Epidemiological Surveillance, Variability, and Evolution of Isolates Belonging to the Spanish Clone of the 4,[5],12:i:- Monophasic Variant of Salmonella enterica Serovar Typhimurium
Abstract
1. Introduction
2. Results
2.1. Incidence and General Properties of Monophasic Isolates of the Spanish Clone During the 2008–2018 Period
2.2. Genome Sequence Analysis
2.3. Comparison of IncC Plasmids from the 2000 to 2003 and 2008 to 2018 Periods
2.4. Genetic Basis of the Monophasic Phenotype
2.5. Genomic Relationships Between the Isolates
3. Discussion
4. Materials and Methods
4.1. Isolate Selection and Preliminary Characterization
4.2. Whole Genome Sequencing and Bioinformatics Analysis
4.3. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M.; International Collaboration on Enteric Disease ‘Burden of Illness Studies’. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Issenhuth-Jeanjean, S.; Roggentin, P.; Mikoleit, M.; Guibourdenche, M.; de Pinna, E.; Nair, S.; Fields, P.I.; Weill, F.X. Supplement 2008-2010 (no. 48) to the White-Kauffmann-Le Minor scheme. Res. Microbiol. 2014, 165, 526–530. [Google Scholar] [CrossRef] [PubMed]
- ECDC (European Food Safety Authority and European Centre for Disease, Prevention and Control). The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, e8442. [Google Scholar] [PubMed]
- EFSA (European Food Safety Authority) Panel on Biological Hazards (BIOHAZ). Scientific Opinion on monitoring and assessment of the public health risk of “Salmonella typhimurium-like” strains. EFSA J. 2010, 8, 1826. [Google Scholar] [CrossRef]
- ECDC (European Food Safety Authority and European Centre for Disease, Prevention and Control). The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e07666. [Google Scholar] [CrossRef]
- Sun, H.; Wan, Y.; Du, P.; Bai, L. The epidemiology of monophasic Salmonella Typhimurium. Foodborne Pathog. Dis. 2020, 17, 87–97. [Google Scholar] [CrossRef]
- Switt, A.I.; Soyer, Y.; Warnick, L.D.; Wiedmann, M. Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i. Foodborne Pathog. Dis. 2009, 6, 407–415. [Google Scholar] [CrossRef]
- Silverman, M.; Simon, M. Phase variation: Genetic analysis of switching mutants. Cell 1980, 19, 845–854. [Google Scholar] [CrossRef]
- Soyer, Y.; Moreno Switt, A.; Davis, M.A.; Maurer, J.; McDonough, P.L.; Schoonmaker-Bopp, D.J.; Dumas, N.B.; Root, T.; Warnick, L.D.; Grohn, Y.T.; et al. Salmonella enterica serotype 4,5,12:i:-, an emerging Salmonella serotype that represents multiple distinct clones. J. Clin. Microbiol. 2009, 47, 3546–3556. [Google Scholar] [CrossRef]
- Antunes, P.; Mourao, J.; Pestana, N.; Peixe, L. Leakage of emerging clinically relevant multidrug-resistant Salmonella clones from pig farms. J. Antimicrob. Chemother. 2011, 66, 2028–2032. [Google Scholar] [CrossRef]
- de la Torre, E.; Zapata, D.; Tello, M.; Mejia, W.; Frias, N.; Garcia Pena, F.J.; Mateu, E.M.; Torre, E. Several Salmonella enterica subsp. enterica serotype 4,5,12:i:- phage types isolated from swine samples originate from serotype Typhimurium DT U302. J. Clin. Microbiol. 2003, 41, 2395–2400. [Google Scholar]
- Echeita, M.A.; Aladuena, A.; Cruchaga, S.; Usera, M.A. Emergence and spread of an atypical Salmonella enterica subsp. enterica serotype 4,5,12:i:- strain in Spain. J. Clin. Microbiol. 1999, 37, 3425. [Google Scholar]
- Echeita, M.A.; Herrera, S.; Usera, M.A. Atypical, fljB-negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:i:- appears to be a monophasic variant of serovar Typhimurium. J. Clin. Microbiol. 2001, 39, 2981–2983. [Google Scholar]
- Garcia, P.; Guerra, B.; Bances, M.; Mendoza, M.C.; Rodicio, M.R. IncA/C plasmids mediate antimicrobial resistance linked to virulence genes in the Spanish clone of the emerging Salmonella enterica serotype 4,[5],12:i. J. Antimicrob. Chemother. 2011, 66, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Garcia, P.; Malorny, B.; Hauser, E.; Mendoza, M.C.; Rodicio, M.R. Genetic types, gene repertoire, and evolution of isolates of the Salmonella enterica serovar 4,5,12:i:- Spanish clone assigned to different phage types. J. Clin. Microbiol. 2013, 51, 973–978. [Google Scholar] [CrossRef]
- Guerra, B.; Laconcha, I.; Soto, S.M.; Gonzalez-Hevia, M.A.; Mendoza, M.C. Molecular characterisation of emergent multiresistant Salmonella enterica serotype [4,5,12:i:-] organisms causing human salmonellosis. FEMS Microbiol. Lett. 2000, 190, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Guerra, B.; Soto, S.M.; Arguelles, J.M.; Mendoza, M.C. Multidrug resistance is mediated by large plasmids carrying a class 1 integron in the emergent Salmonella enterica serotype [4,5,12:i:-]. Antimicrob. Agents Chemother. 2001, 45, 1305–1308. [Google Scholar] [CrossRef] [PubMed]
- Arrieta-Gisasola, A.; Atxaerandio-Landa, A.; Garrido, V.; Grillo, M.J.; Martinez-Ballesteros, I.; Laorden, L.; Garaizar, J.; Bikandi, J. Genotyping study of Salmonella 4,[5],12:i:- monophasic variant of serovar Typhimurium and characterization of the second-phase flagellar deletion by whole genome sequencing. Microorganisms 2020, 8, 2049. [Google Scholar] [CrossRef]
- Garaizar, J.; Porwollik, S.; Echeita, A.; Rementeria, A.; Herrera, S.; Wong, R.M.; Frye, J.; Usera, M.A.; McClelland, M. DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J. Clin. Microbiol. 2002, 40, 2074–2078. [Google Scholar] [CrossRef]
- Laorden, L.; Herrera-Leon, S.; Martinez, I.; Sanchez, A.; Kromidas, L.; Bikandi, J.; Rementeria, A.; Echeita, A.; Garaizar, J. Genetic evolution of the Spanish multidrug-resistant Salmonella enterica 4,5,12:i:- monophasic variant. J. Clin. Microbiol. 2010, 48, 4563–4566. [Google Scholar] [CrossRef]
- Vázquez, X.; García, P.; García, V.; de Toro, M.; Ladero, V.; Heinisch, J.J.; Fernández, J.; Rodicio, R.; Rodicio, M.R. Genomic analysis and phylogenetic position of the complex IncC plasmid found in the Spanish monophasic clone of Salmonella enterica serovar Typhimurium. Sci. Rep. 2021, 11, 11482. [Google Scholar] [CrossRef]
- Hancock, S.J.; Phan, M.D.; Peters, K.M.; Forde, B.M.; Chong, T.M.; Yin, W.F.; Chan, K.G.; Paterson, D.L.; Walsh, T.R.; Beatson, S.A.; et al. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob. Agents Chemother. 2017, 61, e01740-16. [Google Scholar] [CrossRef]
- Nigro, S.J.; Hall, R.M. GIsul2, a genomic island carrying the sul2 sulphonamide resistance gene and the small mobile element CR2 found in the Enterobacter cloacae subspecies cloacae type strain ATCC 13047 from 1890, Shigella flexneri ATCC 700930 from 1954 and Acinetobacter baumannii ATCC 17978 from 1951. J. Antimicrob. Chemother. 2011, 66, 2175–2176. [Google Scholar] [CrossRef]
- McClelland, M.; Sanderson, K.E.; Spieth, J.; Clifton, S.W.; Latreille, P.; Courtney, L.; Porwollik, S.; Ali, J.; Dante, M.; Du, F.; et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 2001, 413, 852–856. [Google Scholar] [CrossRef] [PubMed]
- Branchu, P.; Charity, O.J.; Bawn, M.; Thilliez, G.; Dallman, T.J.; Petrovska, L.; Kingsley, R.A. SGI-4 in monophasic Salmonella typhimurium ST34 is a novel ICE that enhances resistance to copper. Front. Microbiol. 2019, 10, 1118. [Google Scholar] [CrossRef] [PubMed]
- Rabsch, W.; Tschape, H.; Baumler, A.J. Non-typhoidal salmonellosis: Emerging problems. Microbes Infect. 2001, 3, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Tassinari, E.; Bawn, M.; Thilliez, G.; Charity, O.; Acton, L.; Kirkwood, M.; Petrovska, L.; Dallman, T.; Burgess, C.M.; Hall, N.; et al. Whole-genome epidemiology links phage-mediated acquisition of a virulence gene to the clonal expansion of a pandemic Salmonella enterica serovar Typhimurium clone. Microb. Genom. 2020, 6, mgen000456. [Google Scholar] [CrossRef]
- Arrieta-Gisasola, A.; Martinez-Ballesteros, I.; Martinez-Malaxetxebarria, I.; Bikandi, J.; Laorden, L. Detection of mobile genetic elements conferring resistance to heavy metals in Salmonella 4,[5],12:i:- and Salmonella Typhimurium serovars and their association with antibiotic resistance. Int. J. Food Microbiol. 2025, 426, 110890. [Google Scholar] [CrossRef]
- European Parliament and the Council of the European Union. Regulation (EC) No 1831/2003 of the European Parliament and of the Council of 22 September 2003. Off. J. Eur. Union 2003, 4, L 268/29-42. [Google Scholar]
- Cadel-Six, S.; Cherchame, E.; Douarre, P.E.; Tang, Y.; Felten, A.; Barbet, P.; Litrup, E.; Banerji, S.; Simon, S.; Pasquali, F.; et al. The spatiotemporal dynamics and microevolution events that favored the success of the highly clonal multidrug-resistant monophasic Salmonella Typhimurium circulating in Europe. Front. Microbiol. 2021, 12, 651124. [Google Scholar] [CrossRef]
- Mastrorilli, E.; Pietrucci, D.; Barco, L.; Ammendola, S.; Petrin, S.; Longo, A.; Mantovani, C.; Battistoni, A.; Ricci, A.; Desideri, A.; et al. A comparative genomic analysis provides novel insights Into the ecological success of the monophasic Salmonella serovar 4,[5],12:i. Front. Microbiol. 2018, 9, 715. [Google Scholar] [CrossRef]
- Mourao, J.; Novais, C.; Machado, J.; Peixe, L.; Antunes, P. Metal tolerance in emerging clinically relevant multidrug-resistant Salmonella enterica serotype 4,[5],12:i:- clones circulating in Europe. Int. J. Antimicrob. Agents 2015, 45, 610–616. [Google Scholar] [CrossRef]
- Council of the European Union. Council directive 1999/29/EC of 22 April 1999 on the undesirable substances and products in animal nutrition. Counc. Dir. 1999, 9, 32–46. [Google Scholar]
- Harmer, C.J.; Hall, R.M. IS26 and the IS26 family: Versatile resistance gene movers and genome reorganizers. Microbiol. Mol. Biol. Rev. 2024, 88, e0011922. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Hickman, A.B.; Varani, A.M.; Siguier, P.; Chandler, M.; Dekker, J.P.; Dyda, F. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 2015, 6, e00762. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, X.; Fernandez, J.; Heinisch, J.J.; Rodicio, R.; Rodicio, M.R. Insights into the evolution of IncR plasmids found in the Southern European clone of the monophasic variant of Salmonella enterica serovar Typhimurium. Antibiotics 2024, 13, 314. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
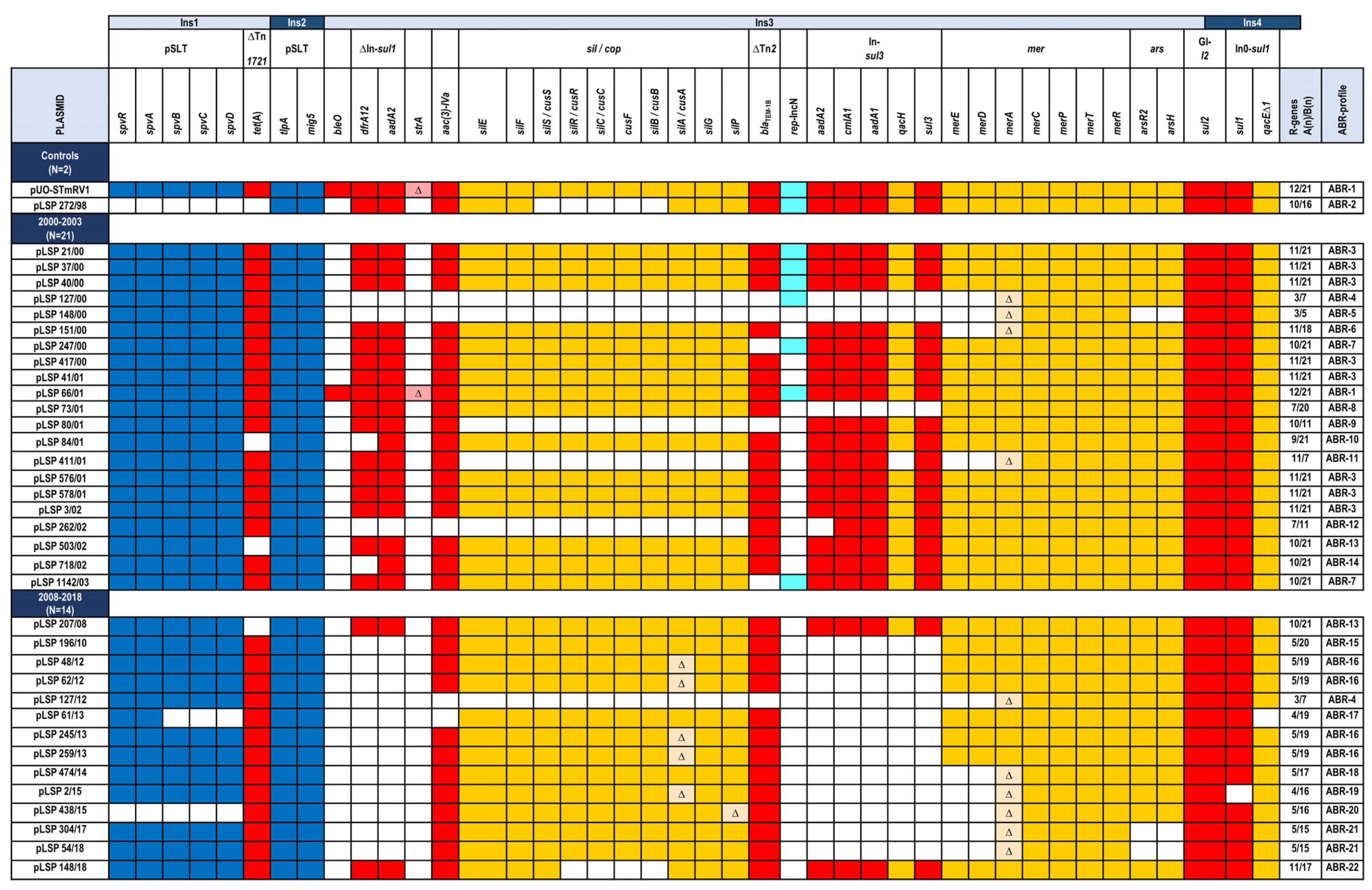
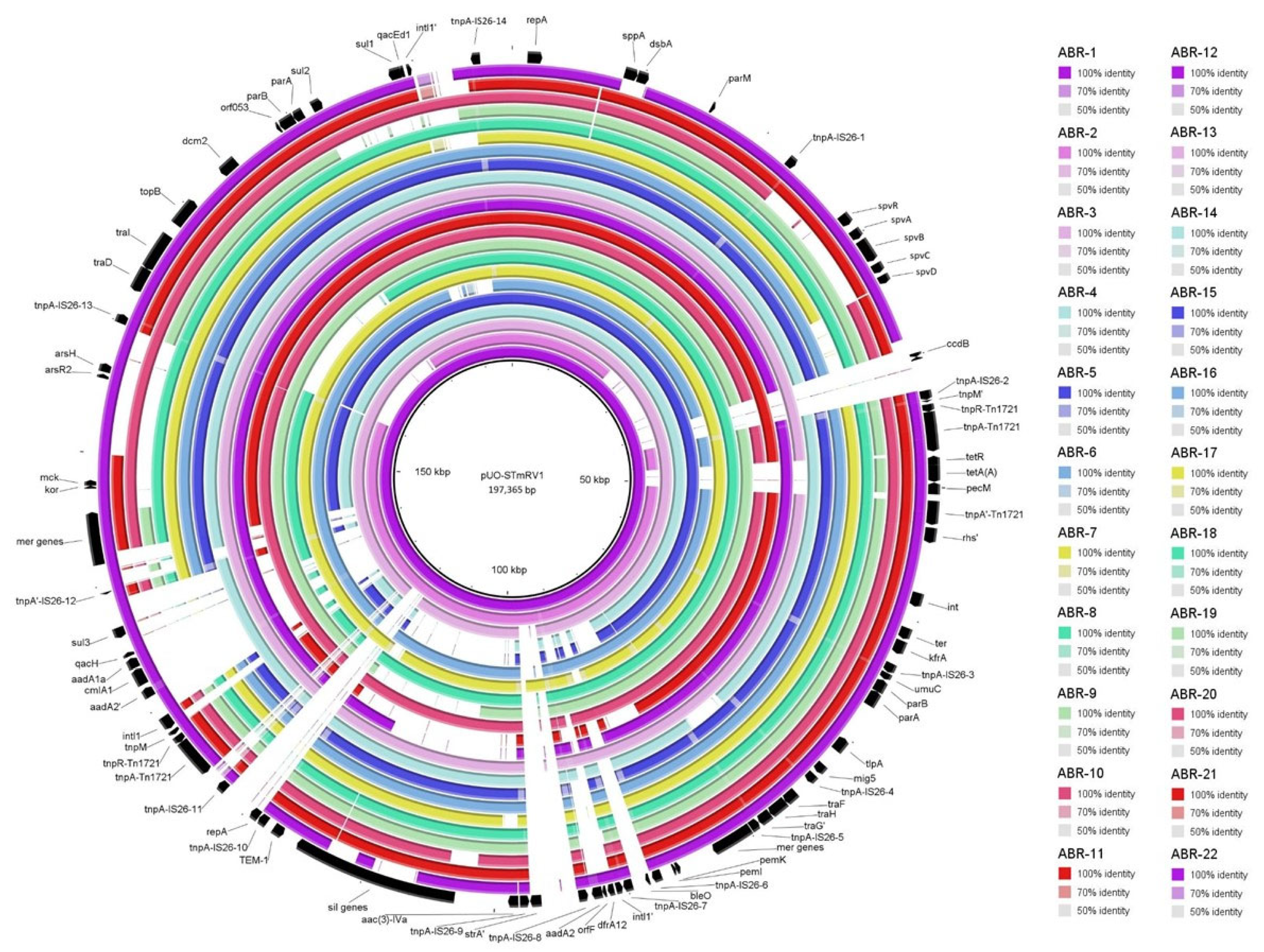
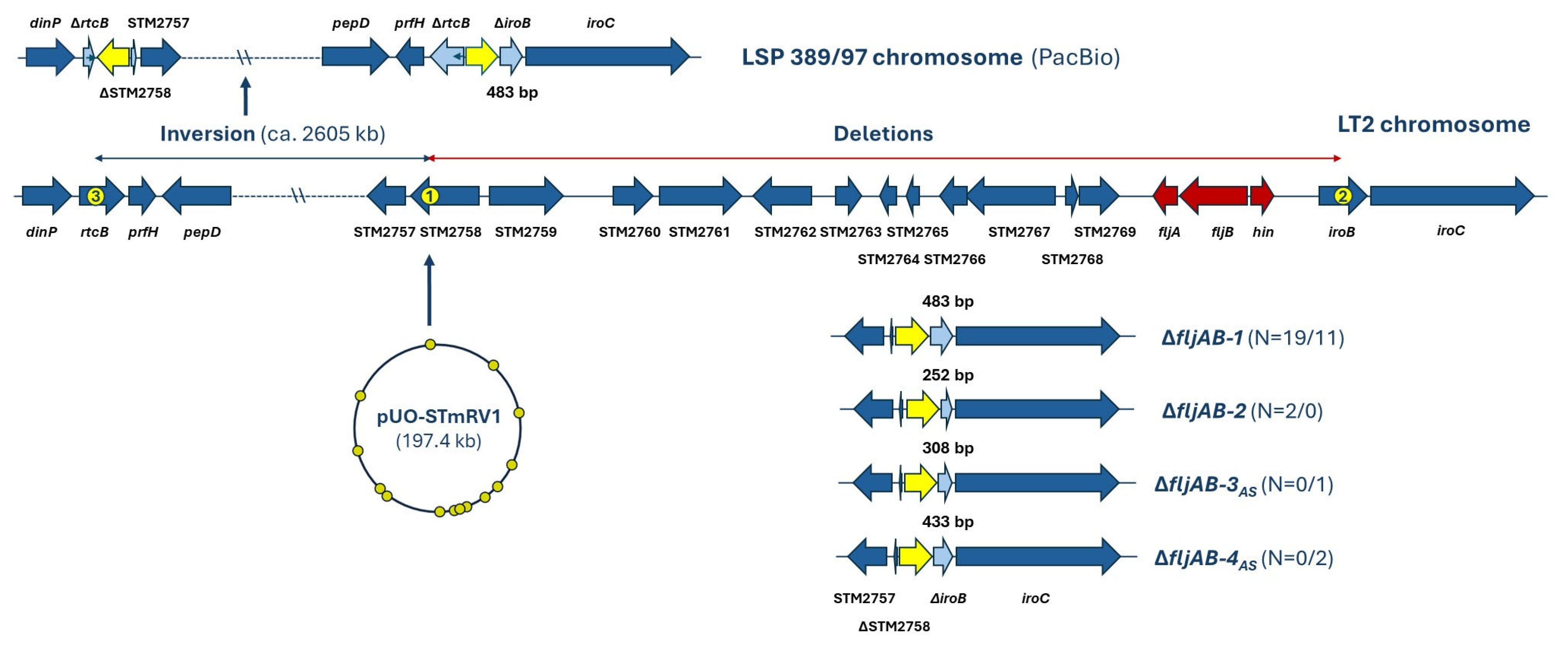

| Isolate a | Patient b Sex/Age Hospital c | AR d Phenotype Encoded by Resistance Plasmids | R-Plasmid (Inc; Size in bp) e Other Plasmids (Size in bp) f |
|---|---|---|---|
| LSP 389/97 | F/17 HUSA | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC/IncN (pUO-STmRV1; 197,365) * |
| LSP 272/98 | M/59 HJ | AMP, AMC, CHL, GEN, TOB, STR, SUL, TMP | IncC/IncN (137,711) unk * (4073), Col(BS512) * (2101) |
| LSP 21/00 | F/1 HCN | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC/IncN (166,540) unk * (4073), Col(BS512) * (2101) |
| LSP 37/00 | F/1 HCN | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC/IncN (167,805) unk * (4073), Col(BS512) * (2101) |
| LSP 40/00 | unk/unk unk | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC/IncN (166,539) unk * (4073), Col(BS512) * (2101) |
| LSP 127/00 | F/1 HUCA | SUL, TET | IncC/IncN (129,018) unk * (4073), Col(BS512) * (2101) |
| LSP 148/00 | F/2 HUC | SUL, TET | IncC (129,267) Col(BS512) * (2101) |
| LSP 151/00 | M/2 HUC | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC (156,873) unk * (4073), unk * (3373), Col(BS512) * (2101), OriColE * (1888) |
| LSP 247/00 | M/11 HUC | CHL, GEN, TOB, STR, SUL, TET, TMP | IncC (173,621) unk * (4073), Col(BS512) * (2101) |
| LSP 417/00 | F/unk HUC | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC (168,460) unk (14,418), unk * (4074), Col(BS512) * (2101) |
| LSP 41/01 | F/55 HCN | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC (168,287) unk (7940), Col(BS512) * (2101) |
| LSP 66/01 | F/4 HCN | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC (176,490) unk * (4073), Col(BS512) * (2101) |
| LSP 73/01 | M/4 HJ | AMP, AMC, GEN, TOB, STR, SUL, TET, TMP | IncC (147,514) unk * (4073), Col(BS512) * (2101) |
| LSP 80/01 | F/1 HUC | CHL, GEN, TOB, STR, SUL, TET, TMP | IncC (167,501) unk * (4074), Col(BS512) * (2101) |
| LSP 84/01 | M/unk HUC | AMP, AMC, CHL, GEN, TOB, STR, SUL | IncC (163,450) unk (10,648), Col(BS512) * (2101) |
| LSP 411/01 | M/unk HCN | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC (125,916) unk * (4073), Col(BS512) * (2101) |
| LSP 576/01 | M/unk HUC | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC (168,372) unk (14,418), unk * (4096), Col(BS512) * (2101) |
| LSP 578/01 | F/3 HUC | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC (161,336) unk * (3373), Col(BS512) * (2101), OriColE * (1888) |
| LSP 3/02 | M/1 HUCA | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC (173,402) unk * (4073), Col(BS512) * (2101), unk * (1989), OriColE * (1888) |
| LSP 262/02 | M/28 HUC | AMP, AMC, CHL, STR, SUL, TET | IncC (165,469) unk * (4074), Col(BS512) * (2101) |
| LSP 503/02 | F/30 HUCA | AMP, AMC, CHL, GEN, TOB, STR, SUL, TMP | IncC (169,183) unk * (4073), Col(BS512) * (2101), OriColE * (1888) |
| LSP 718/02 | F/5 HUCA | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET | IncC (159,273) unk * (4073), unk * (3373), Col(BS512) * (2101), OriColE * (1888) |
| LSP 1142/03 | F/73 HUC | CHL, GEN, TOB, STR, SUL, TET, TMP STR, SUL | IncC/IncN (183,291) IncI1-I(α)/ST154-(96,419) unk * (4073), Col(BS512) * (2101) |
| LSP 207/08 | F/70 HMN | AMP, AMC, CHL, GEN, TOB, STR, SUL, TMP | IncC (164,679) unk (14,518), Col(BS512) + Col8282 (12,901), unk (5052) |
| LSP 196/10 | F/32 HJ | AMP, AMC, GEN, TOB, SUL, TET | IncC (155,741) unk * (4073), Col(BS512) * (2101), oriColE * (1888) |
| LSP 48/12 | F/11 HUCA | AMP, AMC, GEN, TOB, SUL, TET | IncC (157,533) unk (4594), Col(BS512) (3142), unk (2677) |
| LSP 62/12 | F/57 HUC | AMP, AMC, GEN, TOB, SUL, TET | IncC (152,856) Col(BS512) (8718), oriColE (1044) |
| LSP 127/12 | M/73 HUC | SUL, TET | IncC (128,459) Col(pHAD28) * (4418), oriColE * (4230), unk (4203), oriColE * (3830), Col(BS512) (3143) |
| LSP 61/13 | M/4 HUC | AMP, AMC, SUL, TET | IncC (150,970) Col(BS512) (3143), oriColE (2570) |
| LSP 245/13 | F/25 HJ | AMP, AMC, GEN, TOB, SUL, TET | IncC (143,284) unk (14,518), unk (5424), Col(BS512) (3652), unk (2677) |
| LSP 259/13 | F/54 HJ | AMP, AMC, GEN, TOB, SUL, TET | IncC (157,802) unk (5115), Col(BS512) (3651), unk (2677) |
| LSP 474/14 | M/56 HUSA | AMP, AMC, GEN, TOB, SUL, TET | IncC (155,767) unk (7281), unk * (4073), unk (2622) |
| LSP 2/15 | M/75 HUSA | AMP, AMC, GEN, TOB, SUL, TET STR, SUL | IncC (131,910) IncI1-I(α)/ST259 (116,151) unk (4203), unk (3924), oriColE (2667) |
| LSP 438/15 | M/52 HUCA | AMP, AMC, GEN, TOB, SUL, TET | IncC (141,765) unk (4203), unk (2677), Col(BS512) (2622) |
| LSP 304/17 | F/6 HUSA | AMP, AMC, GEN, TOB, SUL, TET | IncC (140,638) Col8282 * (4091), unk * (4073), Col(BS512) (3142), oriColE (2677) |
| LSP 54/18 (LSP 53/18) | F/70 HUC | AMP, AMC, GEN, TOB, SUL, TET | IncC (137,672) unk * (54,805), Col(BS512) (4221), unk * (4091), Col8282 (3142), oriColE (2677) |
| LSP 148/18 | F/unk HUSA | AMP, AMC, CHL, GEN, TOB, STR, SUL, TET, TMP | IncC-(158,125) unk (8840), Col(BS512) (2621) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez, X.; García, P.; Fernández, J.; Ladero, V.; Rodríguez-Lucas, C.; Heinisch, J.J.; Rodicio, R.; Rodicio, M.R. Epidemiological Surveillance, Variability, and Evolution of Isolates Belonging to the Spanish Clone of the 4,[5],12:i:- Monophasic Variant of Salmonella enterica Serovar Typhimurium. Antibiotics 2025, 14, 711. https://doi.org/10.3390/antibiotics14070711
Vázquez X, García P, Fernández J, Ladero V, Rodríguez-Lucas C, Heinisch JJ, Rodicio R, Rodicio MR. Epidemiological Surveillance, Variability, and Evolution of Isolates Belonging to the Spanish Clone of the 4,[5],12:i:- Monophasic Variant of Salmonella enterica Serovar Typhimurium. Antibiotics. 2025; 14(7):711. https://doi.org/10.3390/antibiotics14070711
Chicago/Turabian StyleVázquez, Xenia, Patricia García, Javier Fernández, Víctor Ladero, Carlos Rodríguez-Lucas, Jürgen J. Heinisch, Rosaura Rodicio, and M. Rosario Rodicio. 2025. "Epidemiological Surveillance, Variability, and Evolution of Isolates Belonging to the Spanish Clone of the 4,[5],12:i:- Monophasic Variant of Salmonella enterica Serovar Typhimurium" Antibiotics 14, no. 7: 711. https://doi.org/10.3390/antibiotics14070711
APA StyleVázquez, X., García, P., Fernández, J., Ladero, V., Rodríguez-Lucas, C., Heinisch, J. J., Rodicio, R., & Rodicio, M. R. (2025). Epidemiological Surveillance, Variability, and Evolution of Isolates Belonging to the Spanish Clone of the 4,[5],12:i:- Monophasic Variant of Salmonella enterica Serovar Typhimurium. Antibiotics, 14(7), 711. https://doi.org/10.3390/antibiotics14070711










