Abstract
Background/Objectives: Antimicrobial resistance (AMR) contributes to millions of deaths globally each year, creating an urgent need for new therapeutic agents. Antimicrobial peptides (AMPs) have emerged as promising candidates due to their potential to combat AMR pathogens. This study aimed to evaluate the antimicrobial activity of an AMP from a soil-derived bacterial isolate against Gram-negative bacteria. Method: Soil bacteria were isolated and screened for antimicrobial activity. The bioactive peptide was purified and determined its structure and antimicrobial efficacy. Genomic analysis was conducted to predict the biosynthetic gene clusters (BGCs) responsible for AMP production. Results: Genomic analysis identified the isolate as Paenibacillus sp. Na14, which exhibited low genomic similarity (61.0%) to other known Paenibacillus species, suggesting it may represent a novel species. The AMP from the Na14 strain exhibited heat stability up to 90 °C for 3 h and retained its activity across a broad pH range from 3 to 11. Structural analysis revealed that the Na14 peptide consisted of 14 amino acid residues, adopting an α-helical structure. This peptide exhibited bactericidal activity at concentrations of 2–4 µg/mL within 6–12 h, and its killing rate was concentration-dependent. The peptide was found to disrupt the bacterial membranes. The Na14 peptide shared 64.29% sequence similarity with brevibacillin 2V, an AMP from Brevibacillus sp., which also belongs to the Paenibacillaceae family. Genomic annotation identified BGCs associated with secondary metabolism, with a particular focus on non-ribosomal peptide synthetase (NRPS) gene clusters. Structural modeling of the predicted NRPS enzymes showed high similarity to known NRPS modules in Brevibacillus species. These genomic findings provide evidence supporting the similarity between the Na14 peptide and brevibacillin 2V. Conclusions: This study highlights the discovery of a novel AMP with potent activity against Gram-negative pathogens and provides new insight into conserved AMP biosynthetic enzymes within the Paenibacillaceae family.
1. Introduction
Infections caused by ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) pose a significant global public health threat due to their high morbidity and mortality rates [1]. The World Health Organization (WHO) identifies them as critical concerns because they quickly develop resistance to many antibiotics. From 1990 to 2021, drug-resistant infections, including sepsis, have been linked to an estimated 15 to 20 million deaths worldwide [2]. The WHO has implemented action plans aimed at mitigating the impact of multidrug-resistant infections. These plans emphasize improving awareness of antimicrobial resistance, promoting the rational use of antimicrobial agents, and encouraging the discovery of novel antimicrobial compounds [3].
Antimicrobial peptides (AMPs) are essential natural compounds produced by living organisms to protect themselves against pathogenic infections [4]. These peptides are composed of amino acid sequences that typically combine both polar and non-polar residues, resulting in amphipathic properties that enable them to target and disrupt the membranes of pathogenic cells. AMPs isolated from natural sources are highly diverse, particularly among microbes, leading to a wide variety of AMP classes with distinct mechanisms of action. Their physical interaction with bacterial membranes, involving both polar and hydrophobic regions, allows AMPs to penetrate and destabilize membrane integrity through various models such as the carpet, toroidal pore, barrel-stave, and aggregate mechanisms [5]. The ability of AMPs to physically interact with cell membranes reduces the likelihood of pathogens developing resistance, especially when compared to traditional antibiotics that target specific receptor proteins. This characteristic makes AMPs a promising strategy for combating antibiotic-resistant infections [6].
Soil bacteria have emerged as valuable sources for antibiotic discovery. For example, polymyxins are a class of polypeptide antibiotics derived from Paenibacillus polymyxa, commonly used to treat infections caused by Gram-negative bacteria [7]. Similarly, daptomycin, a cyclic lipopeptide produced by Streptomyces roseosporus, is developed for the treatment of infections caused by susceptible and resistant Gram-positive bacteria [8]. Genomic technology and big data have provided powerful tools for utilizing genetic information to explore the genetic sequences and functions of soil bacteria. Bacterial genomes represent an essential resource for unlocking valuable information from the blueprint of living organisms [9]. Genomic data can reveal functional genes responsible for antimicrobial production, particularly through the identification of biosynthetic gene clusters (BGCs). These discoveries offer significant potential for applications in the biotechnology industry and pharmaceutical development [10].
This research focuses on exploring potential bacterial strains capable of producing AMP with the ability to inhibit specific pathogenic bacteria. This study included the purification and structural analysis of the AMP, along with proposing its mechanism of action against globally significant pathogens. Taxonomic identification and functional gene analysis will be performed using genomic data and bioinformatic tools, aiming to identify the BGCs responsible for AMP production. This work seeks to advance the understanding of novel AMPs and to establish a foundation for the development of new antimicrobial agents for future preclinical and clinical studies.
2. Results
2.1. Isolation and Screening of Soil-Derived Bacteria for Antibacterial Activity
Bacterial strains were isolated from soil samples collected in the south of Thailand. A preliminary study on the cross-streak and agar well diffusion assay revealed that only one isolate, designated Na14, exhibited inhibitory activity against E. coli TISTR 887 and S. aureus TISTR 517 (Figures S1 and S2). The production of bioactive compounds by this bacterial isolate was evaluated by monitoring its growth curve and antibacterial activity. The result showed that the lag phase was observed at the first 4 h of incubation, and the cell-free supernatant (CFS) exhibited no detectable antibacterial activity (Figure 1). The exponential and the stationary phases occurred between 4–24 h and 24–168 h of the incubation period, respectively. Notably, the Na14 isolate produced a significantly higher level of bioactive compounds, with maximal antibacterial activity observed at 24 h. At this point, coinciding with the onset of the stationary phase, the inhibition zones of the CFS were 18.12 ± 0.32 mm for E. coli TISTR 887 and 12.11 ± 0.24 mm for S. aureus TISTR 517. The CFS retained its antibacterial activity without significant variation between 48 and 168 h. Moreover, it was demonstrated that the active substances had higher efficacy against the tested Gram-negative bacteria compared to Gram-positive bacteria.
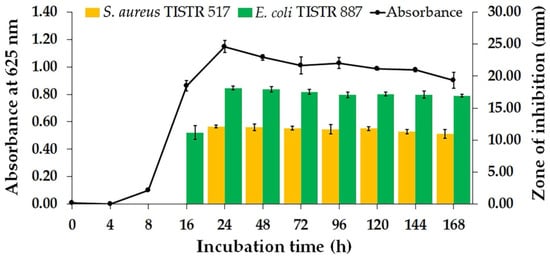
Figure 1.
Bacterial growth curve and kinetics of antibacterial substance production by bacterial isolate Na14.
The results of the production kinetics study revealed that the Na14 isolate produced active compounds with maximal efficacy at 24 h of incubation, indicating this as the optimal incubation period. Subsequently, the CFS obtained at 24 h of cultivation was prepared and evaluated for its antimicrobial activity against additional pathogenic bacteria (Table 1). The bioactive compounds from the Na14 strain exhibited significantly larger inhibition zones against Gram-negative bacteria, ranging from 18.19 ± 0.46 to 18.44 ± 0.26 mm, compared to Gram-positive bacteria, which ranged from 11.33 ± 0.41 to 12.45 ± 0.73 mm. These findings indicated that the Na14 strain predominantly produced compounds with higher efficacy against Gram-negative bacteria. Moreover, colistin and vancomycin were used as positive controls for Gram-negative and Gram-positive bacteria, respectively. The zones of inhibition ranged from 26.52 ± 0.43 to 29.41 ± 0.65 mm for colistin and 28.50 ± 0.22 to 28.96 ± 0.43 mm for vancomycin.

Table 1.
Antimicrobial activity of the CFS derived from the Na14 isolate. The diameter of the inhibition zones observed in the agar well diffusion assay was measured.
2.2. Purification of Bioactive Compounds of the Na14 Isolate
The antimicrobial compounds in the CFS of the Na14 isolate were initially purified using ammonium sulfate precipitation. The fractions obtained at 50–75% saturation exhibited the greatest inhibition zone against E. coli TISTR 887. These active fractions were further purified using cation-exchange chromatography (CIEX), followed by reversed-phase chromatography (RPC). A single peak corresponding to the bioactive compounds was observed during RPC when the concentration of the elution solvent reached 51% (Figure 2a). A purification balance was compiled to quantitatively assess the efficiency at each stage of the purification process (Table 2). As the purification process progressed, the total amount of active peptides decreased, while their specific activity increased. After salt precipitation, the resulting fraction exhibited a specific activity of 60.31 AU/mg, representing a 3.23-fold increase in purity. The specific activity of the Na14 peptide increased from 100.63 to 835.99 AU/mg following sequential purification by CIEX and RPC. Additionally, the peptide purity increased by 5.38-fold and 44.73-fold after each respective purification step.
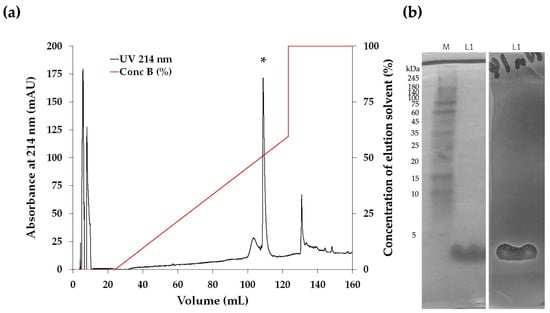
Figure 2.
(a) Reversed-phase HPLC chromatogram of bioactive compounds obtained from the Na14 isolate. The purified fraction was obtained by RPC. Each peak fraction was evaluated for antimicrobial activity against E. coli TISTR 887 using the agar well diffusion method. An asterisk (*) indicates the peak fractions containing the active compounds. (b) SDS-PAGE and gel overlay inhibition assay of the purified Na14 peptide. (Left) Peptides obtained from RPC (L1) were analyzed using 15% SDS-PAGE and visualized by Coomassie blue staining. A protein marker (M) was included for molecular weight comparison. (Right) The SDS-PAGE gel was overlaid with soft agar containing E. coli TISTR 887 and incubated at 37 °C for 18 h. An inhibition zone corresponding to the Na14 peptide was observed.

Table 2.
Purification balance sheet of the Na14 peptide against E. coli TISTR 887.
The purified peptide was subsequently analyzed using 15% SDS-PAGE to estimate its molecular weight. The results showed that fractions with purified active compounds obtained from RPC exhibited a peptide band with a molecular weight below 5 kDa. The corresponding gel was subsequently overlaid with soft agar containing E. coli TISTR 887. An inhibition zone was observed at the same position as the peptide band, indicating that the Na14 peptide exhibited antimicrobial activity (Figure 2b). These findings suggest that the purification process effectively separated the Na14 peptide while retaining its antimicrobial activity. Consequently, the purified peptide was subjected to further characterization.
2.3. Characterization of the Na14 Peptide
The purified Na14 peptide obtained from RPC was analyzed using liquid chromatography–tandem mass spectrometry (LC-MS/MS). The peptides were ionized and subsequently fragmented into several product ions, especially b- and y-ions at peptide bonds. The amino acid sequence was determined using a de novo sequencing approach. The results showed that the Na14 peptide consisted of 14 amino acid residues with a molecular mass of 1555.05 Da. The peptide sequence was identified as LALLVVVKVLKYVV with an average local confidence (ALC) score of 78% (Figure 3). The physicochemical properties of the Na14 peptide were predicted using ProtParam (https://web.expasy.org/protparam; accessed on 26 June 2025) [11]. The predicted isoelectric point (pI) was 9.70. The presence of two lysine (K) residues contributed to a net positive charge of +2 at pH 7.4. The abundance of hydrophobic amino acids, such as valine (V), leucine (L), tyrosine (Y), and alanine (A), resulted in a high hydrophobicity index of 0.958 and a hydrophobic moment of 0.249. It indicated that the Na14 peptide possessed amphipathic characteristics. Moreover, this peptide had the predicted instability index of –4.90, suggesting that it is likely to be stable under in vivo conditions [12].
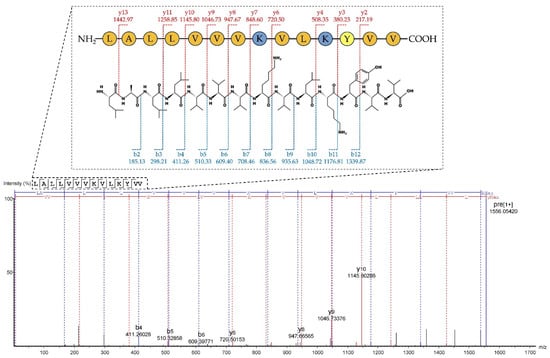
Figure 3.
The de novo sequencing of the Na14 peptide by LC-MS/MS. The fragmentation patterns corresponding to b- and y-ion series were observed, and the resulting mass differences were analyzed to reconstruct the amino acid sequence.
The content of the secondary structure of the Na14 peptide was analyzed based on its circular dichroism (CD) spectrum (Figure 4a). When dissolved in deionized water, the peptide exhibited a negative CD band near 200 nm and low ellipticity above 235 nm, indicating a predominance of random coil (56.8%) and turn (23.7%) conformations (Table 3). In contrast, in the presence of SDS micelles served as a membrane-mimicking agent, the CD spectrum displayed a positive band at 195 nm and two negative bands around 210 nm and 222 nm, suggesting an increase in α-helical content (24.4%) and a corresponding decrease in the proportion of random coil structure (43.9%). The structural model of the Na14 peptide was generated using AlphaFold, revealing that the peptide adopted the α-helical conformation (Figure 4b). The helical wheel projection produced using HeliQuest (https://heliquest.ipmc.cnrs.fr/index.html; accessed on 26 June 2025) indicated that the polar amino acids were positioned on one side of the helix, while the non-polar residues were located on the opposite side (Figure 4c).
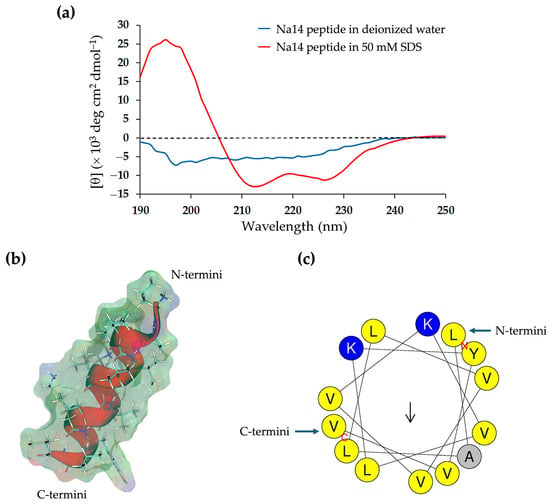
Figure 4.
(a) Circular dichroism spectra of the Na14 peptide. The peptide was dissolved in either deionized water (blue) or a 50 mM SDS solution (red). (b) Structural modeling of the Na14 peptide using AlphaFold. The peptide structure is shown in cartoon style with side chains displayed as stick-and-ball models and overlaid with the molecular surface. The surface is color-coded to indicate polarity: blue for positively charged regions, red for negatively charged regions, and green for non-polar areas. (c) Predicted α-helical conformation of the Na14 peptide, generated by HeliQuest. The helical wheel projection illustrates the amphipathic nature of the peptide using single-letter amino acid codes and color shading: blue for polar residues, yellow for non-polar residues, and grey for small residues.

Table 3.
Predicted secondary structure of the Na14 peptide. The spectra of the purified Na14 peptides in deionized water or 50 mM SDS solution were determined by CD. The CONTINLL method was used to predict the peptide secondary structures.
2.4. Antimicrobial Activity of the Na14 Peptide
The antimicrobial activity of the purified Na14 peptide was evaluated using the broth microdilution assay. The results demonstrated that the Na14 peptide exhibited strong activity against Gram-negative bacteria, including E. coli TISTR 887, P. aeruginosa TISTR 357, and K. pneumoniae TISTR 1383, with minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) values ranging from 2 to 4 µg/mL (Table 4). In contrast, the peptide was ineffective against Gram-positive bacteria such as S. aureus TISTR 517 and MRSA strain 2468, with MIC and MBC values exceeding 128 µg/mL. Furthermore, the antimicrobial potency of the Na14 peptide against Gram-negative bacteria was comparable to that of colistin, which exhibited MIC and MBC values between 1 and 2 µg/mL. In comparison, vancomycin showed superior efficacy against Gram-positive bacteria, with MIC and MBC values of 2 and 4 µg/mL, respectively.

Table 4.
The MIC and MBC values of the Na14 peptide against the tested bacterial strains were determined using the broth microdilution assay. The standard antibiotics, including colistin and vancomycin, were used as controls. All experiments were conducted in triplicate.
2.5. Time–Kill Kinetics of the Na14 Peptide
The time–kill curve demonstrated that the Na14 peptide at 0.5× MIC temporarily suppressed the growth of E. coli TISTR 887 and P. aeruginosa TISTR 357 during the first 3 h of incubation (Figure 5a,b), while the growth of K. pneumoniae TISTR 1383 was inhibited for up to 6 h (Figure 5c). Thereafter, all bacterial strains resumed growth, which continued to increase until the end of the 24 h observation period. In contrast, treatment with higher peptide concentrations effectively killed all tested bacteria. Specifically, the peptide at 1× MIC eliminated E. coli TISTR 887, P. aeruginosa TISTR 357, and K. pneumoniae TISTR 1383 at 12, 6, and 9 h, respectively. Furthermore, the peptide at 2× MIC completely eradicated E. coli TISTR 887 and K. pneumoniae TISTR 1383 at 6 h and P. aeruginosa TISTR 357 at 3 h. The killing rate of the Na14 peptide was calculated to evaluate its effect on cell reduction. The killing rates of the peptide at 1× MIC were 1.33 ± 0.07, 2.47 ± 0.09, and 1.64 ± 0.05 h−1 against E. coli TISTR 887, P. aeruginosa TISTR 357, and K. pneumoniae TISTR 1383, respectively. When treated with 2× MIC of Na14 peptide, the corresponding killing rates increased to 2.68 ± 0.09, 4.99 ± 0.12, and 2.42 ± 0.10 h−1, respectively. These results indicate that higher concentrations of the peptide enhance the bacterial killing rate.
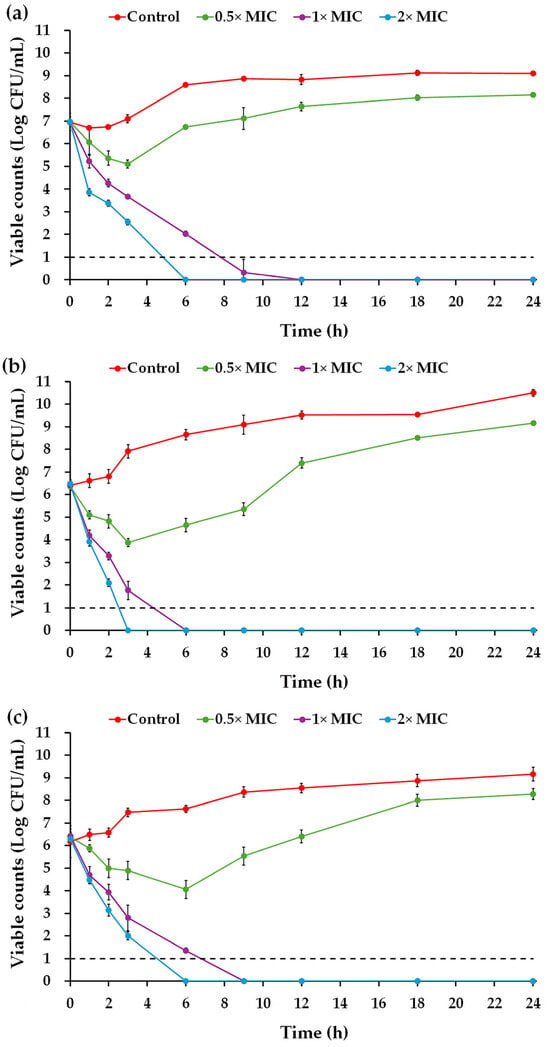
Figure 5.
Time–kill kinetics of the Na14 peptide. The Na14 peptides at different concentrations (0.5×, 1×, and 2× MIC) were tested against (a) E. coli TISTR 887, (b) P. aeruginosa TISTR 357, and (c) K. pneumoniae TISTR 1383. Untreated cells served as controls. At specified time intervals, either undiluted or diluted bacterial suspensions were plated on MH agar. Viable cell counts were determined and expressed on a logarithmic scale. The limit of detection (LOD) of the assay was 10 CFU/mL, indicated by a dashed line.
2.6. Morphological Changes of Bacterial Cells Induced by the Na14 Peptide
The morphologies of E. coli TISTR 887, P. aeruginosa TISTR 357, and K. pneumoniae TISTR 1383 following treatment with the Na14 peptide were examined and compared to those treated with colistin at the same concentration (1× MIC) using a scanning electron microscopy (SEM). In the untreated groups, all bacterial strains retained their rod-shaped morphology, indicating intact cell membranes (Figure 6). After 16 h of exposure to the Na14 peptide, cytoplasmic extrusion was observed, suggesting membrane disruption and signs of cell lysis. Similarly, cells treated with colistin also exhibited apparent evidence of lysis. However, the extent of cell lysis appeared to be more evident in the colistin-treated group compared to those treated with the Na14 peptide.
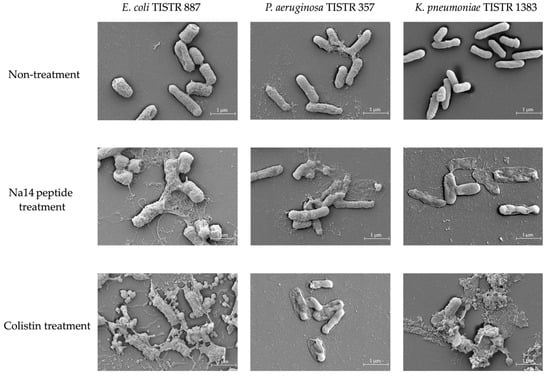
Figure 6.
SEM images showing morphological changes in bacterial cells following treatment with the Na14 peptide and colistin at 1× MIC level, compared to the untreated control. Images were captured at 20,000× magnification.
2.7. Effect of Temperature, pH, and Enzyme on Peptide Stability
The stability of the purified Na14 peptide was evaluated under various conditions, including heat exposure, enzymatic digestion, and pH variation. A concentration of 10× MIC of Na14 peptide was used to assess its antimicrobial activity against the tested pathogens. The remaining activity of the treated samples was compared to that of the untreated control (Table 5). Thermal stability analysis revealed that the peptide retained its antimicrobial activity against all pathogens across a temperature range of 30 °C to 90 °C, with the residual activity ranging from 97.22 ± 1.62% to 100.89 ± 0.39%. The peptide also demonstrated tolerance to a broad pH range between pH 3 and 11, maintaining activity between 97.62 ± 2.08% and 100.85 ± 1.95% throughout the study. Enzymatic stability was evaluated using various proteases. Among these, trypsin treatment exhibited the most significant impact, markedly reducing the antimicrobial activity of the Na14 peptide across all tested bacterial strains, with reductions ranging from 88.72 ± 0.75% to 89.53 ± 1.30%. Treatments with α-chymotrypsin or proteinase K significantly caused moderate reductions in the Na14 peptide activity between 93.23 ± 2.60% and 96.50 ± 1.56%. Furthermore, no significant differences in residual activity were observed across the tested pathogens under any condition, indicating that the Na14 peptide retained consistent antimicrobial activity regardless of the bacterial species.

Table 5.
The residual antimicrobial activity of the Na14 peptide against the tested bacterial strains was evaluated following exposure to various conditions. Results are expressed as the mean ± standard deviation (SD) from three independent experiments.
2.8. Genome Analysis and Bacterial Identification
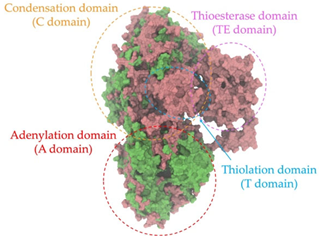
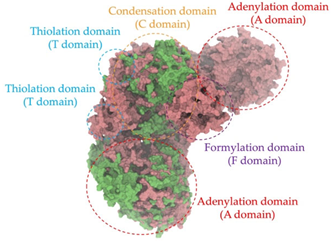
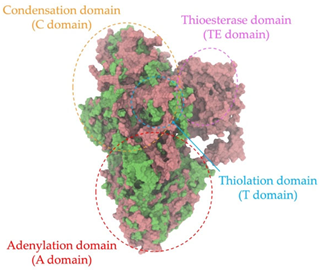
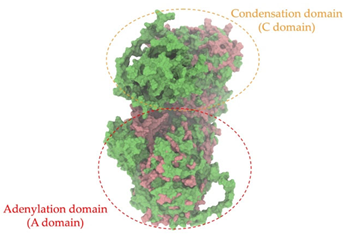
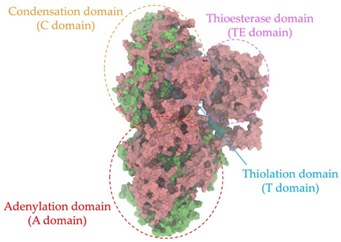
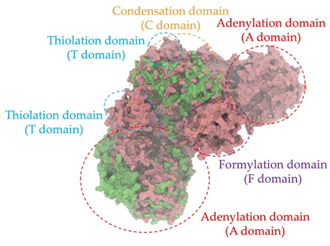
A single colony of the Na14 strain was observed under a light microscope at 1000× magnification, revealing Gram-positive, rod-shaped cells with endospore-forming capability. The genomic sequence of the Na14 strain was obtained using the Illumina HiSeq platform. Paired-end raw reads were quality-checked using FastQC, followed by de novo assembly using SPAdes (version 3.14.1). The assembled genome demonstrated a completeness of 99.14% with a sequencing coverage of 236×. The result revealed a circular genome comprising 7,789,831 base pairs, deposited under the accession number CP186533 at the National Center for Biotechnology Information (NCBI). Species identification was performed using the Type (Strain) Genome Server (TYGS) and constructed phylogenetic tree with MEGA X software (version 10.1.8). The Na14 strain showed the highest similarity to Paenibacillus oleatilyticus SM69, with digital DNA–DNA hybridization (dDDH) values of 79.1% (95% CI: 75.1–82.5) for formula d0, 61.0% (95% CI: 58.1–63.8) for formula d4, and 78.1% (95% CI: 74.7–81.2) for formula d6 (Figure 7a). Taxonomic classification placed the Na14 strain within the genus Paenibacillus, but the dDDH value for formula d4 remained below the 70% threshold, suggesting that Na14 may represent a novel species. The circular genome map of the Na14 strain was visualized using the Proksee platform (CGViewBuilder version 2.0.5, https://proksee.ca, accessed on 10 March 2025), based on gene annotations provided by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP). The annotated genome comprised a total of 7130 genes, including 7054 coding DNA sequences (CDSs) and 76 RNA genes. The RNA genes consisted of 3 rRNAs, 69 tRNAs, and 4 non-coding RNAs (ncRNAs). The potential for secondary metabolite production was assessed using the Antibiotics and Secondary Metabolite Analysis Shell (antiSMASH; version 7.0). A total of 27 biosynthetic gene clusters (BGCs) were identified, including 4 terpene clusters, 1 opine-like metallophore cluster, 11 non-ribosomal peptide synthetase (NRPS) clusters, 7 polyketide synthase (PKS) clusters, 2 ribosomally synthesized and post-translationally modified peptide (RiPP) clusters, and 2 cyclic lactone auto-inducer clusters. The non-ribosomal peptide synthetase (NRPS) modules identified in the genome of the Na14 strain were composed of adenylation (A), condensation (C), thiolation (T), and thioesterase (TE) domains. These domains were visualized on the circular genome map to illustrate the distribution of NRPS-related genes (Figure 7b). In addition, antibiotic resistance genes (ARGs) were identified by comparing the annotated genome against reference sequences from the Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca, accessed on 10 March 2025). A total of 11 antimicrobial resistance genes were detected, including 8 genes associated with glycopeptide resistance, exhibiting 33.75–36.90% identity and 55.48–133.51% coverage; 1 gene related to phenicol resistance, showing 68.06% identity and 99.09% coverage; 1 gene conferring cephalosporin resistance, with 67.45% identity and 100.00% coverage; and 1 gene involved in antibiotic efflux, displaying 90.67% identity and 100.00% coverage. The prediction of antimicrobial production-related genes and antimicrobial resistance genes supports the hypothesis that the Na14 strain possesses both antimicrobial biosynthesis capabilities and self-defense mechanisms.
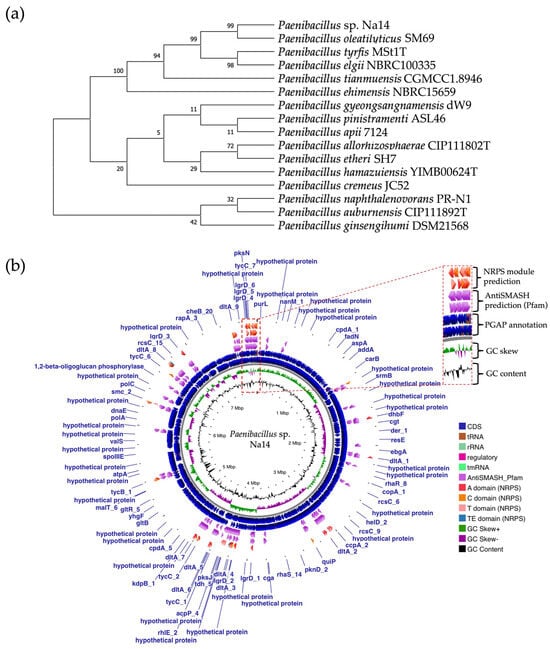
Figure 7.
Genomic identification and taxonomic classification of Paenibacillus sp. Na14. (a) The phylogenetic tree illustrates the relationship of the Na14 strain to other members of the Paenibacillus genus, showing the closest taxonomic affiliation to Paenibacillus oleatilyticus SM69. (b) The circular genome map displays annotated genes along with predicted biosynthetic gene clusters associated with secondary metabolite production. NRPS-encoding regions and their domains are visualized in detail.
2.9. Comparative Analysis of NRPS-Encoding Biosynthetic Gene Clusters Involved in AMP Production
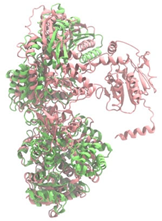
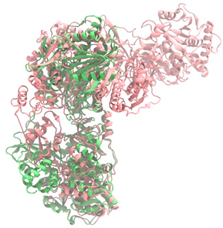
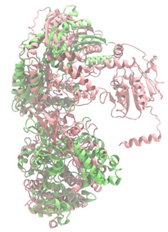
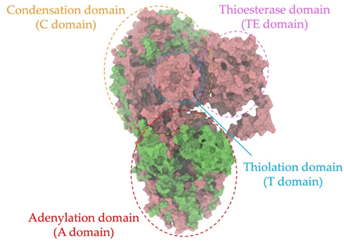
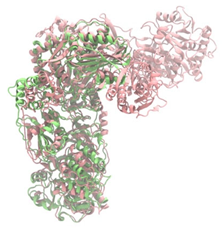
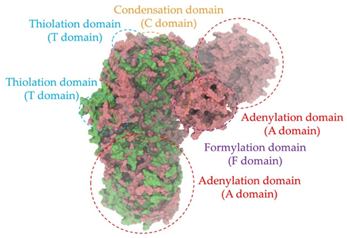
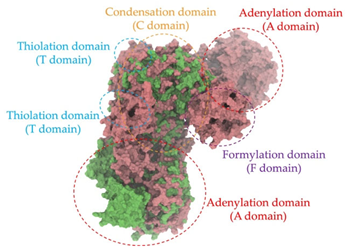
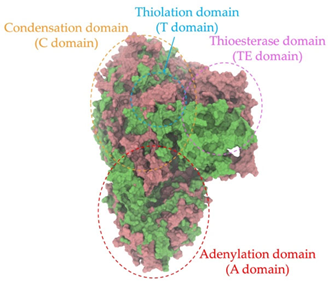
Biosynthetic gene clusters (BGCs) encoding non-ribosomal peptide synthetases (NRPS) play a crucial role in determining the AMP production. The Na14 peptide showed a similarity of amino acid sequence to brevibacillin 2V, produced by the NRPS BGCs of Brevibacillus laterosporus DSM 25 [13]. These findings support further in-depth investigation of the NRPS modules in the Na14 strain to characterize the BGCs involved in AMP biosynthesis and to propose their functional roles in AMP production by Paenibacillus sp. Na14. The amino acid sequences obtained from genomic analysis of NRPS modules by antiSMASH were modeled using the AlphaFold Server (https://alphafoldserver.com, accessed on 20 March 2025). The predicted protein structures were compared to known reference structures using FoldSeek (https://search.foldseek.com, accessed on 20 March 2025), and the best-matching structural homologs were identified based on sequence identity and template modeling (TM) scores. The NRPS architecture was proposed to consist of 1 initial module containing A and T domains, followed by 12 elongation modules composed of C, A, and T domains, and 1 termination module comprising C, A, T, and TE domains. The initial NRPS module in Paenibacillus sp. Na14 exhibited structural similarity to the thiolation-state module of linear gramicidin synthetase subunit A from Brevibacillus parabrevis (PDB ID: 5ES8), with a sequence (seq.) identity of 39.5% and a TM score of 0.83386. The elongation process of the peptide synthesis was predicted to involve 12 modules. It shared structural similarity with the condensation-state module of the same enzyme from Brevibacillus parabrevis (PDB ID: 6MFZ), with the seq. identities ranging from 31.9% to 40.2% and TM-scores between 0.74125 and 0.88822. It was also similar to the termination module of surfactin A synthetase C from Bacillus subtilis (PDB ID: 2VSQ), exhibiting seq. identities ranging from 34.0% to 41.7% and TM-scores between 0.46144 and 0.77661. The termination module of Na14-synthesizing enzyme showed structural similarity to the termination module of surfactin A synthetase C (PDB ID: 2VSQ), with a seq. identity of 33.7% and a TM-score of 0.63631. These structural similarities are supported by the taxonomic classification of Paenibacillus sp. Na14 and Brevibacillus parabrevis, which both belong to the family Paenibacillaceae. Moreover, Paenibacillus sp. Na14 and Bacillus subtilis are members of the order Bacillales. Notably, one of the elongation modules also showed 27.0% seq. identity and a TM-score of 0.77945 to the serine module of the teixobactin-producing NRPS Txo2 from Eleftheria terrae (PDB ID: 6P1J), a bacterium from a distinct phylum (Pseudomonadota). The observed structural overlap between the predicted NRPS modules of the Na14 peptide and known NRPS structures in the Protein Data Bank (PDB) supports a conserved topological organization, suggesting their involvement in antimicrobial peptide biosynthesis within the genome of Paenibacillus sp. Na14 (Table 6).

Table 6.
Structure prediction and structural similarity analysis of the NRPS from the Na14 genome compared to reference protein structures in the PDB.
3. Discussion
Bacterial infections continue to pose a significant global health threat, particularly those caused by antibiotic-resistant strains [14]. Recently, the WHO has revised the list of bacterial priority pathogens in 2024 to guide investments in the development of new and effective antibiotics and to support public health strategies for the surveillance and control of antimicrobial resistance [15]. Several Gram-negative pathogens exhibiting intrinsic or acquired antibiotic resistance are classified among the highest priority levels. For example, E. coli and K. pneumoniae are categorized as critical priority pathogens, while P. aeruginosa is listed as a high-priority group. Numerous research sectors are actively seeking novel antimicrobial agents, with particular emphasis on those derived from soil bacteria.
In this study, a soil bacterium designated as Na14 was isolated and found to exhibit antimicrobial activity against E. coli TISTR 887 and S. aureus TISTR 517. The Na14 isolate produced bioactive compounds exhibiting maximal antimicrobial activity at 24 h of incubation. This time point corresponded to the early stationary phase, a stage in which bacteria commonly produce secondary metabolites, such as antibiotics or antimicrobial peptides, due to various physiological and ecological factors [16]. For example, bacitracin produced by Bacillus paralicheniformis began the synthesis after 5 h of incubation, and its production level was high after 8–10 h of growth, corresponding to the onset of the stationary phase [17]. In addition, the production of surfactin lipopeptide by Bacillus velezensis SK was first detected at the mid-exponential phase at 16 h. Surfactin production reached its maximum level by the end of the stationary phase at 36 h, likely due to nutrient-depleted conditions [18].
The bioactive compound from the Na14 strain was identified as an AMP with the amino acid sequence of LALLVVVKVLKYVV. This peptide exhibited amphipathic characteristics, comprising a cationic region and a hydrophobic domain. Additionally, the Na14 peptide was classified as a valine-rich (42.9%) and leucine-rich (28.6%) AMP, as the content of each amino acid exceeded 25% [19]. The high abundance of these hydrophobic amino acids might exhibit antimicrobial properties through their hydrophobic nature and membrane-disruptive capabilities. A previous study analyzing AMPs in the Antimicrobial Peptide Database (APD) reported that 21% of AMPs met this criterion of amino acid richness. Notably, leucine- and valine-rich AMPs, such as gramicidins B and C, were common among effective AMPs from bacteria [19]. The Na14 peptide was a novel AMP because its sequence was not included in the Database of Antimicrobial Activity and Structure of Peptides (DBAASP) (https://dbaasp.org/search; 23,036 peptides; accessed on 23 June 2025) and the Antimicrobial Peptide Database (APD) (https://aps.unmc.edu/database; 5513 peptides; accessed on 23 June 2025). The Na14 peptide sequence was aligned with other AMPs in the APD database. The highest sequence similarity (64.29%) was observed with brevibacillin 2V from Brevibacillus laterosporus DSM 25 (APD ID: AP03330) [13].
The CD spectrum indicated that the Na14 peptide in water predominantly adopted random coil and turn structures. SDS micelles, commonly used as membrane-mimicking agents, provide a hydrophobic environment that simulates the lipid bilayer of cell membranes. These micelles are often employed to assess changes in peptide conformation and interactions, thereby offering insights into peptide-membrane interactions [20]. In the presence of SDS micelle, the Na14 peptide underwent a conformational transition characterized by an increase in α-helical content, as reflected in its distinct CD spectrum. It is presumed that the Na14 peptide adsorbs onto the micellar surface and undergoes a conformational transition from a random coil to an α-helix [21]. Similar conformational changes in certain AMPs have been reported in previous studies. For example, brevibacillin initially exhibited a random coil structure but underwent a transition to an α-helical structure (43.14%) upon the addition of SDS [21]. This suggested that brevibacillin disrupted the cytoplasmic membrane of bacterial cells, ultimately leading to cell death. In addition, brevibacillin 2V was found to bind to the cell wall synthesis precursor Lipid II, inducing a significant conformational change characterized by an increased α-helical content [13]. Brevibacillin 2V also caused bacterial membrane permeability, supporting its bactericidal mechanism of action. Therefore, the structural transition of the Na14 peptide to an α-helical conformation may represent a mechanism underlying its activity, which requires further characterization.
The Na14 peptide demonstrated potent antimicrobial activity against Gram-negative bacteria, including E. coli TISTR 887, P. aeruginosa TISTR 357, and K. pneumoniae TISTR 1383. Its activity was comparable to that of colistin, exhibiting a bactericidal effect at its 1× MIC level. Brevibacillin 2V, a structurally similar peptide derived from Brevibacillus laterosporus DSM 25, has been reported to display broad-spectrum antimicrobial activity against both Gram-negative and Gram-positive bacteria. Notably, brevibacillin 2V showed MIC values of 16 µg/mL against E. coli ATCC 25922, 64 µg/mL against P. aeruginosa LMG6395, 32 µg/mL against K. pneumoniae LMG20218, and 2 µg/mL against MRSA ATCC 15975 [13]. The time–kill kinetic assay indicated that the Na14 peptide at 1× MIC exerted bactericidal activity, reducing bacterial counts by more than 3 log units. Its efficacy was concentration-dependent, with higher peptide concentrations resulting in an increased bacterial killing rate. The Na14 peptide at 2× MIC completely killed E. coli TISTR 887 and K. pneumoniae TISTR 1383 within 6 h, and P. aeruginosa TISTR 357 within 3 h. In comparison, previous studies have shown that brevibacillin 2V at 10× MIC achieved complete killing of MRSA within 6 h, and a similar killing kinetics was observed for nisin [22]. These findings suggested that certain brevibacillins acted as fast-acting bactericidal peptides through disruption of the bacterial membranes. Additionally, brevicidine derived from Brevibacillus laterosporus DSM25 rapidly eradicated E. coli in 1 h when treated with the peptide at 4× MIC. In contrast, the pathogen remained viable at lower concentrations of brevicidine, ranging from 0.5× to 2× MIC. Brevicidine at higher concentrations exhibited its bactericidal activity by disrupting the outer membrane of Gram-negative bacteria, thereby facilitating its access to and subsequent interaction with inner membrane components [23]. SEM analysis revealed membrane damage in bacterial cells treated with either Na14 peptide or colistin at 1× MIC, showing clear morphological changes such as cell rupture and lysis. Previous SEM studies demonstrated that several AMPs targeted the bacterial membrane by forming pores and causing the extrusion of cellular contents, indicating membrane damage [24,25,26]. The Na14 peptide exhibited a MIC equal to its MBC, indicating bactericidal activity at 1× MIC. Its proposed mechanism involves the positively charged lysine residues facilitating electrostatic binding to the negatively charged bacterial cell wall and membrane. The hydrophobic regions then enable insertion into the lipid bilayer, leading to pore formation and subsequent cell lysis. However, the specific molecular interactions with membrane-integrated components remain to be elucidated and will be the focus of future studies.
Peptide stability is a critical factor influencing the effectiveness of AMP. The Na14 peptide demonstrated strong thermal tolerance across a wide temperature range (30–90 °C). The Na14 peptide is a linear AMP with an amino acid sequence similar to members of the brevibacillin and brevilaterin families, which have been previously isolated from Brevibacillus laterosporus [27]. Previous studies have reported that brevibacillins and brevilaterins exhibit excellent thermal stability, maintaining their antimicrobial activity across a wide temperature range from 40 °C to 100 °C [28,29]. The acid–base stability of Na14 peptide showed broad pH tolerance ranging from pH 3 to 11. In comparison, the structurally related peptides, such as brevibacillin, have been reported to remain stable between pH 5 and 7, while brevibacillin V demonstrated stability over a pH range of 5 to 9 [28]. Proteolytic enzymes pose a significant challenge to the efficacy of AMPs by cleaving peptide bonds. These peptide bonds form the structural backbone of AMPs and are essential for maintaining their stability and function. The Na14 peptide exhibited partial degradation by trypsin, α-chymotrypsin, and proteinase K, while retaining over 88% of its antimicrobial activity. Similarly, partial reductions in activity due to protease susceptibility have been reported for brevilaterins derived from Brevibacillus laterosporus S62-9 and certain AMPs produced by Brevibacillus laterosporus strains BGSP7, BGSP9, and BGSP11, indicating comparable enzymatic sensitivity to that observed in the Na14 peptide [30,31]. The overall activity of the Na14 peptide under various conditions demonstrated that it is a stable AMP, providing strong evidence for its potential in further pharmacokinetic studies and formulation development.
The Na14 strain was identified as Paenibacillus sp. Na14. Paenibacillus spp. are Gram-positive, endospore-forming bacteria capable of surviving in harsh environmental conditions. Genomic analyses have identified several BGCs that are crucial for the production of diverse antimicrobial compounds, many of which show potential for clinical application in the treatment of antimicrobial-resistant infections. Among these, bacteriocins, a class of AMPs produced through ribosomal synthesis, represent an important group of bioactive compounds [32]. Several studies have reported potent antimicrobial agents from Paenibacillus spp., such as Paenibacillus polymyxa OSY-DF, Paenibacillus polymyxa NRRL B-30509, and Paenibacillus sp. strain A3, producing paenibacillin, paenicidin A, and penisin, respectively [33,34,35]. Paenibacillus spp. also produced various AMPs through NRPS system, including members of the polymyxin family such as colistin. These compounds are well-known for their clinical use in treating antibiotic-resistant Gram-negative bacterial infections. Notably, polymyxins have been identified in Paenibacillus polymyxa, Paenibacillus alvei, and Paenibacillus kobensis [36,37]. A truncated structural variant of polymyxins, known as octapeptins, has been reported to exhibit antimicrobial activity against both Gram-positive and Gram-negative bacteria, while demonstrating low toxicity to mammalian cells [38]. In addition to the various BGCs of antimicrobial compounds found in Paenibacillus spp., natural resistance to self-produced antimicrobials plays a vital role in enabling the bacteria to tolerate their antimicrobial compounds. Numerous genes related to antimicrobial resistance have been reported in their genomic data, including ABC transporters and genes conferring resistance to both broad and specific classes of antibiotics. Resistance genes, identified in some Paenibacillus spp. including Paenibacillus sp. Na14, targeted lipopeptide antibiotics, penicillins, and cephalosporins, supporting the idea that these resistance mechanisms serve as self-defense systems against their antimicrobial products [39]. The genomic analysis of Paenibacillus sp. Na14 revealed the presence of 27 BGCs associated with secondary metabolism, including those responsible for the production of bacteriocins and NRPS-synthesizing AMPs. Numerous studies found genes related to secondary metabolite biosynthesis in various Paenibacillus species, supporting the functional potential observed in the Na14 strain. Interestingly, the amino acid sequence of the purified AMP isolated from the Na14 culture showed high similarity to that of brevibacillin 2V, a lipopeptide produced by Brevibacillus laterosporus DSM 25 [13]. Brevibacillus spp. are Gram-positive, endospore-forming bacteria that belong to the same taxonomic family of Paenibacillaceae, including Paenibacillus spp. AMP production in Brevibacillus spp. is predominantly mediated by NRPS systems. Several AMPs have been reported from Brevibacillus spp., including linear gramicidin A, B, and C, which are composed of 15 amino acid residues and are synthesized by 15 NRPS modules, each containing an A domain. The A domain is responsible for the selection and incorporation of specific amino acids during peptide chain synthesis. Linear gramicidin, originally isolated from Brevibacillus brevis, was one of the first AMPs successfully applied in a clinical setting [40,41]. Another group of AMPs produced by Brevibacillus spp. consists of NRPS-synthesized linear lipopeptides. These have been isolated from various species, including bogorols from Brevibacillus laterosporus, BT peptides from Brevibacillus texasporus, and brevibacillin 2V from Brevibacillus laterosporus DSM 25 [42,43,44]. The NRPS module responsible for AMP production is mostly found in Paenibacillus spp. and Brevibacillus spp. [39]. The structural similarity observed between the NRPS modules and AMP products of Paenibacillus sp. Na14 and Brevibacillus spp. supports the presence of conserved homologous features. This similarity is further reinforced by the genotypic and taxonomic relationships shared between the two genera. The evolutionary connection at the family level (Paenibacillaceae) suggests a common ancestral origin, supporting the proposed homology in genetic sequences, encoded proteins, and secondary metabolite biosynthetic pathways [45]. The similarity in protein structure homology between the two genera may originate from horizontal gene transfer, as previously reported in other studies investigating shared antibiotic resistomes between Paenibacillus spp. and Brevibacillus spp. [39].
4. Materials and Methods
4.1. Materials
Ammonium sulfate, sodium acetate, sodium chloride (NaCl), sodium dodecyl sulfate (SDS), sodium hydroxide (NaOH), hydrochloric acid (HCl), methanol, ethanol, and acetonitrile (ACN) were supplied by RCI Labscan Ltd., Bangkok, Thailand. Mueller–Hinton (MH) agar and Luria Bertani (LB) broth were obtained from Titan Biotech Ltd., Rajasthan, India. Colistin, vancomycin, and Coomassie brilliant blue G-250 were procured from Sigma Aldrich Inc., St. Louis, Missouri, USA. Glacial acetic acid and trifluoroacetic acid (TFA) were obtained from Merck KGaA, Darmstadt, Germany. Staphylococcus aureus TISTR 517, Escherichia coli TISTR 887, Pseudomonas aeruginosa TISTR 357, and Klebsiella pneumoniae TISTR 1383 were obtained from the Thailand Institute of Scientific and Technological Research (TISTR), Thailand. MRSA strain 142, 1096, and 2468 were provided by the Medical Technology Laboratory, School of Allied Health Sciences, Walailak University, Thailand.
4.2. Sample Collection and Bacterial Isolation
The samples were obtained from soils at a depth of 10–15 cm in the south of Thailand. They were placed in polyethylene bags and kept in an ice-cooled container. Subsequently, the samples were dried at 50 °C in a hot air oven for two days. The dried samples (10 g) were resuspended in 90 mL of sterile 0.9% NaCl solution. The soil suspension was shaken in an incubator at 150 rpm for 30 min under ambient conditions, followed by incubation at 60 °C for 30 min. Subsequently, the suspension was subjected to 10-fold serial dilutions up to 10−6. The diluted samples (100 µL) from each dilution were spread onto MH agar plates. The plates were incubated at 37 °C for one week, after which individual colonies were restreaked to obtain pure isolates [46].
4.3. Screening of Antibacterial Activity of Soil Bacteria
To identify potential active bacterial isolates, the cross-streak and agar well diffusion method was employed against S. aureus TISTR 517 and E. coli TISTR 887, following the protocol described in a previous study [47]. The production kinetics of antibacterial substances by the active bacterial isolates were investigated. Single colonies were suspended in 0.9% NaCl solution, and the turbidity was adjusted to an optical density (OD) of 0.35 at 625 nm. Two milliliters of the cell suspension were inoculated into 98 mL of LB broth and incubated at 37 °C with shaking at 150 rpm for seven days. At each time interval, bacterial growth was evaluated by measuring the OD at 625 nm using a UV-Vis spectrophotometer (JASCO Corporation, Tokyo, Japan). The cell-free supernatant (CFS) was obtained by centrifuging the culture at 10,000× g at 4 °C for 15 min. Subsequently, 100 µL of the supernatant was loaded into wells on MH agar plates precultured with tested pathogens [46]. Colistin and vancomycin served as positive controls in agar well diffusion assay.
4.4. Purification of the Bioactive Compound
Single colonies of the Na14 isolate were resuspended in 0.9% NaCl solution, and the cell suspension was adjusted to an OD of 0.35 at 625 nm. A 2% aliquot of this suspension was inoculated into 196 mL of LB broth and incubated at 37 °C for 24 h. The CFS was collected by centrifugation at 10,000× g for 15 min at 4 °C. Bioactive compounds were then precipitated using stepwise ammonium sulfate precipitation at 25%, 50%, and 75% saturation levels. The active compounds were purified using cation exchange chromatography (CIEX) followed by reversed-phase chromatography (RPC), as described in our previous study [48]. The purified compounds were concentrated using a refrigerated speed vacuum concentrator. The dried samples were reconstituted in deionized water and subjected to two-fold serial dilutions. An aliquot (100 µL) of each dilution was used to evaluate antimicrobial activity using the agar well diffusion method, employing E. coli TISTR 887 as the tested bacteria. The purification efficiency was assessed by calculating antimicrobial activity at each purification stage using the following equation [49].
where n represents the reciprocal of the highest two-fold dilution that produces a visible inhibition zone, and V denotes the volume of the sample in µL.
4.5. Identification and Analysis of the Na14 Peptide
The purified Na14 peptide was reconstituted in a solvent containing 0.1% formic acid and 1% acetonitrile (ACN) and then analyzed using high-resolution liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) (Thermo Fisher Scientific Inc., Waltham, MA, USA). Peptide separation was carried out on a C18 reversed-phase column. The elution conditions and MS/MS fragmentation parameters followed the protocol described in our previous study [50]. Peptide identification was performed using Peak Studio X. Physicochemical properties, including molecular mass, isoelectric point (pI), net charge, and hydrophobicity, were predicted using the ProtParam and HeliQuest tools [11,51].
The peptides (5 mg) were subjected to analysis using a 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gel was electrophoresed at a constant voltage of 100 V. One half of the gel was stained using Coomassie brilliant blue G-250 to visualize protein bands, with a molecular weight marker included to facilitate band position comparison. The remaining half was fixed in a solution containing 5% acetic acid and 25% methanol for 30 min, followed by a 3 h wash with distilled water. Subsequently, 18 mL of soft MH agar, including E. coli TISTR 887, was overlaid onto the gel to assess antimicrobial activity by observing inhibition zones after incubation at 37 °C for 24 h [52].
To determine the secondary structure composition of the Na14 peptide, peptide samples (0.50 mg/mL) were prepared in either deionized water or 50 mM SDS solution [53]. Circular dichroism (CD) spectra were acquired using a J-815 spectropolarimeter (JASCO Corporation, Tokyo, Japan) at 25 °C over a wavelength range of 190–250 nm. Measurements were conducted in a 1 mm pathlength quartz cuvette. The instrument settings were as follows: a scanning speed of 100 nm/min, a data interval of 0.1 nm, and a bandwidth of 1.0 nm. Spectra were baseline-corrected by subtracting the signal of the corresponding diluent. Secondary structure estimation was performed using the CONTINLL program [54].
4.6. Determination of MIC and MBC Values
The MIC and MBC values of the Na14 peptide were determined by broth microdilution against the tested bacteria, including E. coli TISTR 887, P. aeruginosa TISTR 357, K. pneumoniae TISTR 1383, S. aureus TISTR 517, and MRSA strain 2468. A bacterial suspension was prepared by adjusting its turbidity at OD 625 nm to 0.1 and subsequently made a 20-fold dilution using Cation-Adjusted Mueller–Hinton broth (CAHMB) as a diluent. One hundred microliters of the Na14 peptide in CAHMB, ranging from 0.25 to 128 µg/mL, was transferred into the 96-well plates. The diluted cell suspension (10 µL) was added to each well, and the plates were incubated at 37 °C for 24 h. The CAMHB alone was used as a negative control, while CAMHB containing colistin or vancomycin served as a positive control. The minimum inhibitory concentration (MIC) was defined as the lowest concentration of the Na14 peptide that inhibits visible bacterial growth. To determine the minimum bactericidal concentration (MBC), 100 µL of the samples from each well were plated onto MH agar and incubated at 37 °C for 24 h. The MBC was identified as the lowest concentration of the Na14 peptide at which no bacterial colonies were observed on the agar plate following incubation [55].
4.7. Stability Assessment of the Peptide Under Heat, pH, and Proteolytic Enzyme Conditions
The stability of the peptide was evaluated under various conditions that may influence its antimicrobial activity [30]. The purified Na14 AMP was prepared at a concentration of 10× MIC. Thermal stability was assessed by incubating the peptide at 30 °C, 50 °C, 70 °C, and 90 °C for 3 h. pH stability was evaluated by adjusting the peptide solution to pH 3, 5, 7, 9, and 11 using 1 N HCl or 1 N NaOH, followed by incubation at 37 °C for 3 h and subsequent neutralization to the original pH. To assess enzymatic stability, the peptide was incubated with trypsin, α-chymotrypsin, and proteinase K at a final concentration of 1 mg/mL (Sigma-Aldrich, Warren, MI, USA) at 37 °C for 3 h. The residual antimicrobial activity of each treatment condition was determined using an agar well diffusion assay against E. coli TISTR 887, P. aeruginosa TISTR 357, and K. pneumoniae TISTR 1383 to cross-validate efficacy across multiple pathogens. The untreated peptide served as a control. Statistical significance was analyzed using one-way ANOVA, with comparisons to the control group considered significant at p-value < 0.05. The results are presented as the percentage of residual antimicrobial activity.
4.8. Time–Kill Assay
Bacterial suspensions of E. coli TISTR 887, P. aeruginosa TISTR 357, and K. pneumoniae TISTR 1383 were prepared by adjusting their turbidity to an OD of 0.1 at 625 nm. The cultures were diluted 20-fold using CAMHB. Subsequently, 100 µL of the Na14 peptides in CAMHB, at concentrations corresponding to 0.5×, 1×, and 2× MIC, were dispensed into the 96-well microplates. The diluted bacterial suspension (10 µL) was mixed into each well before incubating the plates at 37 °C. At predetermined time points, 100 µL of either diluted or undiluted samples from each well were plated on MH agar and incubated at 37 °C for 24 h. Bacterial viability was assessed by colony counting and expressed as log CFU/mL [56].
4.9. Morphological Assessment of Bacterial Cells Treated with AMP Using SEM
The morphological effects of the Na14 peptide on E. coli TISTR 887, P. aeruginosa TISTR 357, and K. pneumoniae TISTR 1383 were evaluated using SEM. Overnight cultures (18 h) grown in MH broth were harvested by centrifugation and rinsed twice with sterile 0.9% NaCl. Cell suspensions were then adjusted in CAMHB to an OD of 0.1 at 625 nm. Aliquots were incubated with the AMP at 1× MIC for 16 h. For SEM preparation, treated and control cells were fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) for 24 h, followed by dehydration through a graded ethanol series ranging from 0% to 100%. Residual ethanol was finally removed using a critical point drying machine (Quorum Technologies Ltd., Lewes, East Sussex, UK), after which the specimens were coated with a thin layer of gold. Images were captured at 20,000× magnification [48]. Bacterial morphologies following AMP exposure were compared with those of cells treated with colistin at 1× MIC.
4.10. Genomic Analysis and Taxonomic Classification of the Na14 Strain
A single colony of Na14 strain was cultured on MH agar for 24 h before phenotypic and genotypic identification for taxonomic analysis. Gram staining and endospore-staining were performed, and cells were observed under a compound light microscope at 1000× magnification to determine Gram reaction and endospore formation (Carl Zeiss, Oberkochen, Germany) [57,58]. Genomic DNA was extracted from the bacterial culture and sequenced using the Illumina HiSeq platform with 150 bp paired-end reads (Illumina, San Diego, CA, USA), provided by U2Bio Co., Ltd. (Seoul, Republic of Korea). Raw paired-end sequencing reads underwent quality control and genome assembly using the Galaxy Australia platform (version 23.1) [59]. The quality of raw reads was assessed using FastQC (version 0.12.1) both before and after adapter trimming. Reads shorter than 30 base pairs were filtered out, and adapter sequences were removed using Fastp (version 0.23.4) [60]. De novo genome assembly was performed using the SPAdes genome assembler (version 3.14.1) via the Shovill platform (version 1.1.0). The quality of the assembled genome and its completeness were subsequently assessed using QUAST (version 5.2.0) [61] and CheckM (version 1.0.18), respectively [62]. Taxonomic analysis and genome comparison of the Na14 strain were performed using the Type (Strain) Genome Server (TYGS). Species-level identification was conducted based on digital DNA−DNA hybridization (dDDH) using the Genome BLAST Distance Phylogeny (GBDP) method [63]. The GBDP results were used to construct the phylogenetic tree using MEGA X software (version 10.1.8) [64]. The identified genotypic information of the Na14 strain was deposited in the GenBank database, accompanied by gene annotation performed using the Prokaryotic Genome Annotation Pipeline (PGAP, version 6.10) via the NCBI platform [65,66,67]. The secondary metabolism and antimicrobial peptide production potential of the Na14 strain were assessed through the prediction of biosynthetic gene clusters (BGCs) based on genomic data using the antiSMASH platform (version 7.0) [68]. Antimicrobial resistance genes were predicted using genomic data through the Resistance Gene Identifier (RGI) tool available on the Comprehensive Antibiotic Resistance Database (CARD) [69].
4.11. Comparative Structural Analysis of NRPS Modules from Genomic Data
Insight into the enzymes involved in AMP production was obtained through bacterial genome analysis and comparative approaches using various bioinformatic tools. The amino acid sequences encoded by the NRPS modules, predicted by antiSMASH, were modeled using the AlphaFold Server (https://alphafoldserver.com, accessed on 20 March 2025) [70]. The amino acid sequences and predicted protein structures were compared with reference protein structures deposited in the PDB database to assess structural similarity. The comparison was performed using the FoldSeek server, which identified the best-matched reference proteins based on ranking scores, including root-mean-square deviation (RMSD), template modeling score (TM-score), and sequence identity [71]. The structural comparison of each domain within the predicted NRPS modules was visualized using Visual Molecular Dynamics (VMD, version 1.9.3) and presented by overlaying the predicted protein structures with their corresponding reference structures [72]. The proposed functions of the predicted NRPS modules were assigned based on the annotated functions of reference proteins in the Pfam database, accessed through links provided by the PDB database [73].
5. Conclusions
In response to the urgent need for new therapeutics against antimicrobial-resistant Gram-negative pathogens, this study identified Paenibacillus sp. Na14 as a novel source of a potent AMP. The Na14 peptide displayed typical AMP characteristics, including positive charge and hydrophobicity. It exhibited a rapid killing rate and strong bactericidal activity against Gram-negative bacteria. This peptide demonstrated comparable effectiveness to colistin. Its mode of action was confirmed to involve membrane disruption. Genomic analysis revealed BGCs associated with the NRPS system, providing valuable insights into the AMP biosynthesis pathway. These findings not only expand the current understanding of naturally derived AMPs but also highlight the Na14 peptide as a promising drug candidate for the development of novel therapies targeting multidrug-resistant Gram-negative infections. To advance this discovery toward clinical application, future studies should include chemical synthesis of the peptide to confirm its activity and enable structure–activity relationship (SAR) analysis. Comprehensive in vivo efficacy, toxicity, and safety assessments, followed by clinical evaluation, will be essential to support its therapeutic potential. Furthermore, molecular genetic studies are needed to experimentally validate the predicted AMP biosynthetic enzymes, facilitating the rational development of Na14-derived drug candidates.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antibiotics14080805/s1: Figure S1: Preliminary screening of the antimicrobial activity of the Na14 strain using cross-streak method; Figure S2: Antimicrobial activity of the CFS from the Na14 strain using agar well diffusion assay.
Author Contributions
Conceptualization, N.S. (Nuttapon Songnaka), A.R., and A.A.; methodology, N.S. (Nuttapon Songnaka), A.R., N.S. (Namfa Sermkaew), S.S., S.K., C.A., Y.Y., and A.A.; formal analysis, N.S. (Nuttapon Songnaka), A.R., N.S. (Namfa Sermkaew), S.S., S.K., C.A., Y.Y., and A.A.; investigation, N.S. (Nuttapon Songnaka), A.R., N.S. (Namfa Sermkaew), S.S., S.K., C.A., Y.Y., and A.A.; writing—original draft preparation, N.S. (Nuttapon Songnaka), A.R., N.S. (Namfa Sermkaew), S.S., S.K., C.A., Y.Y., and A.A.; writing—review and editing, N.S. (Nuttapon Songnaka), A.R., N.S. (Namfa Sermkaew), S.S., S.K., C.A., Y.Y., and A.A.; visualization, N.S. (Nuttapon Songnaka), N.S. (Namfa Sermkaew) and A.A.; supervision, A.A.; project administration, A.A.; funding acquisition, A.R. and A.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Walailak University (Contract No. WU59109).
Data Availability Statement
Data are available within the article. Reasonable inquiries for additional information can be directed to the corresponding author.
Acknowledgments
We sincerely acknowledge the Center for Scientific and Technological Equipment, Walailak University for providing the research facilities. Chanat Aonbangkhen would also like to thank the Office of the Ministry of Higher Education, Science, Research, and Innovation.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analysis, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Miller, W.R.; Arias, C.A. ESKAPE pathogens: Antimicrobial resistance, epidemiology, clinical impact and therapeutics. Nat. Rev. Microbiol. 2024, 22, 598–616. [Google Scholar] [CrossRef]
- GBD 2021 Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance 1990–2021: A systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial resistance: A growing serious threat for global public health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Wang, J.; Dou, X.; Song, J.; Lyu, Y.; Zhu, X.; Xu, L.; Li, W.; Shan, A. Antimicrobial peptides: Promising alternatives in the post feeding antibiotic era. Med. Res. Rev. 2019, 39, 831–859. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Yan, Z.B.; Meng, Y.M.; Hong, X.Y.; Shao, G.; Ma, J.J.; Cheng, X.R.; Liu, J.; Kang, J.; Fu, C.Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- Bucataru, C.; Ciobanasu, C. Antimicrobial peptides: Opportunities and challenges in overcoming resistance. Microbiol. Res. 2024, 286, 127822. [Google Scholar] [CrossRef]
- Slingerland, C.J.; Martin, N.I. Recent advances in the development of polymyxin antibiotics: 2010–2023. ACS Infect. Dis. 2024, 10, 1056–1079. [Google Scholar] [CrossRef] [PubMed]
- Rubio, S.; Martínez-Cámara, S.; de la Fuente, J.L.; Rodríguez-Sáiz, M.; Barredo, J.L. Strain improvement program of Streptomyces roseosporus for daptomycin production. Methods Mol. Biol. 2021, 2296, 351–363. [Google Scholar] [CrossRef]
- Kumar, N.; Bhagwat, P.; Singh, S.; Pillai, S. A review on the diversity of antimicrobial peptides and genome mining strategies for their prediction. Biochimie 2024, 227, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.D.T.; Cao, J.; Franco, O.L.; Lu, T.K.; de la Fuente-Nunez, C. Synthetic biology and computer-based frameworks for antimicrobial peptide discovery. ACS Nano 2021, 15, 2143–2164. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the Expasy Server. In The Proteomics Protocols Handbook, 1st ed.; Walker, J.M., Ed.; Humana Press Inc.: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Guruprasad, K.; Reddy, B.V.; Pandit, M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990, 4, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, X.; Shukla, R.; Kumar, R.; Weingarth, M.; Breukink, E.; Kuipers, O.P. Brevibacillin 2V, a novel antimicrobial lipopeptide with an exceptionally low hemolytic activity. Front. Microbiol. 2021, 12, 693725. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Division, World Health Organization. WHO Bacterial Priority Pathogens List, 2024: Bacterial Pathogens of Public Health Importance to Guide Research, Development and Strategies to Prevent and Control Antimicrobial Resistance, 1st ed.; World Health Organization: Geneva, Switzerland, 2024; pp. 1–22.
- Seyedsayamdost, M.R. Toward a global picture of bacterial secondary metabolism. J. Ind. Microbiol. Biotechnol. 2019, 46, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Choyam, S.; Jain, P.M.; Kammara, R. Characterization of a potent new-generation antimicrobial peptide of Bacillus. Front. Microbiol. 2021, 12, 710741. [Google Scholar] [CrossRef] [PubMed]
- Barale, S.S.; Ghane, S.G.; Sonawane, K.D. Purification and characterization of antibacterial surfactin isoforms produced by Bacillus velezensis SK. AMB Express 2022, 12, 7. [Google Scholar] [CrossRef]
- Decker, A.P.; Mechesso, A.F.; Wang, G. Expanding the landscape of amino acid-rich antimicrobial peptides: Definition, deployment in nature, implications for peptide design and therapeutic potential. Int. J. Mol. Sci. 2022, 23, 12874. [Google Scholar] [CrossRef]
- Santoro, A.; Buonocore, M.; Grimaldi, M.; Napolitano, E.; D’Ursi, A.M. Monitoring the conformational changes of the Aβ (25−35) peptide in SDS micelles: A matter of time. Int. J. Mol. Sci. 2023, 24, 971. [Google Scholar] [CrossRef]
- Liu, Y.; Han, P.; Jia, Y.; Chen, Z.; Li, S.; Ma, A. Antibacterial regularity mining beneath the systematic activity database of lipopeptides Brevilaterins: An instructive activity handbook for its food application. Foods 2022, 11, 2991. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, X.; Shukla, R.; Kumar, R.; Weingarth, M.; Breukink, E.; Kuipers, O.P. Brevibacillin 2V exerts its bactericidal activity via binding to lipid II and permeabilizing cellular membranes. Front. Microbiol. 2021, 12, 694847. [Google Scholar] [CrossRef]
- Zhao, X.; Zhong, X.; Yang, S.; Deng, K.; Liu, L.; Song, X.; Zou, Y.; Li, L.; Zhou, X.; Jia, R.; et al. Elucidating the mechanism of action of the Gram-negative-pathogen-selective cyclic antimicrobial lipopeptide Brevicidine. Antimicrob. Agents Chemother. 2023, 67, e0001023. [Google Scholar] [CrossRef]
- Nagarajan, D.; Nagarajan, T.; Roy, N.; Kulkarni, O.; Ravichandran, S.; Mishra, M.; Chakravortty, D.; Chandra, N. Computational antimicrobial peptide design and evaluation against multidrug-resistant clinical isolates of bacteria. J. Biol. Chem. 2018, 293, 3492–3509. [Google Scholar] [CrossRef]
- Liu, J.; Chen, F.; Wang, X.; Peng, H.; Zhang, H.; Wang, K.J. The synergistic effect of mud crab antimicrobial peptides Sphistin and Sph12-38 with antibiotics azithromycin and rifampicin enhances bactericidal activity against Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2020, 10, 572849. [Google Scholar] [CrossRef]
- Lata, M.; Telang, V.; Gupta, P.; Pant, G.; Kalyan, M.; Arockiaraj, J.; Pasupuleti, M. Synthetic short cryptic antimicrobial peptides as templates for the development of novel biotherapeutics against WHO priority pathogen. Int. J. Pept. Res. Ther. 2024, 30, 57. [Google Scholar] [CrossRef]
- Liu, Y.; Zai, X.; Weng, G.; Ma, X.; Deng, D. Brevibacillus laterosporus: A probiotic with important applications in crop and animal production. Microorganisms 2024, 12, 564. [Google Scholar] [CrossRef]
- Wu, Y.; Zhou, L.; Lu, F.; Bie, X.; Zhao, H.; Zhang, C.; Lu, Z.; Lu, Y. Discovery of a novel antimicrobial lipopeptide, Brevibacillin V, from Brevibacillus laterosporus fmb70 and its application on the preservation of skim milk. J. Agric. Food. Chem. 2019, 67, 12452–12460. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, X.; Han, P.; Liu, Y.; Hong, D.; Li, S.; Ma, A.; Jia, Y. Discovery of novel antimicrobial peptides, Brevilaterin V, from Brevibacillus laterosporus S62-9 after regulated by exogenously-added L-valine. LWT 2022, 155, 112962. [Google Scholar] [CrossRef]
- Ning, Y.; Han, P.; Ma, J.; Liu, Y.; Fu, Y.; Wang, Z.; Jia, Y. Characterization of brevilaterins, multiple antimicrobial peptides simultaneously produced by Brevibacillus laterosporus S62-9, and their application in real food system. Food Biosci. 2021, 42, 101091. [Google Scholar] [CrossRef]
- Miljkovic, M.; Jovanovic, S.; O’Connor, P.M.; Mirkovic, N.; Jovcic, B.; Filipic, B.; Dinic, M.; Studholme, D.J.; Fira, D.; Cotter, P.D.; et al. Brevibacillus laterosporus strains BGSP7, BGSP9 and BGSP11 isolated from silage produce broad spectrum multi-antimicrobials. PLoS ONE 2019, 14, e0216773. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Fact. 2016, 15, 203. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.; Yousef, A.E. Biosynthesis of paenibacillin, a lantibiotic with N-terminal acetylation, by Paenibacillus polymyxa. Microbiol. Res. 2015, 181, 15–21. [Google Scholar] [CrossRef]
- Lohans, C.T.; Huang, Z.; Van Belkum, M.J.; Giroud, M.; Sit, C.S.; Steels, E.M.; Zheng, J.; Whittal, R.M.; McMullen, L.M.; Vederas, J.C. Structural characterization of the highly cyclized lantibiotic paenicidin A via a partial desulfurization/reduction strategy. J. Am. Chem. Soc. 2012, 134, 19540–19543. [Google Scholar] [CrossRef]
- Baindara, P.; Chaudhry, V.; Mittal, G.; Liao, L.M.; Matos, C.O.; Khatri, N.; Franco, O.L.; Patil, P.B.; Korpole, S. Characterization of the antimicrobial peptide penisin, a class Ia novel lantibiotic from Paenibacillus sp. strain A3. Antimicrob. Agents Chemother. 2016, 60, 580–591. [Google Scholar] [CrossRef]
- Tambadou, F.; Caradec, T.; Gagez, A.-L.; Bonnet, A.; Sopéna, V.; Bridiau, N.; Thiéry, V.; Didelot, S.; Barthélémy, C.; Chevrot, R. Characterization of the colistin (polymyxin E1 and E2) biosynthetic gene cluster. Arch. Microbiol. 2015, 197, 521–532. [Google Scholar] [CrossRef]
- Martin, N.I.; Hu, H.; Moake, M.M.; Churey, J.J.; Whittal, R.; Worobo, R.W.; Vederas, J.C. Isolation, structural characterization, and properties of mattacin (polymyxin M), a cyclic peptide antibiotic produced by Paenibacillus kobensis M. J. Biol. Chem. 2003, 278, 13124–13132. [Google Scholar] [CrossRef]
- Cochrane, S.A.; Vederas, J.C. Lipopeptides from Bacillus and Paenibacillus spp.: A gold mine of antibiotic candidates. Med. Res. Rev. 2016, 36, 4–31. [Google Scholar] [CrossRef]
- Pawlowski, A.C.; Westman, E.L.; Koteva, K.; Waglechner, N.; Wright, G.D. The complex resistomes of Paenibacillaceae reflect diverse antibiotic chemical ecologies. ISME J. 2018, 12, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Kessler, N.; Schuhmann, H.; Morneweg, S.; Linne, U.; Marahiel, M.A. The linear pentadecapeptide gramicidin is assembled by four multimodular nonribosomal peptide synthetases that comprise 16 modules with 56 catalytic domains. J. Biol. Chem. 2004, 279, 7413–7419. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ballard, J.; Jiang, Y.W. Structure and biosynthesis of the BT peptide antibiotic from Brevibacillus texasporus. Appl. Environ. Microbiol. 2005, 71, 8519–8530. [Google Scholar] [CrossRef]
- Barsby, T.; Kelly, M.T.; Gagné, S.M.; Andersen, R.J. Bogorol A produced in culture by a marine Bacillus sp. reveals a novel template for cationic peptide antibiotics. Org. Lett. 2001, 3, 437–440. [Google Scholar] [CrossRef]
- Yang, X.; Huang, E.; Yuan, C.; Zhang, L.; Yousef, A.E. Isolation and structural elucidation of brevibacillin, an antimicrobial lipopeptide from Brevibacillus laterosporus that combats drug-resistant Gram-positive bacteria. Appl. Environ. Microbiol. 2016, 82, 2763–2772. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.A.; Merrill, B.D.; Breakwell, D.P.; Hope, S.; Grose, J.H. A PCR-based method for distinguishing between two common beehive bacteria, Paenibacillus larvae and Brevibacillus laterosporus. Appl. Environ. Microbiol. 2018, 84, e01886-18. [Google Scholar] [CrossRef] [PubMed]
- Songnaka, N.; Lertcanawanichakul, M.; Atipairin, A. Promising anti-MRSA activity of Brevibacillus sp. isolated from soil and strain improvement by UV mutagenesis. Sci. Pharm. 2021, 89, 1. [Google Scholar] [CrossRef]
- Somsap, O.; Bangrak, P.; Bhoopong, P.; Lertcanawanichakul, M. Antibacterial activity and purification of bacteriocin produced by Brevibacillus laterosporus SA14. Walailak J. Sci. Tech. 2015, 13, 55–65. [Google Scholar] [CrossRef]
- Songnaka, N.; Lertcanawanichakul, M.; Hutapea, A.M.; Krobthong, S.; Yingchutrakul, Y.; Atipairin, A. Purification and characterization of novel anti-MRSA peptides produced by Brevibacillus sp. SPR-20. Molecules 2022, 27, 8452. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Fan, L.; Yan, J.; Jiang, Y.; Doucette, C.; Fillmore, S.; Walker, B. Influence of culture media, pH and temperature on growth and bacteriocin production of bacteriocinogenic lactic acid bacteria. AMB Express 2018, 8, 10. [Google Scholar] [CrossRef]
- Ogunsile, A.; Songnaka, N.; Sawatdee, S.; Lertcanawanichakul, M.; Krobthong, S.; Yingchutrakul, Y.; Uchiyama, J.; Atipairin, A. Anti-methicillin-resistant Staphylococcus aureus and antibiofilm activity of new peptides produced by a Brevibacillus strain. PeerJ 2023, 11, e16143. [Google Scholar] [CrossRef]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific alpha-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Scieuzo, C.; Giglio, F.; Rinaldi, R.; Lekka, M.E.; Cozzolino, F.; Monaco, V.; Monti, M.; Salvia, R.; Falabella, P. In vitro evaluation of the antibacterial activity of the peptide fractions extracted from the hemolymph of Hermetia illucens (Diptera: Stratiomyidae). Insects 2023, 14, 464. [Google Scholar] [CrossRef]
- Radhakrishnan, N.; Kumar, S.D.; Shin, S.Y.; Yang, S. Enhancing selective antimicrobial and antibiofilm activities of melittin through 6-aminohexanoic acid substitution. Biomolecules 2024, 14, 699. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 12th ed.; Clinical and Laboratory Standards Institute: Pittsburgh, PA, USA, 2024. [Google Scholar]
- Kavela, S.; Kakkerla, S.; Thupurani, M.K. Broad-spectrum antimicrobial activity and in vivo efficacy of SK1260 against bacterial pathogens. Front. Microbiol. 2025, 16, 1553693. [Google Scholar] [CrossRef]
- Tripathi, N.; Sapra, A. Gram Staining. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2020; [Updated 14 August 2023]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK562156 (accessed on 28 August 2023).
- Zayaitz, A.; Hussey, M.A. Endospore Staining Protocol. Am. Soc. Microbiol. 2007, 8, 1–11. Available online: https://www.asmscience.org/content/education/protocol/protocol.3112 (accessed on 28 August 2023).
- Galaxy Community. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; O’Neill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef]
- Haft, D.H.; DiCuccio, M.; Badretdin, A.; Brover, V.; Chetvernin, V.; O’Neill, K.; Li, W.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; et al. RefSeq: An update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018, 46, D851–D860. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- van Kempen, M.; Kim, S.S.; Tumescheit, C.; Mirdita, M.; Lee, J.; Gilchrist, C.L.M.; Söding, J.; Steinegger, M. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 2024, 42, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Andreeva, A.; Blum, M.; Chuguransky, S.R.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Llinares-López, F.; Meng-Papaxanthos, L.; et al. The Pfam protein families database: Embracing AI/ML. Nucleic Acids Res. 2025, 53, D523–D534. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).