Resistome and Phylogenomics of Escherichia coli Strains Obtained from Diverse Sources in Jimma, Ethiopia
Abstract
1. Introduction
2. Results
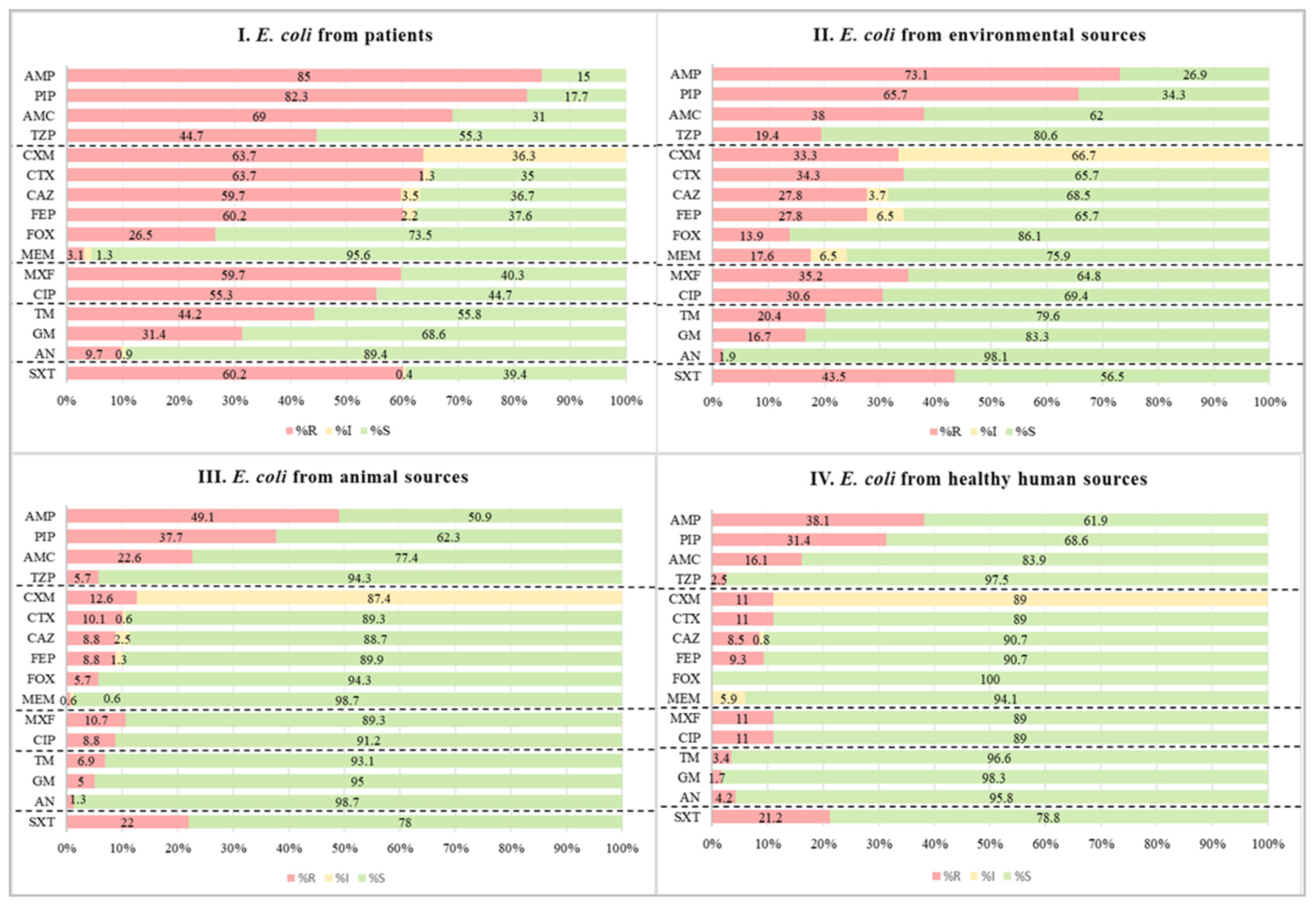
2.1. Phenotypic Antimicrobial Susceptibility Test Results
2.2. Minimal Inhibitory Concentrations (MICs) for Carbapenem-Resistant Isolates
2.3. Distribution and Co-Occurrence of β-Lactamase and Carbapenemase-Encoding Genes
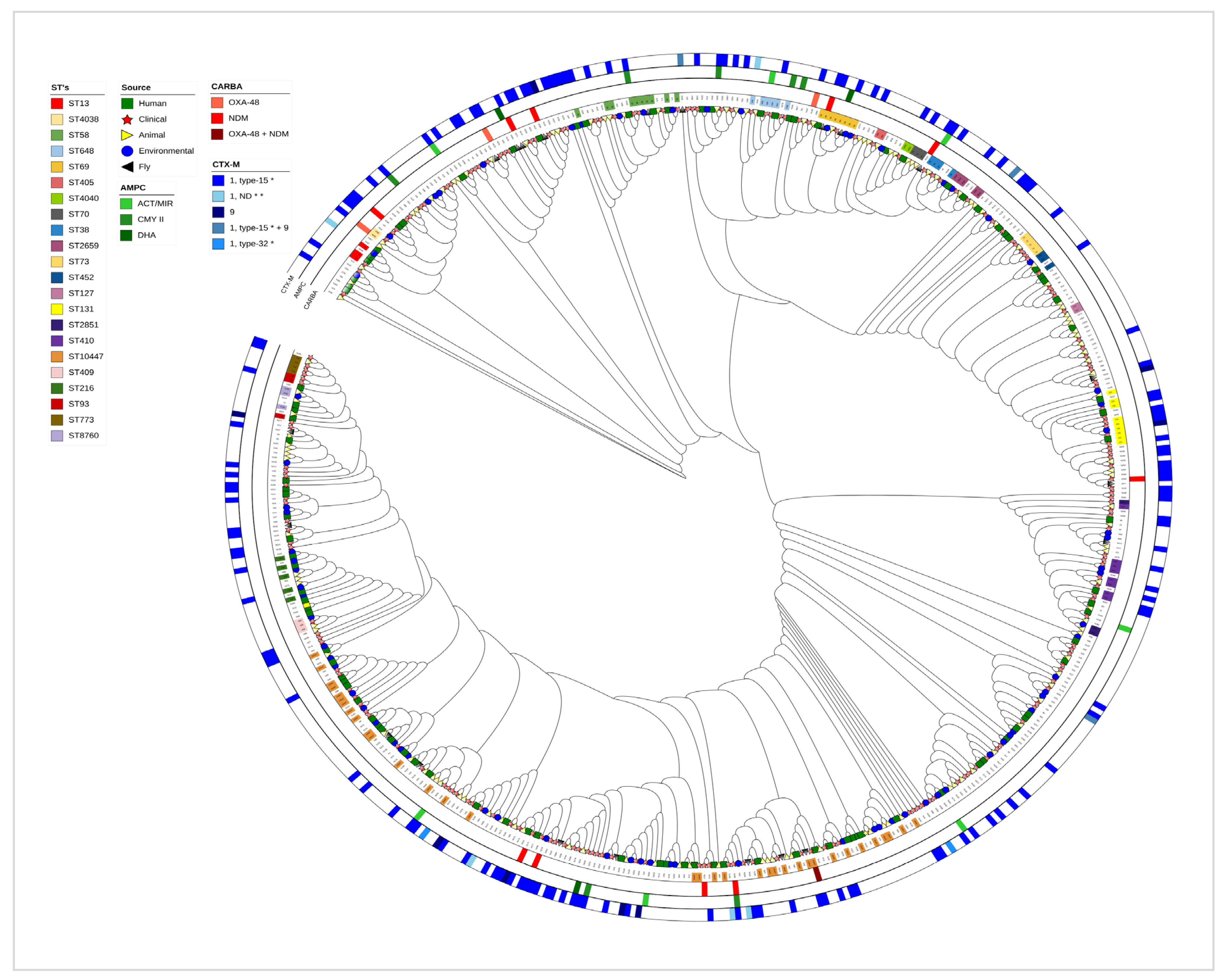
2.4. Multilocus Sequence Type (MLST) Comparison
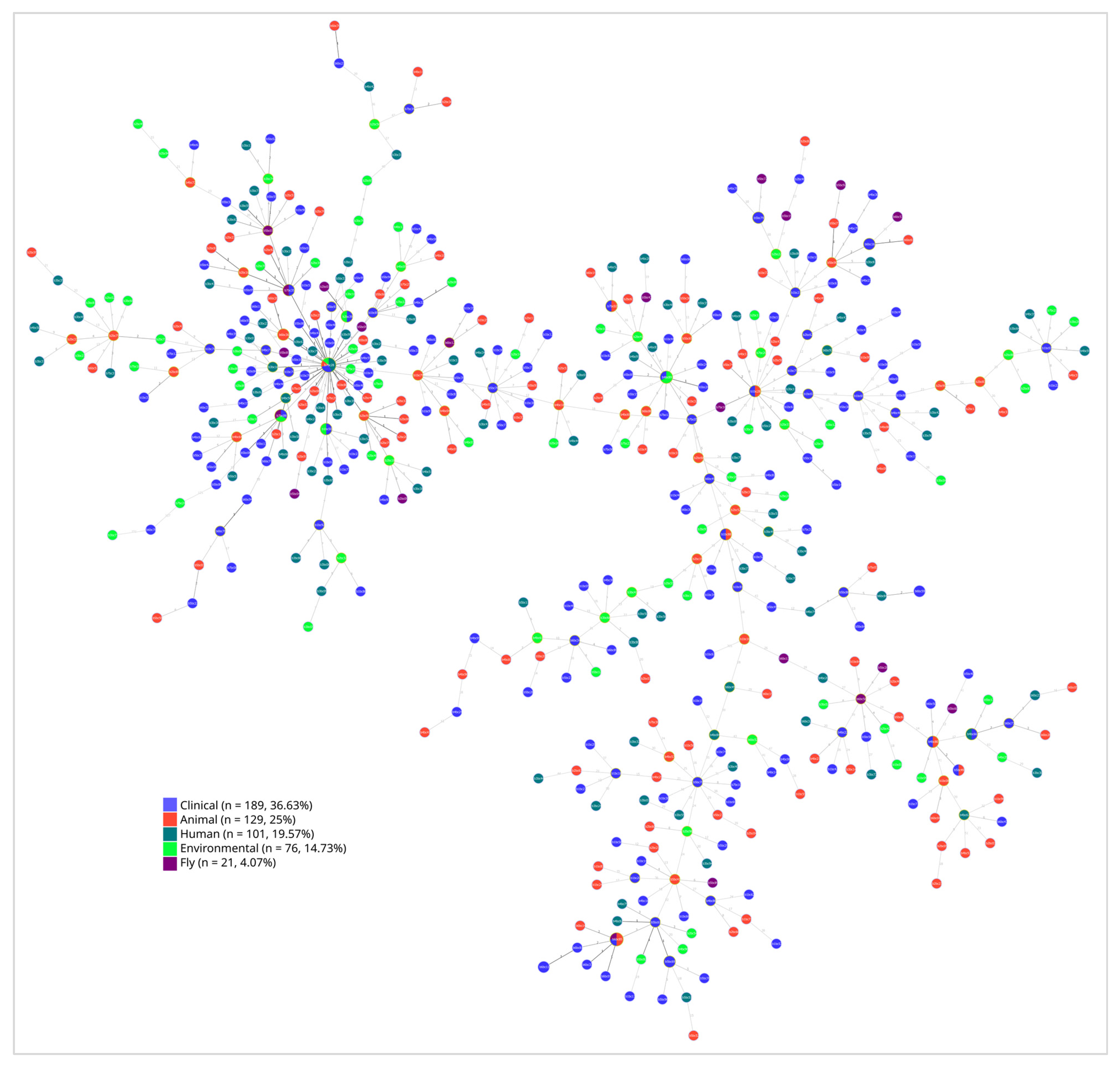
2.5. Phylogenetic Evidence of E. coli Transmission Across Human and Environmental Reservoirs
3. Discussion
Strengths and Limitations of the Study
4. Materials and Methods
4.1. Study Settings and Sample Collections
4.2. Culture and Identification
4.3. Qualitative Phenotypic Antimicrobial Susceptibility Tests
4.4. Quantitative Phenotypic Carbapenem Resistance
4.5. DNA Extraction
4.6. Characterization of ESBL- and Carbapenem-Resistant Isolates
4.7. Library Preparation and Multilocus Sequence Typing (MLST)
4.8. Bioinformatic Analysis
4.9. Quality Assurance
4.10. Ethical Considerations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, M.; Jiang, M.; Wang, Z.; Chen, R.; Zhuge, X.; Dai, J. Characterization of antimicrobial resistance in chicken-source phylogroup F Escherichia coli: Similar populations and resistance spectrums between E. coli recovered from chicken colibacillosis tissues and retail raw meats in Eastern China. Poult. Sci. 2021, 100, 101370. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Kong, L.; Liao, Y.; Tian, Y.; Wu, Q.; Liu, H.; Wang, X. Mini-Review: Antibiotic-Resistant Escherichia coli from Farm Animal-Associated Sources. Antibiotics 2022, 11, 1535. [Google Scholar] [CrossRef] [PubMed]
- Geurtsen, J.; de Been, M.; Weerdenburg, E.; Zomer, A.; McNally, A.; Poolman, J. Genomics and pathotypes of the many faces of Escherichia coli. FEMS Microbiol. Rev. 2022, 46, fuac031. [Google Scholar] [CrossRef] [PubMed]
- Puligundla, P.; Lim, S. Biocontrol Approaches against Escherichia coli O157:H7 in Foods. Foods 2022, 11, 756. [Google Scholar] [CrossRef]
- Gambushe, S.M.; Zishiri, O.T.; El Zowalaty, M.E. Review of Escherichia coli O157:H7 Prevalence, Pathogenicity, Heavy Metal and Antimicrobial Resistance, African Perspective. Infect. Drug Resist. 2022, 15, 4645–4673. [Google Scholar] [CrossRef]
- Ali, S.; Birhane, M.; Bekele, S.; Kibru, G.; Teshager, L.; Ylima, Y.; Ahmed, Y.; Fentahun, N.; Assefa, H.; Gashaw, M.; et al. Healthcare associated infection and its risk factors among patients admitted to a tertiary hospital in Ethiopia: Longitudinal study. Antimicrob. Resist. Infect. Control 2018, 7, 2. [Google Scholar] [CrossRef]
- van Duin, D.; Paterson, D.L. Multidrug-Resistant Bacteria in the Community: An Update. Infect. Dis. Clin. N. Am. 2020, 34, 709–722. [Google Scholar] [CrossRef]
- Arbab, S.; Ullah, H.; Wang, W.; Zhang, J. Antimicrobial drug resistance against Escherichia coli and its harmful effect on animal health. Vet. Med. Sci. 2022, 8, 1780–1786. [Google Scholar] [CrossRef]
- Castillo, A.K.; Espinoza, K.; Chaves, A.F.; Guibert, F.; Ruiz, J.; Pons, M.J. Antibiotic susceptibility among non-clinical Escherichia coli as a marker of antibiotic pressure in Peru (2009–2019): One health approach. Heliyon 2022, 8, e10573. [Google Scholar] [CrossRef]
- Fuga, B.; Sellera, F.P.; Cerdeira, L.; Esposito, F.; Cardoso, B.; Fontana, H.; Moura, Q.; Cardenas-Arias, A.; Sano, E.; Ribas, R.M.; et al. WHO critical priority Escherichia coli as One Health challenge for a post-pandemic scenario: Genomic surveillance and analysis of current trends in Brazil. Microbiol. Spectr. 2022, 10, e0125621. [Google Scholar] [CrossRef]
- Bindayna, K.M.; Joji, R.M.; Ezzat, H.; Jahrami, H.A. Antibiotic-resistance genes in E. coli strains in GCC countries: A meta-analysis. Saudi J. Med. Med. Sci. 2022, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Gniadkowski, M.; Giske, C.G.; Poirel, L.; Woodford, N.; Miriagou, V.; European Network on Carbapenemases. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Infect. 2012, 18, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Rosenkilde, C.E.; Munck, C.; Porse, A.; Linkevicius, M.; Andersson, D.I.; Sommer, M.O. Collateral sensitivity constrains resistance evolution of the CTX-M-15 β-lactamase. Nat. Commun. 2019, 10, 618. [Google Scholar] [CrossRef]
- Gashaw, M.; Berhane, M.; Bekele, S.; Kibru, G.; Teshager, L.; Yilma, Y.; Ahmed, Y.; Fentahun, N.; Assefa, H.; Wieser, A.; et al. Emergence of high drug resistant bacterial isolates from patients with health care associated infections at Jimma University medical center: A cross sectional study. Antimicrob. Resist. Infect. Control. 2018, 7, 138. [Google Scholar] [CrossRef]
- Fujita, A.W.; Werner, K.; Jacob, J.T.; Tschopp, R.; Mamo, G.; Mihret, A.; Abdissa, A.; Kempker, R.; Rebolledo, P.A. Antimicrobial resistance through the lens of one health in Ethiopia: A review of the literature among humans, animals, and the environment. Int. J. Infect. Dis. 2022, 119, 120–129. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbath, S.; Hindler, J.F.; Kahlmeter, G.; Liljequist, B.O.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2021. Available online: http://www.eucast.org (accessed on 16 October 2024).
- Chelkeba, L.; Melaku, T.; Mega, T.A. Gram-Negative Bacteria Isolates and Their Antibiotic-Resistance Patterns in Patients with Wound Infection in Ethiopia: A Systematic Review and Meta-Analysis. Infect. Drug Resist. 2021, 14, 277–302. [Google Scholar] [CrossRef]
- Chelkeba, L.; Fanta, K.; Mulugeta, T.; Melaku, T. Bacterial profile and antimicrobial resistance patterns of common bacteria among pregnant women with bacteriuria in Ethiopia: A systematic review and meta-analysis. Arch. Gynecol. Obstet. 2022, 306, 663–686. [Google Scholar] [CrossRef]
- Toy, T.; Pak, G.D.; Duc, T.P.; Campbell, J.I.; El Tayeb, M.A.; Kalckreuth, V.V.; Im, J.; Panzner, U.; Espinoza, L.M.C.; Eibach, D.; et al. Multicountry Distribution and Characterization of Extended-spectrum β-Lactamase-associated Gram-negative Bacteria from Bloodstream Infections in Sub-Saharan Africa. Clin. Infect. Dis. 2019, 69 (Suppl. 6), S449–S458. [Google Scholar] [CrossRef]
- Sewunet, T.; Asrat, D.; Woldeamanuel, Y.; Ny, S.; Westerlund, F.; Aseffa, A.; Giske, C.G. Polyclonal spread of blaCTX-M-15 through high-risk clones of Escherichia coli at a tertiary hospital in Ethiopia. J. Glob. Antimicrob. Resist. 2022, 29, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Zeynudin, A.; Pritsch, M.; Schubert, S.; Messerer, M.; Liegl, G.; Hoelscher, M.; Belachew, T.; Wieser, A. Prevalence and antibiotic susceptibility pattern of CTX-M type extended-spectrum β-lactamases among clinical isolates of gram-negative bacilli in Jimma, Ethiopia. BMC Infect. Dis. 2018, 18, 524. [Google Scholar] [CrossRef] [PubMed]
- Mahazu, S.; Sato, W.; Ayibieke, A.; Prah, I.; Hayashi, T.; Suzuki, T.; Iwanaga, S.; Ablordey, A.; Saito, R. Insights and genetic features of extended-spectrum beta-lactamase producing Escherichia coli isolates from two hospitals in Ghana. Sci. Rep. 2022, 12, 1843. [Google Scholar] [CrossRef]
- Olorunleke, S.O.; Kirchner, M.; Duggett, N.; AbuOun, M.; Okorie-Kanu, O.J.; Stevens, K.; Card, R.M.; Chah, K.F.; Nwanta, J.A.; Brunton, L.A.; et al. Molecular characterization of extended spectrum cephalosporin resistant Escherichia coli isolated from livestock and in-contact humans in Southeast Nigeria. Front. Microbiol. 2022, 13, 937968. [Google Scholar] [CrossRef] [PubMed]
- Freire, S.; Grilo, T.; Teixeira, M.L.; Fernandes, E.; Poirel, L.; Aires-de-Sousa, M. Screening and Characterization of Multidrug-Resistant Enterobacterales among Hospitalized Patients in the African Archipelago of Cape Verde. Microorganisms 2022, 10, 1426. [Google Scholar] [CrossRef]
- Khan, A.U.; Maryam, L.; Zarrilli, R. Structure, Genetics and Worldwide Spread of New Delhi Metallo-β-lactamase (NDM): A threat to public health. BMC Microbiol. 2017, 17, 101. [Google Scholar] [CrossRef]
- Wu, W.; Feng, Y.; Tang, G.; Qiao, F.; McNally, A.; Zong, Z. NDM Metallo-β-Lactamases and Their Bacterial Producers in Health Care Settings. Clin. Microbiol. Rev. 2019, 32, e00115-18. [Google Scholar] [CrossRef]
- Pritsch, M.; Zeynudin, A.; Messerer, M.; Baumer, S.; Liegl, G.; Schubert, S.; Löscher, T.; Hoelscher, M.; Belachew, T.; Rachow, A.; et al. First report on bla (NDM-1)-producing Acinetobacter baumannii in three clinical isolates from Ethiopia. BMC Infect. Dis. 2017, 17, 180. [Google Scholar] [CrossRef]
- Tian, D.; Wang, B.; Zhang, H.; Pan, F.; Wang, C.; Shi, Y.; Sun, Y. Dissemination of the bla(NDM-5) Gene via IncX3-Type Plasmid among Enterobacteriaceae in Children. mSphere 2020, 5, e00699-19. [Google Scholar] [CrossRef]
- Toledano-Tableros, J.E.; Gayosso-Vázquez, C.; Jarillo-Quijada, M.D.; Fernández-Vázquez, J.L.; Morfin-Otero, R.; Rodríguez-Noriega, E.; Giono-Cerezo, S.; Gutkind, G.; Conza, J.D.; Santos-Preciado, J.I.; et al. Dissemination of bla (NDM-) (1) Gene Among Several Klebsiella pneumoniae Sequence Types in Mexico Associated with Horizontal Transfer Mediated by IncF-Like Plasmids. Front. Microbiol. 2021, 12, 611274. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chen, W.Y.; Hsu, W.Y.; Tang, H.J.; Chou, Y.; Chang, Y.H.; Chen, C.C.; Chuang, Y.C.; Chang, T.H. Distribution of β-lactamases and emergence of carbapenemases co-occurring Enterobacterales isolates with high-level antibiotic resistance identified from patients with intra-abdominal infection in the Asia-Pacific region, 2015–2018. J. Microbiol. Immunol. Infect. 2022, 55 Pt 2, 1263–1272. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.H.; Tuckman, M.; Keeney, D.; Ruzin, A.; Bradford, P.A. Characterization and sequence analysis of extended-spectrum-{beta}-lactamase-encoding genes from Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates collected during tigecycline phase 3 clinical trials. Antimicrob. Agents Chemother. 2009, 53, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Martak, D.; Henriot, C.P.; Hocquet, D. Environment, animals, and food as reservoirs of antibiotic-resistant bacteria for humans: One health or more? Infect. Dis. Now. 2024, 54, 104895. [Google Scholar] [CrossRef]
- Gashaw, M.; Gudina, E.K.; Tadesse, W.; Froeschl, G.; Ali, S.; Seeholzer, T.; Kroidl, A.; Wieser, A. Hospital wastes as potential sources for multi-drug-resistant ESBL-producing bacteria at a tertiary hospital in Ethiopia. Antibiotics 2024, 13, 374. [Google Scholar] [CrossRef]
- Pai, M.; Gandra, S.; Thapa, P.; Carmona, S. Tackling antimicrobial resistance: Recognising the proposed five blind spots can accelerate progress. Lancet Microbe. 2025, 6, 100968. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Marialouis, X.A.; Al-Ansari, M.M.; Al-Humaid, L.; Santhanam, A.; Obulisamy, P.K. Multilocus sequence typing and ERIC-PCR fingerprinting of virulent clinical isolates of uropathogenic multidrug resistant Escherichia coli. J. King Saud Univ. Sci. 2022, 34, 101874. [Google Scholar] [CrossRef]
- Gupta, A.; Jordan, I.K.; Rishishwar, L. stringMLST: A fast k-mer based tool for multilocus sequence typing. Bioinformatics 2016, 33, 119–121. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.i.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Francisco, A.P.; Bugalho, M.; Ramirez, M.; Carriço, J.A. Global optimal eBURST analysis of multilocus typing data using a graphic matroid approach. BMC Bioinform. 2009, 10, 152. [Google Scholar] [CrossRef] [PubMed]
- Feil, E.J.; Li, B.C.; Aanensen, D.M.; Hanage, W.P.; Spratt, B.G. eBURST: Inferring Patterns of Evolutionary Descent among Clusters of Related Bacterial Genotypes from Multilocus Sequence Typing Data. J. Bacteriol. 2004, 186, 1518–1530. [Google Scholar] [CrossRef] [PubMed]



| Types of Antimicrobial Resistance Genes | Sources of E. coli Strains | Total | |||
|---|---|---|---|---|---|
| Patients % (n = 226) | Healthy Humans % (n = 118) | Animals % (n = 159) | Environment % (n = 108) | % (n = 611) | |
| Carbapenemase-encoding genes | 3.5 (8) | 0 | 0.6 (1) | 5.6 (6) | 2.5 (15) |
| NDM | 2.2 (5) | 0 | 0 | 5.6 (6) | 1.8 (11) |
| OXA-48 | 0.9 (2) | 0 | 0.6 (1) | 0 | 0.5 (3) |
| OXA-48 + NDM | 0.4 (1) | 0 | 0 | 0 | 0.2 (1) |
| ESBL-encoding genes | 59.7 (135) | 11.0 (13) | 9.4 (15) | 37.9 (41) | 33.4 (204) |
| CTX-M-15 | 55.3 (125) | 9.3 (11) | 7.5 (12) | 30.6 (33) | 29.6 (181) |
| CTX-M-9 | 1.3 (3) | 0.9 (1) | 1.3 (2) | 2.7 (3) | 1.5 (9) |
| CTX-M, ND | 1.8 (4) | 0 | 0.6 (1) | 2.8 (3) | 1.3 (8) |
| CTX-M-32 | 1.3 (3) | 0 | 0 | 0 | 0.5 (3) |
| CTX-M-15 + 9 | 0 | 0.9 (1) | 0 | 1.8 (2) | 0.5 (3) |
| AMPC-encoding genes | 7.1 (16) | 0 | 3.1 (5) | 5.6 (6) | 4.4 (27) |
| CMY II (n = 11) | 4.0 (9) | 0 | 0.6 (1) | 0.9 (1) | 1.8 (11) |
| ACT/MIR (n = 10) | 2.2 (5) | 0 | 1.3 (2) | 2.8 (3) | 1.6 (10) |
| DHA (n = 5) | 0.9 (2) | 0 | 0.6 (1) | 1.9 (2) | 0.8 (5) |
| ACT/MIR + DHA (n = 1) | 0 | 0 | 0.6 (1) | 0 | 0.2 (1) |
| TEM/SHV-encoding genes | 41.2 (93) | 5.1 (6) | 8.2 (13) | 34.3 (37) | 24.4 (149) |
| TEM (WT) (n = 144) | 39.4 (89) | 5.1 (6) | 8.2 (13) | 33.4 (36) | 23.6 (144) |
| SHV (WT) (n = 4) | 1.8 (4) | 0 | 0 | 0 | 0.6 (4) |
| TEM-104K + 164C (n = 1) | 0 | 0 | 0 | 0.9 (1) | 0.2 (1) |
| Total | 66.8 (151) | 11.9 (14) | 12.6 (20) | 54.6 (59) | 39.9 (244) |
| Types of Antimicrobial Resistance Genes | Sources of the Resistant Strains | Total | |
|---|---|---|---|
| Patients (n = 151) | Others (n = 93) | % (n = 244) | |
| Co-existing in carbapenemase-producing strains | 5.3 (8) | 7.5 (7) | 6.2 (15) |
| NDM + CTX-M-15 + TEM | 1.3 (2) | 2.2 (2) | 1.6 (4) |
| NDM + CTX-M-15 | 0.66 (1) | 3.2 (3) | 1.6 (4) |
| OXA-48 + CTX-M-15 + TEM | 1.3 (2) | 1.1 (1) | 1.2 (3) |
| NDM + CMY II + CTX-M-15 + TEM | 0.66 (1) | 0 | 0.4 (1) |
| OXA-48 + NDM + CTX-M-15 + TEM | 0.66 (1) | 0 | 0.4 (1) |
| NDM + CTX-M, ND + TEM | 0 | 1.1 (1) | 0.4 (1) |
| NDM | 0.66 (1) | 0 | 0.4 (1) |
| ESBL-producing strains | 85.4 (129) | 65.6 (61) | 77.9 (190) |
| CTX-M-15 + TEM | 39.1 (59) | 28.0 (26) | 34.8 (85) |
| CTX-M-15 | 31.1 (47) | 20.4 (19) | 27.1 (66) |
| CTX-M-9 | 2.0 (3) | 5.4 (5) | 3.3 (8) |
| CTX-M-15 + CMY II + TEM | 2.6 (4) | 0 | 1.6 (4) |
| CTX-M-15 + CMY II | 1.3 (2) | 2.2 (2) | 1.6 (4) |
| CTX-M group 1, ND + TEM | 2.0 (3) | 0 | 1.2 (3) |
| CTX-M-15 + TEM + SHV | 2.0 (3) | 0 | 1.2 (3) |
| CTX-M-32 + TEM | 2.0 (3) | 0 | 1.2 (3) |
| CTX-M group 1, ND | 0.66 (1) | 2.2 (2) | 1.2 (3) |
| CTX-M-15 + 9 + TEM | 0 | 2.2 (2) | 0.8 (2) |
| CTX-M-15 + ACT/MIR | 1.3 (2) | 0 | 0.8 (2) |
| CTX-M-15 + SHV | 0.66 (1) | 0 | 0.4 (1) |
| CTX-M-15 + DHA | 0.66 (1) | 0 | 0.4 (1) |
| CTX-M group 1, ND + ACT/MIR + DHA | 0 | 1.1 (1) | 0.4 (1) |
| CTX-M-15 + ACT/MIR + TEM | 0 | 1.1 (1) | 0.4 (1) |
| CTX-M-9 + ACT/MIR + TEM | 0 | 1.1 (1) | 0.4 (1) |
| CTX-M-15 + 9 | 0 | 1.1 (1) | 0.4 (1) |
| CTX-M-15 + DHA + TEM-104K + 164C | 0 | 1.1 (1) | 0.4 (1) |
| AMPC-encoding genes | 4.0 (6) | 5.4 (5) | 4.5 (11) |
| ACT/MIR | 2.0 (3) | 3.2 (3) | 2.4 (6) |
| CMY II + TEM | 1.3 (2) | 0 | 0.8 (2) |
| DHA | 0 | 2.2 (2) | 0.8 (2) |
| DHA + TEM | 0.66 (1) | 0 | 0.4 (1) |
| TEM-encoding genes | 5.3 (8) | 21.5 (20) | 11.5 (28) |
| TEM- (WT) | 5.3 (8) | 21.5 (20) | 11.5 (28) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gashaw, M.; Gudina, E.K.; Froeschl, G.; Matar, R.; Ali, S.; Gabriele, L.; Hohensee, A.; Seeholzer, T.; Kroidl, A.; Wieser, A. Resistome and Phylogenomics of Escherichia coli Strains Obtained from Diverse Sources in Jimma, Ethiopia. Antibiotics 2025, 14, 706. https://doi.org/10.3390/antibiotics14070706
Gashaw M, Gudina EK, Froeschl G, Matar R, Ali S, Gabriele L, Hohensee A, Seeholzer T, Kroidl A, Wieser A. Resistome and Phylogenomics of Escherichia coli Strains Obtained from Diverse Sources in Jimma, Ethiopia. Antibiotics. 2025; 14(7):706. https://doi.org/10.3390/antibiotics14070706
Chicago/Turabian StyleGashaw, Mulatu, Esayas Kebede Gudina, Guenter Froeschl, Ralph Matar, Solomon Ali, Liegl Gabriele, Amelie Hohensee, Thomas Seeholzer, Arne Kroidl, and Andreas Wieser. 2025. "Resistome and Phylogenomics of Escherichia coli Strains Obtained from Diverse Sources in Jimma, Ethiopia" Antibiotics 14, no. 7: 706. https://doi.org/10.3390/antibiotics14070706
APA StyleGashaw, M., Gudina, E. K., Froeschl, G., Matar, R., Ali, S., Gabriele, L., Hohensee, A., Seeholzer, T., Kroidl, A., & Wieser, A. (2025). Resistome and Phylogenomics of Escherichia coli Strains Obtained from Diverse Sources in Jimma, Ethiopia. Antibiotics, 14(7), 706. https://doi.org/10.3390/antibiotics14070706






