023 Antibacterial Clinical Pipeline Analyses. Lancet Microbe 2025, 6, 100992. [Google Scholar] [CrossRef] [PubMed]
- Husna, A.; Rahman, M.M.; Badruzzaman, A.T.M.; Sikder, M.H.; Islam, M.R.; Rahman, M.T.; Alam, J.; Ashour, H.M. Extended-Spectrum Beta-Lactamases (Esbl): Challenges and Opportunities. Biomedicines 2023, 11, 2937. [Google Scholar] [CrossRef]
- Philippon, A.; Slama, P.; Deny, P.; Labia, R. A Structure-Based Classification of Class a Beta-Lactamases, a Broadly Diverse Family of Enzymes. Clin. Microbiol. Rev. 2016, 29, 29–57. [Google Scholar] [CrossRef]
- Naas, T.; Aubert, D.; Ozcan, A.; Nordmann, P. Chromosome-Encoded Narrow-Spectrum Ambler Class a Beta-Lactamase Gil-1 from Citrobacter gillenii. Antimicrob. Agents Chemother. 2007, 51, 1365–1372. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ortiz de la Rosa, J.M.; Bouvier, M.; Poirel, L.; Greub, G.; Blanc, D.; Nordmann, P. Cross-Reaction of Naturally-Produced Beta-Lactamases from Citrobacter farmeri and Citrobacter amalonaticus with Immunological Detection of Ctx-M Enzymes. Diagn. Microbiol. Infect. Dis. 2022, 104, 115760. [Google Scholar] [CrossRef]
- Underwood, S.; Avison, M.B. Citrobacter koseri and Citrobacter amalonaticus Isolates Carry Highly Divergent Beta-Lactamase Genes Despite Having High Levels of Biochemical Similarity and 16s rRNA Sequence Homology. J. Antimicrob. Chemother. 2004, 53, 1076–1080. [Google Scholar] [CrossRef]
- Bartowsky, E.; Normark, S. Purification and Mutant Analysis of Citrobacter freundii Ampr, the Regulator for Chromosomal Ampc Beta-Lactamase. Mol. Microbiol. 1991, 5, 1715–1725. [Google Scholar] [CrossRef]
- Guerin, F.; Isnard, C.; Cattoir, V.; Giard, J.C. Complex Regulation Pathways of Ampc-Mediated B-Lactam Resistance in Enterobacter cloacae Complex. Antimicrob. Agents Chemother. 2015, 59, 7753–7761. [Google Scholar] [CrossRef] [PubMed]
- Avison, M.B.; Underwood, S.; Okazaki, A.; Walsh, T.R.; Bennett, P.M. Analysis of Ampc Beta-Lactamase Expression and Sequence in Biochemically Atypical Ceftazidime-Resistant Enterobacteriaceae from Paediatric Patients. J. Antimicrob. Chemother. 2004, 53, 584–591. [Google Scholar] [CrossRef]
- Kuga, A.; Okamoto, R.; Inoue, M. Ampr Gene Mutations That Greatly Increase Class C B-Lactamase Activity in Enterobacter cloacae. Antimicrob. Agents Chemother. 2000, 44, 561–567. [Google Scholar] [CrossRef]
- Nakano, R.; Nakano, A.; Yano, H.; Okamoto, R. Role of Ampr in the High Expression of the Plasmid-Encoded Ampc Beta-Lactamase Cfe-1. mSphere 2017, 2, e00192-17. [Google Scholar] [CrossRef] [PubMed]
- Caille, O.; Zincke, D.; Merighi, M.; Balasubramanian, D.; Kumari, H.; Kong, K.F.; Silva-Herzog, E.; Narasimhan, G.; Schneper, L.; Lory, S.; et al. Structural and Functional Characterization of Pseudomonas aeruginosa Global Regulator Ampr. J. Bacteriol. 2014, 196, 3890–3902. [Google Scholar] [CrossRef] [PubMed]
- Juan, C.; Macia, M.D.; Gutierrez, O.; Vidal, C.; Perez, J.L.; Oliver, A. Molecular Mechanisms of Beta-Lactam Resistance Mediated by Ampc Hyperproduction in Pseudomonas aeruginosa Clinical Strains. Antimicrob. Agents Chemother. 2005, 49, 4733–4738. [Google Scholar] [CrossRef]
- Kaneko, K.; Okamoto, R.; Nakano, R.; Kawakami, S.; Inoue, M. Gene Mutations Responsible for Overexpression of Ampc B-Lactamase in Some Clinical Isolates of Enterobacter cloacae. J. Clin. Microbiol. 2005, 43, 2955–2958. [Google Scholar] [CrossRef]
- Balcewich, M.D.; Reeve, T.M.; Orlikow, E.A.; Donald, L.J.; Vocadlo, D.J.; Mark, B.L. Crystal Structure of the Ampr Effector Binding Domain Provides Insight into the Molecular Regulation of Inducible Ampc Beta-Lactamase. J. Mol. Biol. 2010, 400, 998–1010. [Google Scholar] [CrossRef]
- Petrella, S.; Clermont, D.; Casin, I.; Jarlier, V.; Sougakoff, W. Novel Class a Beta-Lactamase Sed-1 from Citrobacter sedlakii: Genetic Diversity of Beta-Lactamases within the Citrobacter Genus. Antimicrob. Agents Chemother. 2001, 45, 2287–2298. [Google Scholar] [CrossRef]
- Tasnim, Y.; Stanley, C.; Rahman, M.K.; Awosile, B. Bla Sed-1 Beta-Lactamase-Producing Citrobacter Sedlakii Isolated from Horses and Genomic Comparison with Human-Derived Isolates. J. Appl. Microbiol. 2024, 135, lxae278. [Google Scholar] [CrossRef]
- Matagne, A.; Lamotte-Brasseur, J.; Frere, J.M. Catalytic Properties of Class a Beta-Lactamases: Efficiency and Diversity. Biochem. J. 1998, 330 Pt 2, 581–598. [Google Scholar] [CrossRef]
- Bennett, P.M.; Chopra, I. Molecular Basis of Beta-Lactamase Induction in Bacteria. Antimicrob. Agents Chemother. 1993, 37, 153–158. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maddocks, S.E.; Oyston, P.C.F. Structure and Function of the Lysr-Type Transcriptional Regulator (Lttr) Family Proteins. Microbiology (Reading) 2008, 154 Pt 12, 3609–3623. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.F.; Jayawardena, S.R.; Indulkar, S.D.; Del Puerto, A.; Koh, C.L.; Hoiby, N.; Mathee, K. Pseudomonas aeruginosa Ampr Is a Global Transcriptional Factor that Regulates Expression of Ampc and Poxb Beta-Lactamases, Proteases, Quorum Sensing, and Other Virulence Factors. Antimicrob. Agents Chemother. 2005, 49, 4567–4575. [Google Scholar] [CrossRef]
- Stapleton, P.; Shannon, K.; Phillips, I. DNA Sequence Differences of Ampd Mutants of Citrobacter freundii. Antimicrob. Agents Chemother. 1995, 39, 2494–2498. [Google Scholar] [CrossRef]
- Kohlmann, R.; Bahr, T.; Gatermann, S.G. Species-Specific Mutation Rates for Ampc Derepression in Enterobacterales with Chromosomally Encoded Inducible Ampc Beta-Lactamase. J. Antimicrob. Chemother. 2018, 73, 1530–1536. [Google Scholar] [CrossRef]
- Arpin, C.; Labia, R.; Andre, C.; Frigo, C.; El Harrif, Z.; Quentin, C. Shv-16, a Beta-Lactamase with a Pentapeptide Duplication in the Omega Loop. Antimicrob. Agents Chemother. 2001, 45, 2480–2485. [Google Scholar] [CrossRef]
- Hayes, F.; Hallet, B.; Cao, Y. Insertion Mutagenesis as a Tool in the Modification of Protein Function. Extended Substrate Specificity Conferred by Pentapeptide Insertions in the Omega-Loop of Tem-1 Beta-Lactamase. J. Biol. Chem. 1997, 272, 28833–28836. [Google Scholar] [CrossRef]
- Vakulenko, S.B.; Taibi-Tronche, P.; Toth, M.; Massova, I.; Lerner, S.A.; Mobashery, S. Effects on Substrate Profile by Mutational Substitutions at Positions 164 and 179 of the Class a Tem (Puc19) Beta-Lactamase from Escherichia coli. J. Biol. Chem. 1999, 274, 23052–23060. [Google Scholar] [CrossRef] [PubMed]
- Laubacher, M.E.; Ades, S.E. The Rcs Phosphorelay Is a Cell Envelope Stress Response Activated by Peptidoglycan Stress and Contributes to Intrinsic Antibiotic Resistance. J. Bacteriol. 2008, 190, 2065–2074. [Google Scholar] [CrossRef] [PubMed]
- Sutterlin, H.A.; Zhang, S.; Silhavy, T.J. Accumulation of Phosphatidic Acid Increases Vancomycin Resistance in Escherichia coli. J. Bacteriol. 2014, 196, 3214–3220. [Google Scholar] [CrossRef]
- Negrete-Gonzalez, C.; Turrubiartes-Martinez, E.; Briano-Macias, M.; Noyola, D.; Perez-Gonzalez, L.F.; Gonzalez-Amaro, R.; Nino-Moreno, P. Plasmid Carrying Blactx-M-15, Blaper-1, and Blatem-1 Genes in Citrobacter spp. from Regional Hospital in Mexico. Infect. Dis. 2022, 15, 11786337211065750. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-Spectrum Beta-Lactamases: A Clinical Update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Liakopoulos, A.; Mevius, D.; Ceccarelli, D. A Review of Shv Extended-Spectrum Beta-Lactamases: Neglected yet Ubiquitous. Front. Microbiol. 2016, 7, 1374. [Google Scholar] [CrossRef]
- Richter, M.; Rossello-Mora, R.; Oliver Glockner, F.; Peplies, J. Jspeciesws: A Web Server for Prokaryotic Species Circumscription Based on Pairwise Genome Comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Nakano, R.; Okamoto, R.; Nakano, Y.; Kaneko, K.; Okitsu, N.; Hosaka, Y.; Inoue, M. Cfe-1, a Novel Plasmid-Encoded Ampc B-Lactamase with an Ampr Gene Originating from Citrobacter freundii. Antimicrob. Agents Chemother. 2004, 48, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 35th ed; Clsi Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2025. [Google Scholar]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA Hybridization Values and Their Relationship to Whole-Genome Sequence Similarities. Int. J. Syst. Evol. Microbiol. 2007, 57 Pt 1, 81–91. [Google Scholar] [CrossRef]
- Chun, J.; Rainey, F.A. Integrating Genomics into the Taxonomy and Systematics of the Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 2, 316–324. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. Resfinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Ambler, R.P.; Coulson, A.F.; Frere, J.M.; Ghuysen, J.M.; Joris, B.; Forsman, M.; Levesque, R.C.; Tiraby, G.; Waley, S.G. A Standard Numbering Scheme for the Class a Beta-Lactamases. Biochem. J. 1991, 276 Pt 1, 269–270. [Google Scholar] [CrossRef]
- Bai, L.; Xia, S.; Lan, R.; Liu, L.; Ye, C.; Wang, Y.; Jin, D.; Cui, Z.; Jing, H.; Xiong, Y.; et al. Isolation and Characterization of Cytotoxic, Aggregative Citrobacter freundii. PLoS ONE 2012, 7, e33054. [Google Scholar] [CrossRef]
- Waley, S.G. A Spectrophotometric Assay of Beta-Lactamase Action on Penicillins. Biochem. J. 1974, 139, 789–790. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Nakano, R.; Yamada, Y.; Nakano, A.; Suzuki, Y.; Saito, K.; Sakata, R.; Ogawa, M.; Narita, K.; Kuga, A.; Suwabe, A.; et al. The Role of Nmcr, Ampr, and Ampd in the Regulation of the Class a Carbapenemase Nmca in Enterobacter ludwigii. Front. Microbiol. 2021, 12, 794134. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Tanaka, H.; Kurisu, G.; Nakano, R.; Yano, H.; Sakai, H. Structural Insights into the Substrate Specificity of Imp-6 and Imp-1 Metallo-B-Lactamases. J. Biochem. 2022, 173, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Laraki, N.; Franceschini, N.; Rossolini, G.M.; Santucci, P.; Meunier, C.; de Pauw, E.; Amicosante, G.; Frere, J.M.; Galleni, M. Biochemical Characterization of the Pseudomonas aeruginosa 101/1477 Metallo-B-Lactamase Imp-1 Produced by Escherichia coli. Antimicrob. Agents Chemother. 1999, 43, 902–906. [Google Scholar] [CrossRef] [PubMed]

| Species | β-Lactamase (Class) | Regulator Gene | Expression | |
|---|---|---|---|---|
| C. freundii complex | ||||
| C. braakii | AmpC (C) | AmpR | Inducible | |
| C. freundii | AmpC (C) | AmpR | Inducible | |
| C. murliniae | AmpC (C) | AmpR | Inducible | |
| C. youngae | AmpC (C) | AmpR | Inducible | |
| C. werkmanii | AmpC (C) | AmpR | Inducible | |
| C. portucalensis | AmpC (C) | AmpR | Inducible | |
| C. gillenii | GIL-1 (A) | - | Constitutive | |
| C. sedlakii | Sed-1 (A) | SedR | Inducible | |
| C. farmeri | Sed-1 (A) | SedR | Inducible | |
| C. rodentium | Sed-1 (A) | SedR | Inducible | |
| C. amalonaticus | CdiA (A) | CdiR | Inducible | |
| C. koseri | CKO-1 (A) | - | Constitutive | |
| C. cronae | AmpC (C) | AmpR | Inducible | |
| C. pasteurii | AmpC (C) | AmpR | Inducible | |
| Species | Strain | Genes | AmpD | MIC (µg/mL) a | Relative β-Lactamase Activity (U/mg Protein) c | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PIP | CTX | CTX/CLA b | CAZ | FEP | CMZ | CFX | ATM | IPM | Basal | Induced d | Induced/ Basal | ||||
| C. sedlakii | NR2807 | blaSed-1, sedR | mutant | 256 | 32 | 0.5 | 16 | 4 | 2 | 8 | 64 | 0.125 | 8.39 | 8.54 | 1.02 |
| C. sedlakii | ATCC51115 | blaSed-1, sedR | wild-type | 8 | 0.5 | 0.25 | 2 | 0.125 | 2 | 8 | 1 | 0.25 | 0.13 | 0.40 | 3.11 |
| E. coli | pCR2807/ML4947 | blaSed-1, sedR | wild-type | 256 | 8 | ≤0.06 | 4 | 1 | 1 | 4 | 16 | 0.125 | 2.78 | 3.68 | 1.32 |
| E. coli | pCR2807/ML4953 | blaSed-1, sedR | mutant | >256 | 64 | 1 | 16 | 4 | 1 | 4 | 64 | 0.125 | 23.50 | 36.66 | 1.56 |
| E. coli | pCR2807ΔSedR/ML4947 | blaSed-1 | wild-type | 64 | 2 | ≤0.06 | 2 | 0.25 | 1 | 4 | 8 | 0.125 | 1.44 | NT e | - |
| E. coli | pCR2807ΔSedR/ML4953 | blaSed-1 | mutant | 256 | 4 | ≤0.06 | 2 | 0.5 | 2 | 4 | 8 | 0.125 | 3.42 | NT | - |
| E. coli | pCR2807ΔSedR, pAmpR135D/ML4953 | blaSed-1, ampRwt | mutant | 64 | 4 | ≤0.06 | 4 | 1 | 2 | 4 | 16 | 0.25 | 2.07 | 6.15 | 2.98 |
| E. coli | pCR2807ΔSedR, pAmpR135A/ML4953 | blaSed-1, ampRmt | mutant | >256 | 64 | 0.125 | 16 | 4 | 1 | 4 | 64 | 0.25 | 20.08 | 23.50 | 1.17 |
| E. coli | pCR51115/ML4947 | blaSed-1, sedR | wild-type | 128 | 4 | ≤0.06 | 4 | 1 | 2 | 4 | 16 | 0.25 | 2.40 | 3.79 | 1.58 |
| E. coli | pCR51115/ML4953 | blaSed-1, sedR | mutant | 256 | 16 | 0.5 | 16 | 4 | 1 | 4 | 64 | 0.25 | 16.39 | 22.41 | 1.37 |
| E. coli | pCR51115ΔSedR/ML4947 | blaSed-1 | wild-type | 64 | 2 | ≤0.06 | 2 | 0.25 | 1 | 4 | 8 | 0.25 | 1.48 | NT | - |
| E. coli | pCR51115ΔSedR/ML4953 | blaSed-1 | mutant | 256 | 2 | ≤0.06 | 2 | 0.5 | 1 | 4 | 8 | 0.125 | 3.35 | NT | - |
| E. coli | pCR51115ΔSedR, pAmpR135D/ML4953 | blaSed-1, ampRwt | mutant | 64 | 2 | ≤0.06 | 4 | 1 | 1 | 2 | 8 | 0.25 | 2.85 | 4.14 | 1.45 |
| E. coli | pCR51115ΔSedR, pAmpR135A/ML4953 | blaSed-1, ampRmt | mutant | >256 | 32 | ≤0.06 | 16 | 4 | 1 | 2 | 32 | 0.25 | 14.85 | 16.71 | 1.13 |
| E. coli | ML4947 | - | wild-type | 2 | ≤0.06 | ≤0.06 | 0.25 | ≤0.06 | 1 | 0.5 | ≤0.06 | ≤0.06 | <0.01 | NT | - |
| E. coli | ML4953 | - | mutant | 4 | ≤0.06 | ≤0.06 | 0.5 | ≤0.06 | 1 | 0.5 | ≤0.06 | ≤0.06 | <0.01 | NT | - |
| Species | Strains | Selection a,b | Mutation c | MIC (µg/mL) a | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sed-1 d | SedR | AmpD | Other Mutated Genes | PIP | CTX | CTX/CLA e | CAZ | FEP | CMZ | CFX | ATM | IPM | |||
| C. sedlakii | NR2807 | - | - | - | - | - | 256 | 32 | 0.5 | 16 | 4 | 2 | 8 | 64 | 0.125 |
| C. sedlakii | NR4573 | CAZ 64 | P167Q | - | - | - | 128 | 8 | 0.25 | 256 | 1 | 2 | 8 | 128 | 0.125 |
| C. sedlakii | NR4574 | CAZ 64 | D179G | - | - | - | 128 | 4 | 0.25 | >256 | 1 | 2 | 8 | 16 | 0.125 |
| C. sedlakii | NR4575 | CAZ 64 | I173M, P174A, 174_175insS | - | - | - | 64 | 8 | 0.25 | >256 | 2 | 2 | 8 | 32 | 0.125 |
| C. sedlakii | NR4586 | FEP 8 | - | - | - | citC, cdsA, ispH | >256 | 256 | 0.25 | 16 | 16 | 2 | 4 | 256 | 0.5 |
| C. sedlakii | NR4062 | CMZ 8 | - | - | - | rseA | 256 | 64 | 16 | 16 | 16 | 32 | 64 | 128 | 0.25 |
| C. sedlakii | NR5701 | ATM 256 | - | - | - | tsuA, cdsA, ubiD | >256 | 128 | 0.25 | 16 | 8 | 1 | 4 | 256 | 0.5 |
| C. sedlakii | NR4584 | IPM 0.5 | - | - | - | pbpA, cdsA | 256 | 32 | 0.5 | 16 | 4 | 2 | 8 | 64 | 1 |
| E. coli | pCR4573/ML4947 | - | P167Q | - | - | - | 64 | 2 | ≤0.06 | 64 | 0.5 | 1 | 4 | 16 | 0.125 |
| E. coli | pCR2807/ML4947 | - | - | - | - | - | 256 | 8 | ≤0.06 | 4 | 1 | 4 | 4 | 16 | 0.125 |
| β-Lactamase and Parameter | Value for Antibiotic a | |||
|---|---|---|---|---|
| Piperacillin | Cefotaxime | Ceftazidime | Aztreonam b,c | |
| Wild-type (NR2807) | ||||
| kcat (s−1) | 624 ± 31.3 | 230 ± 21.3 | 4.55 ± 0.612 | 9.26 ± 0.133 |
| Km (μM) | 326 ± 29.5 | 264 ± 30.2 | 3484 ± 486 | 29.6 ± 2.65 |
| kcat/Km (s−1·mM−1) | 1910 | 871 | 1.31 | 313 |
| P167Q mutant (NR4573) | ||||
| kcat (s−1) | 163 ± 1.24 | 172 ± 3.12 | 0.649 ± 0.0542 | 4.42 ± 0.0496 |
| Km (μM) | 43.0 ± 1.56 | 417 ± 9.19 | 117 ± 11.9 | 55.1 ± 2.77 |
| kcat/Km (s−1·mM−1) | 3790 | 413 | 5.55 | 80.2 |
| D179G mutant (NR4574) | ||||
| kcat (s−1) | 2.57 ± 0.0551 | 0.213 ± 0.0017 | 0.0436 ± 0.0013 | NH |
| Km (μM) | 10.5 ± 1.22 | 5.34 ± 0.273 | 2.87 ± 0.46 | NH |
| kcat/Km (s−1·mM−1) | 245 | 39.9 | 15.2 | ND |
| I173M, P174A, 174_175insS mutant (NR4575) | ||||
| kcat (s−1) | 62.5 ± 2.13 | 6.06 ± 0.262 | 7.94 ± 0.862 | NH |
| Km (μM) | 84.0 ± 6.55 | 13.6 ± 1.89 | 196 ± 24.1 | NH |
| kcat/Km (s−1·mM−1) | 744 | 446 | 40.5 | ND |
| Bacterial Strains or Plasmids | Characteristics a |
|---|---|
| Strains | |
| NR2807 | Clinical isolate of C. sedlakii from Japan |
| ATCC51115 | Reference strain of C. sedlakii purchased from NITE Biological Resource Center |
| NR4573 | Ceftazidime-resistant mutant of C. sedlakii NR2807 with Sed-1 mutation (P167Q) |
| NR4574 | Ceftazidime-resistant mutant of C. sedlakii NR2807 with Sed-1 mutation (D179G) |
| NR4575 | Ceftazidime-resistant mutant of C. sedlakii NR2807 with Sed-1 mutation (I173M, P174A, 174_175insS) |
| ML4947 | E. coli (F− galK2 galT22 hsdR hsdM lacY1 metB1 relA supE44 Rif r), cloning host with AmpD wild type |
| ML4953 | E. coli (F− galK2 galT22 hsdR hsdM lacY1 metB1 relA supE44 Rif r ampD9), cloning host with AmpD mutant |
| TOP10 | E. coli (F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara leu)7697 galE15 galK16 rpsL(StrR) endA1 nupG, cloning host for analyzing the Sed-1 β-lactamase production |
| BL21(DE3) | E. coli (F− ompT hsdSB(rB− mB−) gal dcm (DE3)), cloning host for analyzing enzyme kinetic |
| Plasmids | |
| pCR2807 | pCR-Blunt II-TOPO containing blased-1 and sedR from NR2807 amplified using Sed-1down and SedRdown |
| pCR2807ΔSedR | pCR-Blunt II-TOPO containing blased-1 from NR2807 amplified using Sed-1down and SedRUpR |
| pCR51115 | pCR-Blunt II-TOPO containing blased-1 and sedR from pCR51115 amplified using Sed-1down and SedRdown |
| pCR51115ΔSedR | pCR-Blunt II-TOPO containing blased-1 from pCR51115 amplified using Sed-1down and SedRUpR |
| pCR4573 | pCR-Blunt II-TOPO containing blased-1 and sedR from NR4573 amplified using Sed-1down and SedRdown |
| pAmpR135D | pMW219 containing ampR fragment of wild type (AmpR135Asp) obtained from [12] |
| pAmpR135A | pMW219 containing ampR fragment of mutant (AmpR135Ala) obtained from [12] |
| pCR-Blunt II-TOPO | Cloning vector purchased from Thermo Fisher Scientific, Km r Zeo r |
| pMW219 | Cloning vector purchased from Nippon Gene, Km r |
| pET-28a (+) | Protein expression vector purchased from Novagen, Km r |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watanabe, M.; Nakano, R.; Yamamoto, K.; Nakano, A.; Suzuki, Y.; Saito, K.; Nakashima, S.; Endo, K.; Narita, K.; Yano, H. Unique Regulation of Sed-1 β-Lactamase in Citrobacter sedlakii: Insights on Resistance to Third-Generation Cephalosporin. Antibiotics 2025, 14, 823. https://doi.org/10.3390/antibiotics14080823
Watanabe M, Nakano R, Yamamoto K, Nakano A, Suzuki Y, Saito K, Nakashima S, Endo K, Narita K, Yano H. Unique Regulation of Sed-1 β-Lactamase in Citrobacter sedlakii: Insights on Resistance to Third-Generation Cephalosporin. Antibiotics. 2025; 14(8):823. https://doi.org/10.3390/antibiotics14080823
Chicago/Turabian StyleWatanabe, Mako, Ryuichi Nakano, Keizo Yamamoto, Akiyo Nakano, Yuki Suzuki, Kai Saito, Satoko Nakashima, Kentaro Endo, Kazuya Narita, and Hisakazu Yano. 2025. "Unique Regulation of Sed-1 β-Lactamase in Citrobacter sedlakii: Insights on Resistance to Third-Generation Cephalosporin" Antibiotics 14, no. 8: 823. https://doi.org/10.3390/antibiotics14080823
APA StyleWatanabe, M., Nakano, R., Yamamoto, K., Nakano, A., Suzuki, Y., Saito, K., Nakashima, S., Endo, K., Narita, K., & Yano, H. (2025). Unique Regulation of Sed-1 β-Lactamase in Citrobacter sedlakii: Insights on Resistance to Third-Generation Cephalosporin. Antibiotics, 14(8), 823. https://doi.org/10.3390/antibiotics14080823







