Genomic Insights of Emerging Multidrug-Resistant OXA-48-Producing ST135 Proteus mirabilis
Abstract
1. Introduction
2. Results
2.1. Isolation of Bacteria, Antibiotic Susceptibility Testing, and Characterization of β-Lactamases
2.2. WGS and In Silico Prediction of MGEs, ARGs, and VFs
2.3. MLST, Virulence ST, Phylogenetic, and Phylogenomic Analyses of Strain Pm GR-1
2.4. Comparative Genomics of Strain Pm GR-1
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Identification, Antimicrobial Susceptibility Testing, and Detection of β-Lactamases
5.2. MLST STs, WGS, and Bioinformatic Analyses
5.3. In Silico Prediction of MGEs, ARGs, and VFs
5.4. Phylogenetic, Phylogenomic, and Comparative Genomic Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement:
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ARG | antimicrobial resistance gene |
| GI | genomic island |
| MDR | multidrug-resistant |
| MGE | mobile genetic element |
| MLST | multilocus-sequence typing |
| SNP | single-nucleotide polymorphism |
| ST | sequence type |
| VF | virulence factor |
References
- Armbruster, C.E.; Mobley, H.L.T.; Pearson, M.M. Pathogenesis of Proteus mirabilis infection. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef]
- Schaffer, J.N.; Pearson, M.M. Proteus mirabilis and urinary tract infections. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Bonnin, R.A.; Dortet, L.; Naas, T. Genetics of acquired antibiotic resistance genes in Proteus spp. Front. Microbiol. 2020, 11, 256. [Google Scholar] [CrossRef]
- Al-Qurashi, E.; Elbnna, K.; Ahmad, I.; Abulreesh, H.H. Antibiotic resistance in Proteus mirabilis: Mechanism, status, and public health significance. J. Pure Appl. Microbiol. 2022, 16, 1550–1561. [Google Scholar] [CrossRef]
- Boyd, S.E.; Holmes, A.; Peck, R.; Livermore, D.M.; Hope, W. OXA-48-like β-lactamases: Global epidemiology, treatment options, and development pipeline. Antimicrob. Agents Chemother. 2022, 66, e0021622. [Google Scholar] [CrossRef]
- Castanheira, M.; Doyle, T.B.; Collingsworth, T.D.; Sader, H.S.; Mendes, R.E. Increasing frequency of OXA-48-producing Enterobacterales worldwide and activity of ceftazidime/avibactam, meropenem/vaborbactam and comparators against these isolates. J. Antimicrob. Chemother. 2021, 76, 3125–3134. [Google Scholar] [CrossRef]
- Chen, L.; Al Laham, N.; Chavda, K.D.; Mediavilla, J.R.; Jacobs, M.R.; Bonomo, R.A.; Kreiswirth, B.N. First report of an OXA-48-producing multidrug-resistant Proteus mirabilis strain from Gaza, Palestine. Antimicrob. Agents Chemother. 2015, 59, 4305–4307. [Google Scholar] [CrossRef]
- Pedraza, R.; Kieffer, N.; Guzmán-Puche, J.; Artacho, M.J.; Pitart, C.; Hernández-García, M.; Vila, J.; Cantón, R.; Martinez-Martinez, L. Hidden dissemination of carbapenem-susceptible OXA-48-producing Proteus mirabilis. J. Antimicrob. Chemother. 2022, 77, 3009–3015. [Google Scholar] [CrossRef]
- Sattler, J.; Noster, J.; Stelzer, Y.; Spille, M.; Schäfer, S.; Xanthopoulou, K.; Sommer, J.; Jantsch, J.; Peter, S.; Göttig, S.; et al. OXA-48-like carbapenemases in Proteus mirabilis-novel genetic environments and a challenge for detection. Emerg. Microbes Infect. 2024, 13, 2353310. [Google Scholar] [CrossRef]
- Sattler, J.; Tsvetkov, T.; Stelzer, Y.; Schäfer, S.; Sommer, J.; Noster, J.; Göttig, S.; Hamprecht, A. Emergence of Tn1999.7, a new transposon in blaOXA-48-harboring plasmids associated with increased plasmid stability. Antimicrob. Agents Chemother. 2022, 66, e0078722. [Google Scholar] [CrossRef]
- Vourli, S.; Tsorlini, H.; Katsifa, H.; Polemis, M.; Tzouvelekis, L.S.; Kontodimou, A.; Vatopoulos, A.C. Emergence of Proteus mirabilis carrying the blaVIM-1 metallo-beta-lactamase gene. Clin. Microbiol. Infect. 2006, 12, 691–694. [Google Scholar] [CrossRef]
- Tsakris, A.; Ikonomidis, A.; Poulou, A.; Spanakis, N.; Pournaras, S.; Markou, F. Transmission in the community of clonal Proteus mirabilis carrying VIM-1 metallo-beta-lactamase. J. Antimicrob. Chemother. 2007, 60, 136–139. [Google Scholar] [CrossRef]
- Protonotariou, E.; Poulou, A.; Politi, L.; Meletis, G.; Chatzopoulou, F.; Malousi, A.; Metallidis, S.; Tsakris, A.; Skoura, L. Clonal outbreak caused by VIM-4-producing Proteus mirabilis in a Greek tertiary-care hospital. Int. J. Antimicrob. Agents 2020, 56, 106060. [Google Scholar] [CrossRef]
- Meletis, G.; Malousi, A.; Tychala, A.; Kassomenaki, A.; Vlachodimou, N.; Mantzana, P.; Metallidis, S.; Skoura, L.; Protonotariou, E. Probable three-species in vivo transfer of blaNDM-1 in a single patient in Greece: Occurrence of NDM-1-producing Klebsiella pneumoniae, Proteus mirabilis, and Morganella morganii. Antibiotics 2023, 12, 1206. [Google Scholar] [CrossRef]
- Yang, A.; Tian, Y.; Li, X. Unveiling the hidden arsenal: New insights into Proteus mirabilis virulence in UTIs. Front. Cell. Infect. Microbiol. 2024, 14, 1465460. [Google Scholar] [CrossRef] [PubMed]
- Pearson, M.M.; Sebaihia, M.; Churcher, C.; Quail, M.A.; Seshasayee, A.S.; Luscombe, N.M.; Abdellah, Z.; Arrosmith, C.; Atkin, B.; Chillingworth, T.; et al. Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 2008, 190, 4027–4037. [Google Scholar] [CrossRef] [PubMed]
- Lian, S.; Liu, Y.; Hu, S.; Shen, C.; Ma, Y.; Yin, P.; He, Z. Genomic insights on cgMLST markers, drug resistance, and urease cluster of Proteus mirabilis strains. Microbiol. Spectr. 2025, 13, e0099224. [Google Scholar] [CrossRef]
- Jaidane, N.; Tilouche, L.; Oueslati, S.; Girlich, D.; Azaiez, S.; Jacquemin, A.; Dortet, L.; Naija, W.; Trabelsi, A.; Naas, T.; et al. Clonal Dissemination of NDM-producing Proteus mirabilis in a teaching hospital in Sousse, Tunisia. Pathogens 2025, 14, 298. [Google Scholar] [CrossRef]
- Peters, J.E. Targeted transposition with Tn7 elements: Safe sites, mobile plasmids, CRISPR/Cas and beyond. Mol. Microbiol. 2019, 112, 1635–1644. [Google Scholar] [CrossRef]
- Wei, Q.; Hu, Q.; Li, S.; Lu, H.; Chen, G.; Shen, B.; Zhang, P.; Zhou, Y. A novel functional class 2 integron in clinical Proteus mirabilis isolates. J. Antimicrob. Chemother. 2014, 69, 973–976. [Google Scholar] [CrossRef]
- Yu, X.; Torzewska, A.; Zhang, X.; Yin, Z.; Drzewiecka, D.; Cao, H.; Liu, B.; Knirel, Y.A.; Rozalski, A.; Wang, L.; et al. Genetic diversity of the O antigens of Proteus species and the development of a suspension array for molecular serotyping. PLoS ONE 2017, 12, e0183267. [Google Scholar] [CrossRef]
- Chakkour, M.; Hammoud, Z.; Farhat, S.; El Roz, A.; Ezzeddine, Z.; Ghssein, G. Overview of Proteus mirabilis pathogenicity and virulence. Insights into the role of metals. Front Microbiol. 2024, 15, 1383618. [Google Scholar] [CrossRef] [PubMed]
- Bode, N.J.; Debnath, I.; Kuan, L.; Schulfer, A.; Ty, M.; Pearson, M.M. Transcriptional analysis of the MrpJ network: Modulation of diverse virulence-associated genes and direct regulation of mrp fimbrial and flhDC flagellar operons in Proteus mirabilis. Infect Immun. 2015, 83, 2542–2556. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Feng, J.; Sachs, G. Helicobacter pylori 5′ureB-sRNA, a cis-encoded antisense small RNA, negatively regulates ureAB expression by transcription termination. J. Bacteriol. 2013, 195, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, M.J.; Pearson, M.M.; Mobley, H.L.T. Proteus mirabilis UreR coordinates cellular functions required for urease activity. J. Bacteriol. 2024, 206, e00031-24. [Google Scholar] [CrossRef]
- EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters Version 15.0, Valid from 2025-01-01. Available online: https://www.eucast.org/ (accessed on 20 February 2025).
- Kieffer, N.; Poirel, L.; Nordmann, P. Rapid immunochromatography- based detection of carbapenemase producers. Infection 2019, 47, 673–675. [Google Scholar] [CrossRef]
- Bernabeu, S.; Ratnam, K.C.; Boutal, H.; Gonzalez, C.; Vogel, A.; Devilliers, K.; Plaisance, M.; Oueslati, S.; Malhotra-Kumar, S.; Dortet, L.; et al. A lateral flow immunoassay for the rapid identification of CTX-M-producing Enterobacterales from culture plates and positive blood cultures. Diagnostics 2020, 10, 764. [Google Scholar] [CrossRef]
- Mavroidi, A.; Katsiari, M.; Likousi, S.; Palla, E.; Roussou, Z.; Nikolaou, C.; Mathas, C.; Merkouri, E.; Platsouka, E.D. Changing characteristics and in vitro susceptibility to ceftazidime/avibactam of bloodstream extensively drug-resistant Klebsiella pneumoniae from a Greek Intensive Care Unit. Microb. Drug Resist. 2020, 26, 28–37. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Dai, H.; Lu, B.; Li, Z.; Huang, Z.; Cai, H.; Yu, K.; Wang, D. Multilocus sequence analysis for the taxonomic updating and identification of the genus Proteus and reclassification of Proteus genospecies 5 O’Hara et al. 2000, Proteus cibarius Hyun et al. 2016 as later heterotypic synonyms of Proteus terrae Behrendt et al. 2015. BMC Microbiol. 2020, 20, 152. [Google Scholar] [CrossRef]
- Algammal, A.M.; Hashem, H.R.; Alfifi, K.J.; Hetta, H.F.; Sheraba, N.S.; Ramadan, H.; El-Tarabili, R.M. atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci. Rep. 2021, 11, 9476. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. “Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification”. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Garcillán-Barcia, M.P.; Redondo-Salvo, S.; Vielva, L.; de la Cruz, F. MOBscan: Automated Annotation of MOB Relaxases. Methods Mol. Biol. 2020, 2075, 295–308. [Google Scholar] [CrossRef]
- Li, X.; Xie, Y.; Liu, M.; Tai, C.; Sun, J.; Deng, Z.; Ou, H.Y. oriTfinder: A web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 2018, 46, W229–W234. [Google Scholar] [CrossRef]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Weigel, L.M.; Anderson, G.J.; Tenover, F.C. DNA gyrase and topoisomerase IV mutations associated with fluoroquinolone resistance in Proteus mirabilis. Antimicrob. Agents Chemother. 2002, 46, 2582–2587. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB. A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Bertelli, C.; Gray, K.L.; Woods, N.; Lim, A.C.; Tilley, K.E.; Winsor, G.L.; Hoad, G.R.; Roudgar, A.; Spencer, A.; Peltier, J.; et al. Enabling genomic island prediction and comparison in multiple genomes to investigate bacterial evolution and outbreaks. Microb. Genom. 2022, 8, mgen000818. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

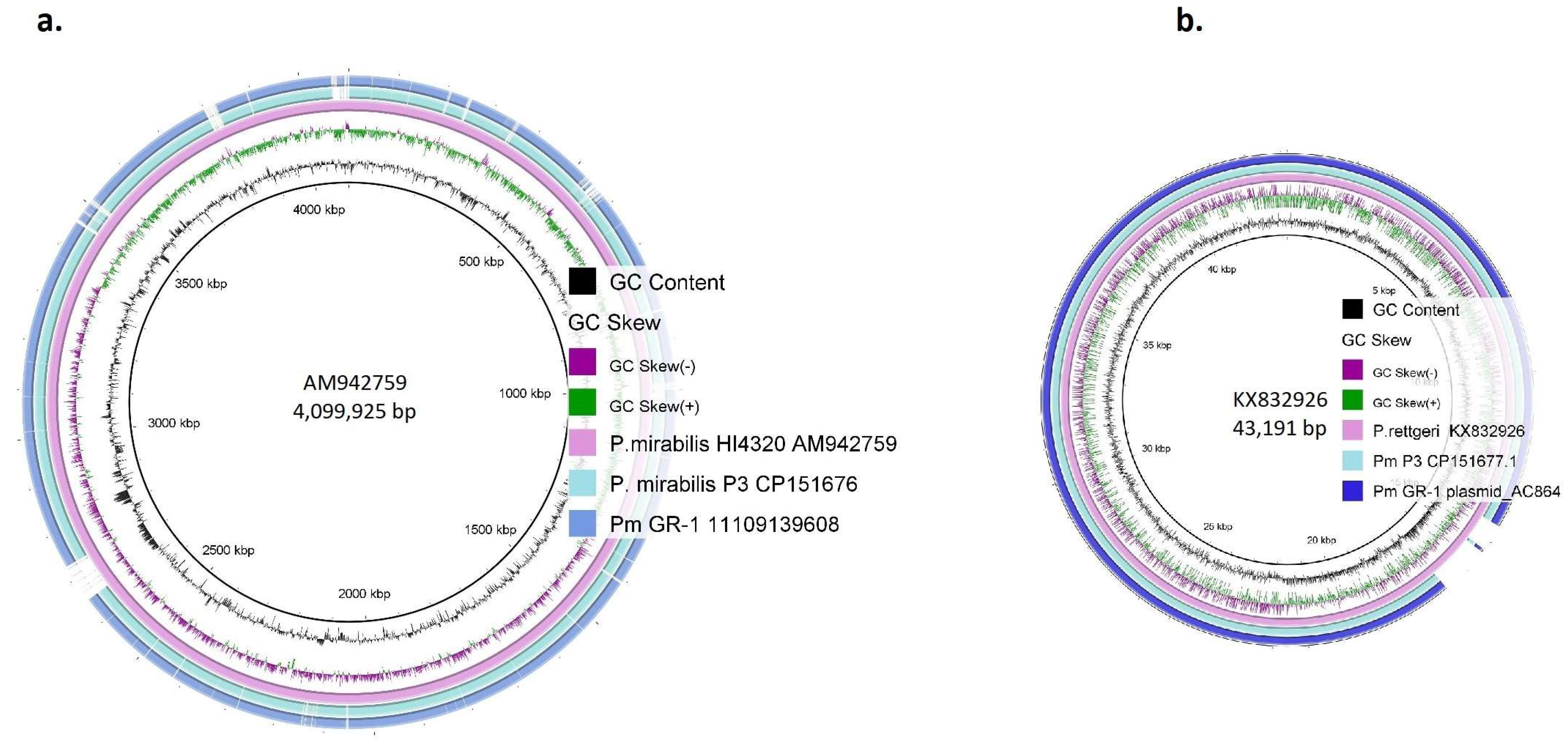
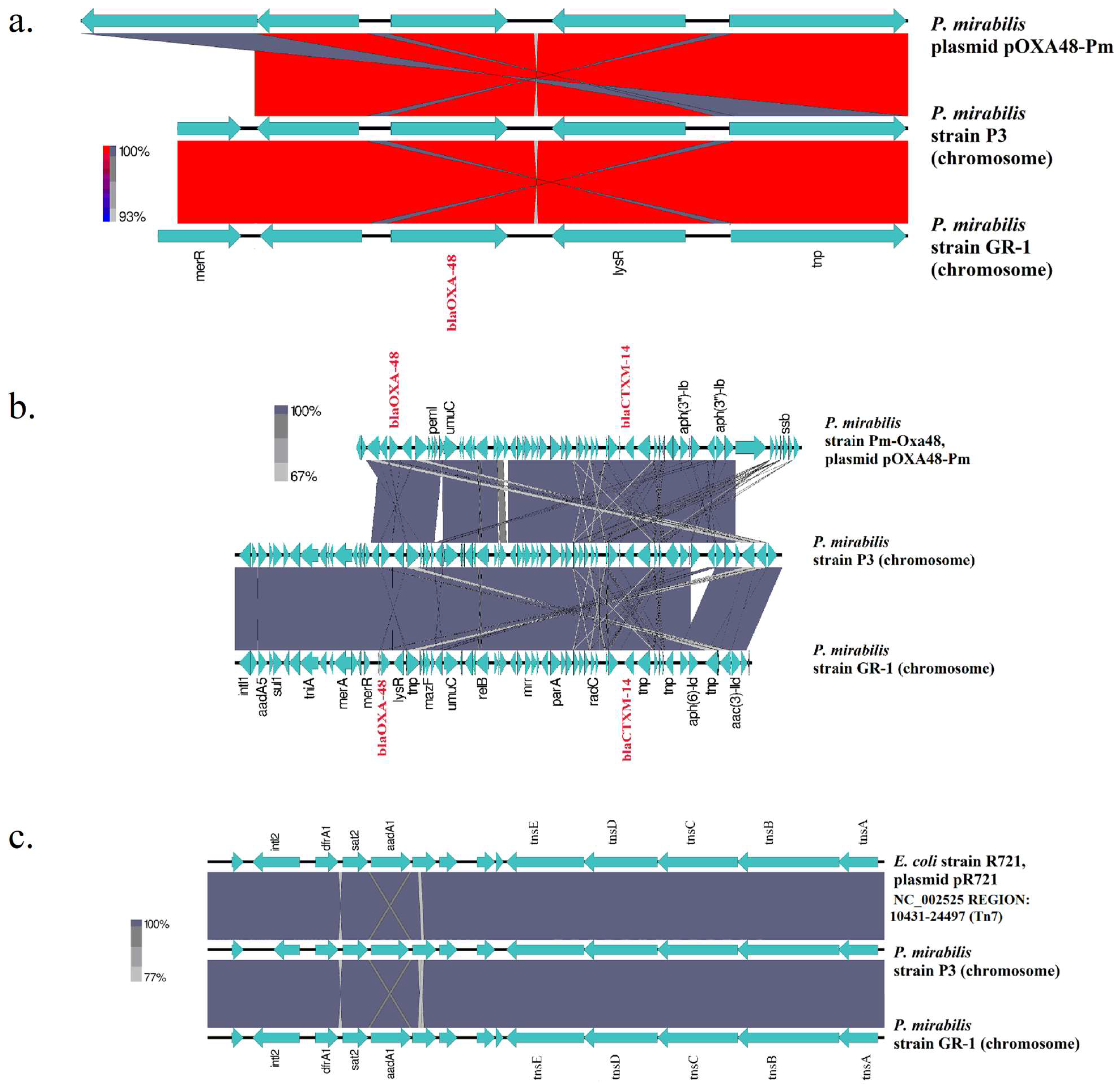
| Pm GR-1 11109139608_(Nanopore) | Molecule Type | Size (bp) | GC (%) | MGEs | ARGs |
|---|---|---|---|---|---|
| contig_0001 | chromosome | 1,905,607 | 39.72 | Tn7, IS26 | aadA5, cat, dfrA17, qacEdelta1, sul1, merA, merC, merD, merE, merP, merR, merT |
| contig_0002 | chromosome | 915,722 | 40.26 | ISVsa5, Tn4352, IS26, IS629, IS5, cn_2401_ISVsa5, cn_3556_IS26 | blaOXA-48, blaCTX-M-14, blaTEM-1, aac(3)-IId, aadA5, aph(3′)-Ia, aph(3″)-Ib, aph(6)-Id, catA1, dfrA17, qacEdelta1, sat2, sul1, tet(J), merA, merC, merD, merE, merP, merR, merT, terD, terZ |
| contig_0003 | chromosome | 820,194 | 38.30 | ||
| contig_0004 | chromosome | 308,558 | 37.36 | ||
| contig_0005 | chromosome | 295,079 | 38.20 | ||
| contig_0006 | chromosome | 45,133 | 40.69 | ||
| contig_0007 | plasmid AC864 * | 40,918 | 36.21 | blaTEM-2 | |
| contig_0008 | chromosome | 5453 | 39.57 | ||
| contig_0009 | chromosome | 4956 | 38.03 |
| Virulence Factors | Length (bp) | Pm GR-1_11109139608 Contig | Coordinates |
|---|---|---|---|
| vST138 allelic profile | |||
| cheB (allele 1) | 1053 | contig_0003 | 510,332–511,384 |
| cheY (allele 1) | 390 | contig_0003 | 509,883–510,272 |
| flgG (allele 52) | 783 | contig_0003 | 497,099–497,881 |
| flgH (allele 46) | 744 | contig_0003 | 496,293–497,036 |
| fliI (allele 79) | 1374 | contig_0003 | 483,487–484,860 |
| flip (allele 56) | 771 | contig_0003 | 489,299–490,069 |
| flN (allele 1) | 411 | contig_0003 | 488,439–488,849 |
| gmhA (allele 46) | 579 | contig_0001 | 607,440–608,018 |
| KdsA (allele 66) | 855 | contig_0004 | 134,233–135,087 |
| lpxC (allele 57) | 918 | contig_0005 | 118,044–118,961 |
| lpxD (allele 55) | 1029 | contig_0002 | 161,072–162,100 |
| luxS (allele-43) | 516 | contig_0001 | 573,194–573,709 |
| haemolysin genes | |||
| hpmA, B | 4733 | contig_0005 | 126,318–132,770 |
| zapA | 1475 | contig_0001 | 772,637–774,112 |
| O-antigen locus | 19,356 | contig_0001 | 1,638,693–1,658,049 |
| flagella locus | 53,161 | contig_0003 | 468,664–521,825 |
| MR/P fimbriae operon | 9500 | contig_0001 | 784,087–793,586 |
| urease gene cluster | 4195 | contig_0001 | 1,121,504–1,125,699 |
| hydrogenase system (hybOABCDE) | 5081 | contig_0001 | 1,028,369–1,034,350 |
| molybdate-binding Protein (modA) | 770 | contig_0003 | 410,098–410,868 |
| sigma factor RpoE (rpoE) | 548 | contig_0001 | 1,098,303–1,098,851 |
| polyphosphate kinase 1 (ppk1) | 2081 | contig_0003 | 413,425–415,506 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mavroidi, A.; Froukala, E.; Spanakis, N.; Michelaki, A.; Orfanidou, M.; Koumaki, V.; Tsakris, A. Genomic Insights of Emerging Multidrug-Resistant OXA-48-Producing ST135 Proteus mirabilis. Antibiotics 2025, 14, 750. https://doi.org/10.3390/antibiotics14080750
Mavroidi A, Froukala E, Spanakis N, Michelaki A, Orfanidou M, Koumaki V, Tsakris A. Genomic Insights of Emerging Multidrug-Resistant OXA-48-Producing ST135 Proteus mirabilis. Antibiotics. 2025; 14(8):750. https://doi.org/10.3390/antibiotics14080750
Chicago/Turabian StyleMavroidi, Angeliki, Elisavet Froukala, Nick Spanakis, Aikaterini Michelaki, Maria Orfanidou, Vasiliki Koumaki, and Athanasios Tsakris. 2025. "Genomic Insights of Emerging Multidrug-Resistant OXA-48-Producing ST135 Proteus mirabilis" Antibiotics 14, no. 8: 750. https://doi.org/10.3390/antibiotics14080750
APA StyleMavroidi, A., Froukala, E., Spanakis, N., Michelaki, A., Orfanidou, M., Koumaki, V., & Tsakris, A. (2025). Genomic Insights of Emerging Multidrug-Resistant OXA-48-Producing ST135 Proteus mirabilis. Antibiotics, 14(8), 750. https://doi.org/10.3390/antibiotics14080750








