Whole Genome Sequence Analysis of Multidrug-Resistant Staphylococcus aureus and Staphylococcus pseudintermedius Isolated from Superficial Pyoderma in Dogs and Cats
Abstract
1. Introduction
2. Results
2.1. Isolation and Identification of Staphylococci
2.2. Antibiotic Susceptibility and MDR Patterns
2.3. Detection of Antibiotic-Resistant Genes
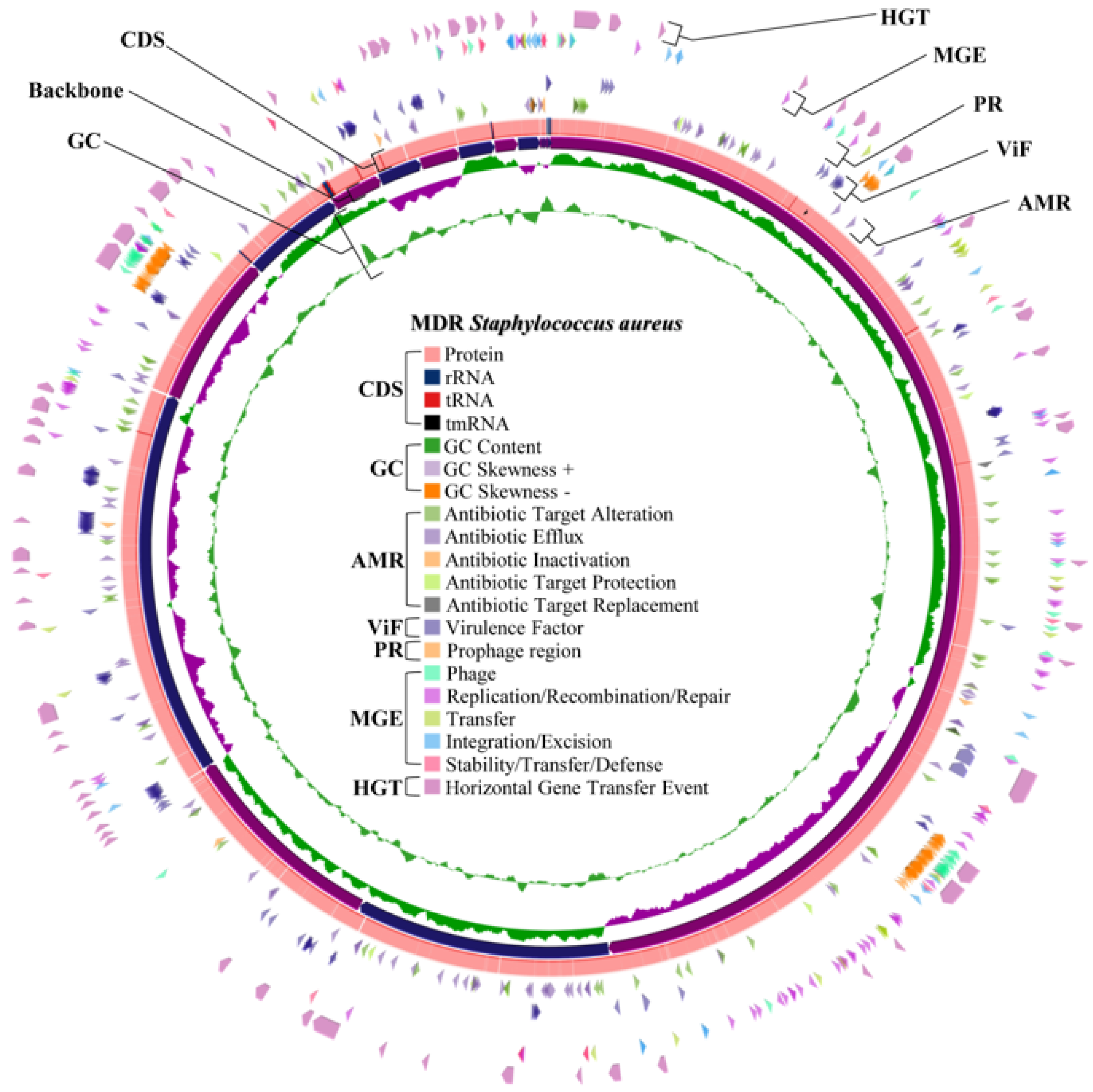
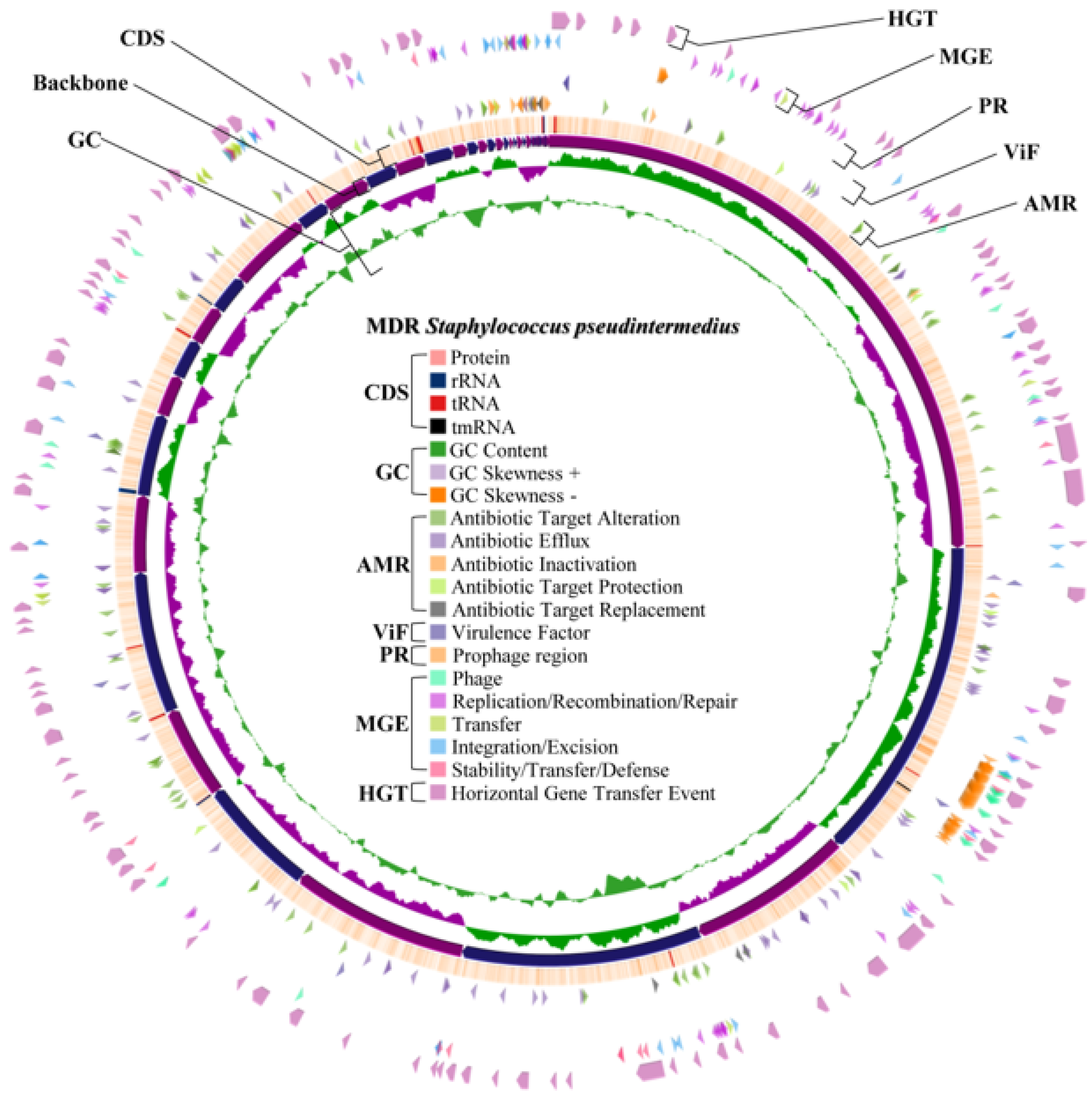
2.4. Whole Genome Characteristics of S. aureus WU1-1 and S. pseudintermedius WU48-1
2.5. Multilocus Sequence Typing of S. aureus WU1-1 and S. pseudintermedius WU48-1
2.6. Virulence Gene Determination in Whole Genome Sequences of S. aureus WU1-1 and S. pseudintermedius WU48-1
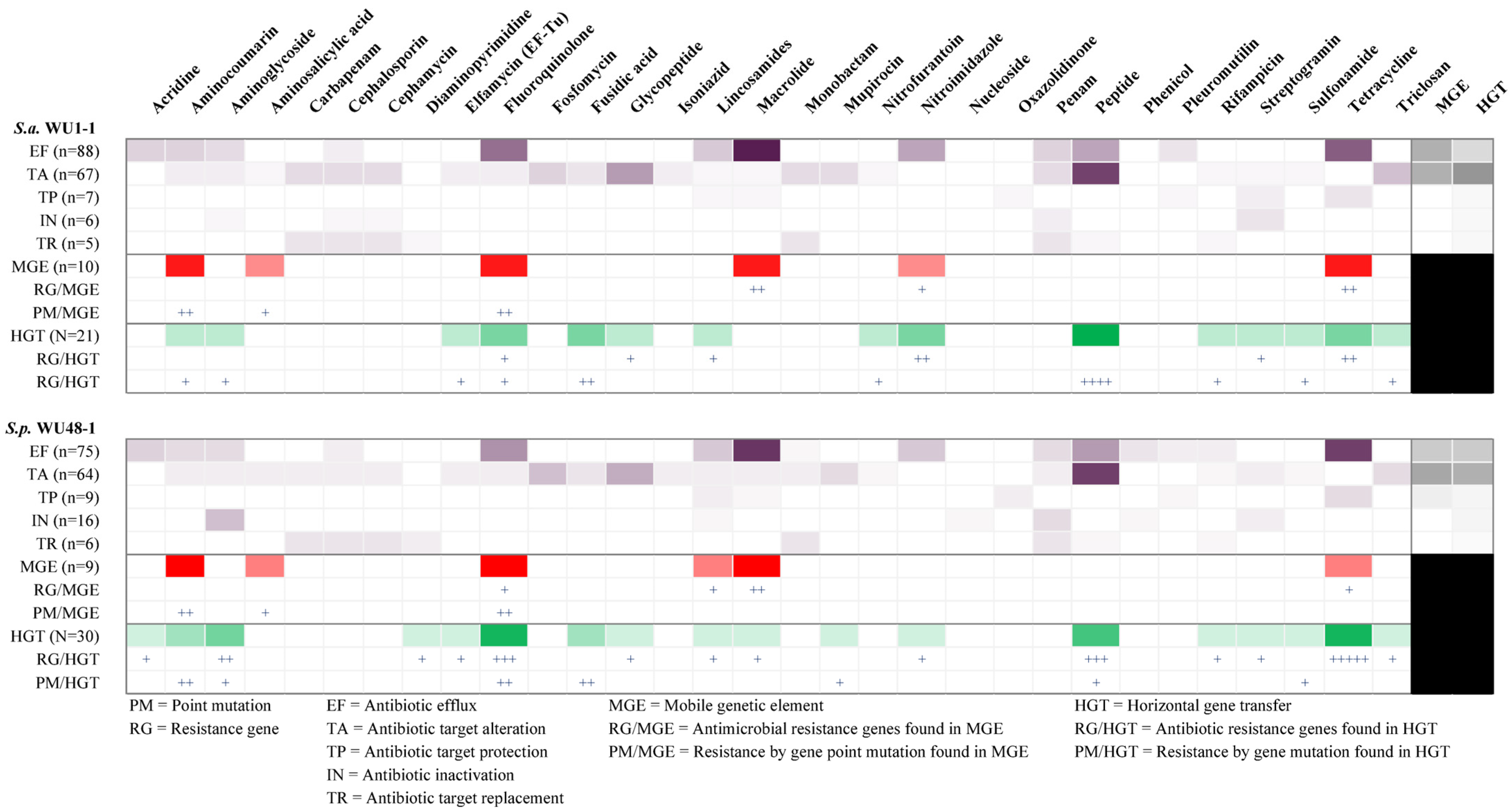
2.7. Antimicrobial Resistance Gene in Whole Genome Sequence Determination of S. aureus WU1-1 and S. pseudintermedius WU48-1
2.8. Horizontal Gene Transfer and Mobile Genetic Element Prediction of Antimicrobial Resistance Genes in S. aureus WU1-1 and S. pseudintermedius WU48-1
2.9. Prediction of the Prophage Insertion Region of S. aureus WU1-1 and S. pseudintermedius WU48-1
3. Discussion
4. Materials and Methods
4.1. Ethical Approval
4.2. Isolation and Identification of Bacteria
4.3. Antibiotic Susceptibility
4.4. Bacterial DNA Extraction
4.5. Detection of Antibiotic Resistance Genes
4.6. Whole Genome Sequencing and Bioinformatics
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Faccin, M.; Wiener, D.J.; Rech, R.R.; Santoro, D.; Rodrigues Hoffmann, A. Common Superficial and Deep Cutaneous Bacterial Infections in Domestic Animals: A Review. Vet. Pathol. 2023, 60, 796–811. [Google Scholar] [CrossRef]
- Cuny, C.; Layer-Nicolaou, F.; Weber, R.; Köck, R.; Witte, W. Colonization of Dogs and Their Owners with Staphylococcus aureus and Staphylococcus pseudintermedius in Households, Veterinary Practices, and Healthcare Facilities. Microorganisms 2022, 10, 677. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, A.; Lloyd, D.H. What Has Changed in Canine Pyoderma? A Narrative Review. Vet. J. 2018, 235, 73–82. [Google Scholar] [CrossRef]
- Rodrigues Hoffmann, A.; Patterson, A.P.; Diesel, A.; Lawhon, S.D.; Ly, H.J.; Elkins Stephenson, C.; Mansell, J.; Steiner, J.M.; Dowd, S.E.; Olivry, T.; et al. The Skin Microbiome in Healthy and Allergic Dogs. PLoS ONE 2014, 9, e83197. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.T.; Madhavan, V.; Kumar, P.; Muniraj, G.; Sivakumar, N.; Kannan, J. Epidemiology and Zoonotic Potential of Livestock-Associated Staphylococcus aureus Isolated at Tamil Nadu, India. BMC Microbiol. 2023, 23, 326. [Google Scholar] [CrossRef] [PubMed]
- Summers, J.F.; Brodbelt, D.C.; Forsythe, P.J.; Loeffler, A.; Hendricks, A. The Effectiveness of Systemic Antimicrobial Treatment in Canine Superficial and Deep Pyoderma: A Systematic Review. Vet. Dermatol. 2012, 23, 305–329, e61. [Google Scholar] [CrossRef] [PubMed]
- Couto, I.; Sanches, I.S.; Sá-Leão, R.; de Lencastre, H. Molecular Characterization of Staphylococcus sciuri Strains Isolated from Humans. J. Clin. Microbiol. 2000, 38, 1136–1143. [Google Scholar] [CrossRef]
- Östholm Balkhed, Å.; Söderlund, R.; Gunnarsson, L.; Wikström, C.; Ljung, H.; Claesson, C.; Börjesson, S. An Investigation of Household Dogs as the Source in a Case of Human Bacteraemia Caused by Staphylococcus pseudintermedius. Infect. Ecol. Epidemiol. 2023, 13, 2229578. [Google Scholar] [CrossRef]
- Stepanović, S.; Dimitrijević, V.; Vuković, D.; Dakić, I.; Savić, B.; Svabic-Vlahović, M. Staphylococcus sciuri as a Part of Skin, Nasal and Oral Flora in Healthy Dogs. Vet. Microbiol. 2001, 82, 177–185. [Google Scholar] [CrossRef]
- Somayaji, R.; Priyantha, M.A.R.; Rubin, J.E.; Church, D. Human Infections Due to Staphylococcus pseudintermedius, an Emerging Zoonosis of Canine Origin: Report of 24 Cases. Diagn. Microbiol. Infect. Dis. 2016, 85, 471–476. [Google Scholar] [CrossRef]
- Andrade, M.; Oliveira, K.; Morais, C.; Abrantes, P.; Pomba, C.; Rosato, A.E.; Couto, I.; Costa, S.S. Virulence Potential of Biofilm-Producing Staphylococcus pseudintermedius, Staphylococcus aureus and Staphylococcus coagulans Causing Skin Infections in Companion Animals. Antibiotics 2022, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.; Correia, E.; Pereira, J.E.; González-Machado, C.; Capita, R.; Alonso-Calleja, C.; Igrejas, G.; Poeta, P. Exploring the Biofilm Formation Capacity in S. pseudintermedius and Coagulase-Negative Staphylococci species. Pathogens 2022, 11, 689. [Google Scholar] [CrossRef] [PubMed]
- Götz, F. Staphylococcus and Biofilms. Mol. Microbiol. 2002, 43, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Chuprom, J.; Kidsin, K.; Sangkanu, S.; Nissapatorn, V.; Wiart, C.; de Lourdes Pereira, M.; Wongtawan, T.; Daus, M.; Sotthibandhu, D.S.; Tipmanee, V.; et al. Knema Retusa is Antibacterial and Antibiofilm against Antibiotic Resistant Staphylococcus aureus and S. haemolyticus Isolated in Bovine Mastitis. Vet. Res. Commun. 2022, 47, 523–538. [Google Scholar] [CrossRef]
- May, E.R. Bacterial Skin Diseases: Current Thoughts on Pathogenesis and Management. Vet. Clin. N. Am. Small Anim. Pract. 2006, 36, 185–202. [Google Scholar] [CrossRef]
- Tanveer, M.; Ntakiyisumba, E.; Hirwa, F.; Yoon, H.; Oh, S.-I.; Kim, C.; Kim, M.H.; Yoon, J.-S.; Won, G. Prevalence of Bacterial Pathogens Isolated from Canines with Pyoderma and Otitis Externa in Korea: A Systematic Review and Meta-Analysis. Vet. Sci. 2024, 11, 656. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Argudín, M.A.; Deplano, A.; Meghraoui, A.; Dodémont, M.; Heinrichs, A.; Denis, O.; Nonhoff, C.; Roisin, S. Bacteria from Animals as a Pool of Antimicrobial Resistance Genes. Antibiotics 2017, 6, 12. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic Resistance-the Need for Global Solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef]
- Velazquez-Meza, M.E.; Galarde-López, M.; Carrillo-Quiróz, B.; Alpuche-Aranda, C.M. Antimicrobial Resistance: One Health Approach. Vet. World 2022, 15, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Sanz, E.; Torres, C.; Lozano, C.; Zarazaga, M. High Diversity of Staphylococcus aureus and Staphylococcus 0seudintermedius Lineages and Toxigenic Traits in Healthy Pet-Owning Household Members. Underestimating Normal Household Contact? Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 83–94. [Google Scholar] [CrossRef]
- Khairullah, A.R.; Sudjarwo, S.A.; Effendi, M.H.; Ramandinianto, S.C.; Gelolodo, M.A.; Widodo, A.; Riwu, K.H.P.; Kurniawati, D.A. Pet Animals as Reservoirs for Spreading Methicillin-Resistant Staphylococcus aureus to Human Health. J. Adv. Vet. Anim. Res. 2023, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Stull, J.W.; Brophy, J.; Weese, J.S. Reducing the Risk of Pet-Associated Zoonotic Infections. Can. Med. Assoc. J. 2015, 187, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, L.; García-Fonticoba, R.; Pérez, D.; Viñes, J.; Fàbregas, N.; Madroñero, S.; Meroni, G.; Martino, P.A.; Martínez, S.; Maté, M.L.; et al. Whole Genome Sequencing and de Novo Assembly of Staphylococcus pseudintermedius: A Pangenome Approach to Unravelling Pathogenesis of Canine Pyoderma. Vet. Dermatol. 2021, 32, 654–663. [Google Scholar] [CrossRef]
- Little, S.V.; Bryan, L.K.; Hillhouse, A.E.; Konganti, K.; Lawhon, S.D. Whole-Genome Sequences of Staphylococcus pseudintermedius Isolates from Canine and Human Bacteremia Infections. Microbiol. Resour. Announc. 2019, 8, e00735-19. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Otto, M. Virulence Mechanisms of Staphylococcal Animal Pathogens. Int. J. Mol. Sci. 2023, 24, 14587. [Google Scholar] [CrossRef]
- Putriningsih, P.A.S.; Phuektes, P.; Jittimanee, S.; Kampa, J. Methicillin-Resistant Staphylococci in Canine Pyoderma in Thailand. Vet. World 2023, 16, 2340–2348. [Google Scholar] [CrossRef]
- Jantorn, P.; Heemmamad, H.; Soimala, T.; Indoung, S.; Saising, J.; Chokpaisarn, J.; Wanna, W.; Tipmanee, V.; Saeloh, D. Antibiotic Resistance Profile and Biofilm Production of Staphylococcus pseudintermedius Isolated from Dogs in Thailand. Pharmaceuticals 2021, 14, 592. [Google Scholar] [CrossRef]
- Chueahiran, S.; Yindee, J.; Boonkham, P.; Suanpairintr, N.; Chanchaithong, P. Methicillin-Resistant Staphylococcus aureus Clonal Complex 398 as a Major MRSA Lineage in Dogs and Cats in Thailand. Antibiotics 2021, 10, 243. [Google Scholar] [CrossRef]
- Lynch, S.A.; Helbig, K.J. The Complex Diseases of Staphylococcus pseudintermedius in Canines: Where to Next? Vet. Sci. 2021, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Ross Fitzgerald, J. The Staphylococcus intermedius Group of Bacterial Pathogens: Species Re-Classification, Pathogenesis and the Emergence of Meticillin Resistance. Vet. Dermatol. 2009, 20, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Bardiau, M.; Yamazaki, K.; Ote, I.; Misawa, N.; Mainil, J.G. Characterization of Methicillin-Resistant Staphylococcus pseudintermedius Isolated from Dogs and Cats. Microbiol. Immunol. 2013, 57, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Moses, I.B.; Santos, F.F.; Gales, A.C. Human Colonization and Infection by Staphylococcus pseudintermedius: An Emerging and Underestimated Zoonotic Pathogen. Microorganisms 2023, 11, 581. [Google Scholar] [CrossRef]
- Rana, E.A.; Nizami, T.A.; Islam, M.S.; Sarker, S.; Rahman, H.; Hoque, A.; Rahman, M. Antimicrobial Resistance and Virulence Profiling of Staphylococcus pseudintermedius Isolated from Cats, Bangladesh. Vet. Q. 2024, 44, 1–11. [Google Scholar] [CrossRef]
- Ben Chehida, F.; Tombari, W.; Gharsa, H.; Rabia, Y.; Ferhi, S.; Jrad, M.; Messadi, L. New Insights into Molecular Characterization, Antimicrobial Resistance and Virulence Factors of Methicillin-Sensitive Coagulase-Positive Staphylococcus spp. from Dogs with Pyoderma and Otitis Externa. Microbiol. Res. 2024, 15, 1208–1224. [Google Scholar] [CrossRef]
- Menandro, M.L.; Dotto, G.; Mondin, A.; Martini, M.; Ceglie, L.; Pasotto, D. Prevalence and Characterization of Methicillin-Resistant Staphylococcus pseudintermedius from Symptomatic Companion Animals in Northern Italy: Clonal Diversity and Novel Sequence Types. Comp. Immunol. Microbiol. Infect. Dis. 2019, 66, 101331. [Google Scholar] [CrossRef]
- Nocera, F.P.; De Martino, L. Methicillin-Resistant Staphylococcus pseudintermedius: Epidemiological Changes, Antibiotic Resistance, and Alternative Therapeutic Strategies. Vet. Res. Commun. 2024, 48, 3505–3515. [Google Scholar] [CrossRef]
- Jin, M.; Osman, M.; Green, B.A.; Yang, Y.; Ahuja, A.; Lu, Z.; Cazer, C.L. Evidence for the Transmission of Antimicrobial Resistant Bacteria between Humans and Companion Animals: A Scoping Review. One Health 2023, 17, 100593. [Google Scholar] [CrossRef]
- Rodrigues Hoffmann, A.; Proctor, L.M.; Surette, M.G.; Suchodolski, J.S. The Microbiome: The Trillions of Microorganisms That Maintain Health and Cause Disease in Humans and Companion Animals. Vet. Pathol. 2016, 53, 10–21. [Google Scholar] [CrossRef]
- Abdolghanizadeh, S.; Salmeh, E.; Mirzakhani, F.; Soroush, E.; Siadat, S.D.; Tarashi, S. Microbiota Insights into Pet Ownership and Human Health. Res. Vet. Sci. 2024, 171, 105220. [Google Scholar] [CrossRef] [PubMed]
- WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance. Ranking of Medically Important Antimicrobials for Risk Management of Antimicrobial Resistance Due to Non-Human Use; World Health Organization: Geneva, Switzerland, 2018; ISBN 9789241515528. [Google Scholar]
- Sahin-Tóth, J.; Kovács, E.; Tóthpál, A.; Juhász, J.; Forró, B.; Bányai, K.; Havril, K.; Horváth, A.; Ghidán, Á.; Dobay, O. Whole Genome Sequencing of Coagulase Positive Staphylococci from a Dog-and-Owner Screening Survey. PLoS ONE 2021, 16, e0245351. [Google Scholar] [CrossRef] [PubMed]
- Altwiley, D.; Brignoli, T.; Edwards, A.; Recker, M.; Lee, J.C.; Massey, R.C. A Functional Menadione Biosynthesis Pathway is Required for Capsule Production by Staphylococcus aureus. Microbiology 2021, 167, 001108. [Google Scholar] [CrossRef] [PubMed]
- Zaatout, N.; Ayachi, A.; Kecha, M. Staphylococcus aureus Persistence Properties Associated with Bovine Mastitis and Alternative Therapeutic Modalities. J. Appl. Microbiol. 2020, 129, 1102–1119. [Google Scholar] [CrossRef]
- Ridder, M.J.; Daly, S.M.; Hall, P.R.; Bose, J.L. Quantitative Hemolysis Assays. Methods Mol. Biol. 2021, 2341, 25–30. [Google Scholar]
- Zhu, Z.; Hu, Z.; Li, S.; Fang, R.; Ono, H.K.; Hu, D.-L. Molecular Characteristics and Pathogenicity of Staphylococcus aureus Exotoxins. Int. J. Mol. Sci. 2023, 25, 395. [Google Scholar] [CrossRef]
- Song, L.; Hobaugh, M.R.; Shustak, C.; Cheley, S.; Bayley, H.; Gouaux, J.E. Structure of Staphylococcal Alpha-Hemolysin, a Heptameric Transmembrane Pore. Science 1996, 274, 1859–1866. [Google Scholar] [CrossRef]
- Lemjabbar, H.; Basbaum, C. Platelet-Activating Factor Receptor and ADAM10 Mediate Responses to Staphylococcus aureus in Epithelial Cells. Nat. Med. 2002, 8, 41–46. [Google Scholar] [CrossRef]
- Maretzky, T.; Reiss, K.; Ludwig, A.; Buchholz, J.; Scholz, F.; Proksch, E.; de Strooper, B.; Hartmann, D.; Saftig, P. ADAM10 Mediates E-Cadherin Shedding and Regulates Epithelial Cell-Cell Adhesion, Migration, and Beta-Catenin Translocation. Proc. Natl. Acad. Sci. USA 2005, 102, 9182–9187. [Google Scholar] [CrossRef]
- Bhakdi, S.; Tranum-Jensen, J. Alpha-Toxin of Staphylococcus aureus. Microbiol. Rev. 1991, 55, 733–751. [Google Scholar] [CrossRef]
- Huseby, M.; Shi, K.; Brown, C.K.; Digre, J.; Mengistu, F.; Seo, K.S.; Bohach, G.A.; Schlievert, P.M.; Ohlendorf, D.H.; Earhart, C.A. Structure and Biological Activities of Beta Toxin from Staphylococcus aureus. J. Bacteriol. 2007, 189, 8719–8726. [Google Scholar] [CrossRef] [PubMed]
- Kmieciak, W.; Szewczyk, E.M.; Ciszewski, M. Searching for Beta-Haemolysin Hlb Gene in Staphylococcus pseudintermedius with Species-Specific Primers. Curr. Microbiol. 2016, 73, 148–152. [Google Scholar] [CrossRef]
- Jia, Y.; Guan, Z.; Liu, C.; Huang, M.; Li, J.; Feng, J.; Shen, B.; Yang, G. Staphylococcus aureus β-Hemolysin Causes Skin Inflammation by Acting as an Agonist of Epidermal Growth Factor Receptor. Microbiol. Spectr. 2024, 12, e0222723. [Google Scholar] [CrossRef]
- Tajima, A.; Iwase, T.; Shinji, H.; Seki, K.; Mizunoe, Y. Inhibition of Endothelial Interleukin-8 Production and Neutrophil Transmigration by Staphylococcus aureus Beta-Hemolysin. Infect. Immun. 2009, 77, 327–334. [Google Scholar] [CrossRef]
- Katayama, Y.; Baba, T.; Sekine, M.; Fukuda, M.; Hiramatsu, K. Beta-Hemolysin Promotes Skin Colonization by Staphylococcus aureus. J. Bacteriol. 2013, 195, 1194–1203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, S.; Zhang, X.-K.; Wang, Y.; Yang, L.-Y.; Zeng, H.; Yan, D.-P.; Zou, Q.-M.; Zuo, Q.-F. Mechanisms of Fibronectin-Binding Protein A (FnBPA(110-263)) Vaccine Efficacy in Staphylococcus aureus Sepsis versus Skin Infection. Clin. Immunol. 2018, 194, 1–8. [Google Scholar] [CrossRef]
- Cho, S.H.; Strickland, I.; Boguniewicz, M.; Leung, D.Y. Fibronectin and Fibrinogen Contribute to the Enhanced Binding of Staphylococcus aureus to Atopic Skin. J. Allergy Clin. Immunol. 2001, 108, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Bowman, L.; Palmer, T. The Type VII Secretion System of Staphylococcus. Annu. Rev. Microbiol. 2021, 75, 471–494. [Google Scholar] [CrossRef]
- Brdová, D.; Ruml, T.; Viktorová, J. Mechanism of Staphylococcal Resistance to Clinically Relevant Antibiotics. Drug Resist. Updat. Rev. Comment. Antimicrob. Anticancer Chemother. 2024, 77, 101147. [Google Scholar] [CrossRef]
- Vandendriessche, S.; Vanderhaeghen, W.; Soares, F.V.; Hallin, M.; Catry, B.; Hermans, K.; Butaye, P.; Haesebrouck, F.; Struelens, M.J.; Denis, O. Prevalence, Risk Factors and Genetic Diversity of Methicillin-Resistant Staphylococcus aureus Carried by Humans and Animals across Livestock Production Sectors. J. Antimicrob. Chemother. 2013, 68, 1510–1516. [Google Scholar] [CrossRef]
- Dashtbani-Roozbehani, A.; Brown, M.H. Efflux Pump Mediated Antimicrobial Resistance by Staphylococci in Health-Related Environments: Challenges and the Quest for Inhibition. Antibiotics 2021, 10, 1502. [Google Scholar] [CrossRef]
- Lindsay, J.A.; Holden, M.T.G. Staphylococcus aureus: Superbug, Super Genome? Trends Microbiol. 2004, 12, 378–385. [Google Scholar] [CrossRef]
- Malachowa, N.; DeLeo, F.R. Mobile Genetic Elements of Staphylococcus aureus. Cell. Mol. Life Sci. 2010, 67, 3057–3071. [Google Scholar] [CrossRef] [PubMed]
- Alibayov, B.; Baba-Moussa, L.; Sina, H.; Zdeňková, K.; Demnerová, K. Staphylococcus aureus Mobile Genetic Elements. Mol. Biol. Rep. 2014, 41, 5005–5018. [Google Scholar] [CrossRef] [PubMed]
- Feßler, A.T.; Wang, Y.; Wu, C.; Schwarz, S. Mobile Macrolide Resistance Genes in Staphylococci. Plasmid 2018, 99, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, J.A. Staphylococcus aureus Genomics and the Impact of Horizontal Gene Transfer. Int. J. Med. Microbiol. 2014, 304, 103–109. [Google Scholar] [CrossRef]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic Resistance Genes in Bacteria: Occurrence, Spread, and Control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef]
- Gu, Y.; Shen, S.; Han, B.; Tian, X.; Yang, F.; Zhang, K. Family Livestock Waste: An Ignored Pollutant Resource of Antibiotic Resistance Genes. Ecotoxicol. Environ. Saf. 2020, 197, 110567. [Google Scholar] [CrossRef]
- Broaders, E.; Gahan, C.G.M.; Marchesi, J.R. Mobile Genetic Elements of the Human Gastrointestinal Tract: Potential for Spread of Antibiotic Resistance Genes. Gut Microbes 2013, 4, 271–280. [Google Scholar] [CrossRef]
- Deshamukhya, C.; Bhowmik, D.; Dhar Chanda, D.; Bhattacharjee, A. Tn5406, a Staphylococcal Transposon Associated with Macrolide-Lincosamide-Streptogramin(b) Resistance in Clinical Isolates of Staphylococcus aureus. Indian J. Med. Microbiol. 2023, 42, 30–33. [Google Scholar] [CrossRef]
- Markhali, H.F.; Habibollahi, H.; Safari Motlagh, M.R.; Kaviani, B.; Roque-Borda, C.A. Genetic Diversity of Tn916 Transposon and Its Association with Tetracycline Resistance in Staphylococcus aureus Isolates. Ecol. Genet. Genom. 2024, 33, 100306. [Google Scholar] [CrossRef]
- Noble, W.C.; Rahman, M.; Karadec, T.; Schwarz, S. Gentamicin Resistance Gene Transfer from Enterococcus faecalis and E. faecium to Staphylococcus aureus, S. intermedius and S. hyicus. Vet. Microbiol. 1996, 52, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Pertics, B.Z.; Szénásy, D.; Dunai, D.; Born, Y.; Fieseler, L.; Kovács, T.; Schneider, G. Isolation of a Novel Lytic Bacteriophage against a Nosocomial Methicillin-Resistant Staphylococcus aureus Belonging to ST45. BioMed Res. Int. 2020, 2020, 5463801. [Google Scholar] [CrossRef] [PubMed]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Clinical & Laboratory Standards Institute (CLSI). Supplement M100. In Performance Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2020. [Google Scholar]
- Qu, Y.; Zhao, H.; Nobrega, D.B.; Cobo, E.R.; Han, B.; Zhao, Z.; Li, S.; Li, M.; Barkema, H.W.; Gao, J. Molecular Epidemiology and Distribution of Antimicrobial Resistance Genes of Staphylococcus Species Isolated from Chinese Dairy Cows with Clinical Mastitis. J. Dairy Sci. 2019, 102, 1571–1583. [Google Scholar] [CrossRef]
- Pournajaf, A.; Ardebili, A.; Goudarzi, L.; Khodabandeh, M.; Narimani, T.; Abbaszadeh, H. PCR-Based Identification of Methicillin-Resistant Staphylococcus aureus Strains and Their Antibiotic Resistance Profiles. Asian Pac. J. Trop. Biomed. 2014, 4, S293–S297. [Google Scholar] [CrossRef]
- Strompfová, V.; Štempelová, L.; Bujňáková, D.; Karahutová, L.; Nagyová, M.; Siegfried, L. Virulence Determinants and Antibiotic Resistance in Staphylococci Isolated from the Skin of Captive Bred Reptiles. Vet. Res. Commun. 2024, 48, 1471–1480. [Google Scholar] [CrossRef]
- Gharaibeh, M.H.; Mahafzah, T.A.; Abu-Qatouseh, L.F.; Khanfar, M.; Abdulmawjood, A. Molecular Characterization and Antimicrobial-Resistance Gene Profile of Staphylococcus aureus Strains Isolated from Ovine Mastitis in Jordan. Vet. World 2025, 18, 270–279. [Google Scholar] [CrossRef]
- Youn, J.-H.; Park, Y.H.; Hang’ombe, B.; Sugimoto, C. Prevalence and Characterization of Staphylococcus aureus and Staphylococcus pseudintermedius Isolated from Companion Animals and Environment in the Veterinary Teaching Hospital in Zambia, Africa. Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 123–130. [Google Scholar] [CrossRef]
- El-Razik, K.A.A.; Arafa, A.A.; Hedia, R.H.; Ibrahim, E.S. Tetracycline Resistance Phenotypes and Genotypes of Coagulase-Negative Staphylococcal Isolates from Bubaline Mastitis in Egypt. Vet. World 2017, 10, 702–710. [Google Scholar] [CrossRef]
- Li, X.; Huang, T.; Xu, K.; Li, C.; Li, Y. Molecular Characteristics and Virulence Gene Profiles of Staphylococcus aureus Isolates in Hainan, China. BMC Infect. Dis. 2019, 19, 873. [Google Scholar] [CrossRef] [PubMed]

| Animals | No. of Isolates | No. of Staphylococci | p-Value |
|---|---|---|---|
| Canine (n = 42) | 75 (76.53%) | 46 (46.94%) | 0.93 ns |
| Feline (n = 14) | 23 (23.47%) | 15 (15.31%) | |
| Total | 98 (100%) | 61 (62.24%) |
| Groups of Microorganisms | No. of Isolates | Percentage (%) |
|---|---|---|
| 1. Coagulase positive staphylococci | ||
| Staphylococcus aureus | 5 | 5.10% |
| Staphylococcus delphini | 1 | 1.02% |
| Staphylococcus intermedius | 13 | 13.26% |
| Staphylococcus pseudintermedius | 3 | 3.06% |
| 2. Coagulase negative staphylococci | ||
| Staphylococcus arlettae | 1 | 1.02% |
| Staphylococcus cohnii | 8 | 8.16% |
| Staphylococcus epidermidis | 2 | 2.04% |
| Staphylococcus felis | 2 | 2.04% |
| Staphylococcus pasteuri | 3 | 3.06% |
| Staphylococcus saprophyticus | 13 | 13.26% |
| Staphylococcus sciuri | 8 | 8.16% |
| Staphylococcus simulans | 2 | 2.04% |
| 3. Non-staphylococci | ||
| Brevibacterium casei | 7 | 7.14% |
| Micrococcus luteus | 11 | 11.22% |
| Enterococcus faecalis | 1 | 1.02% |
| Enterococcus faecium | 2 | 2.04% |
| Enterococcus hirae | 2 | 2.04% |
| Rothia nasimurium | 2 | 2.04% |
| 4. Yeast | ||
| Candida tropicalis | 8 | 8.16% |
| 5. No organism identification | 4 | 4.08% |
| Total | 98 | 100% |
| Antibiotics | Susceptibility (n = 61) | Reference Strains (S. aureus) | ||||
|---|---|---|---|---|---|---|
| Class | Name | S | I | R | ATCC25923 | DMST4775 |
| Aminopenicillin | Penicillin G | 39 | 4 | 18 | S | S |
| Ampicillin | 47 | 0 | 14 | S | S | |
| Methicillin | 59 | 1 | 1 | S | S | |
| Macrolide | Erythromycin | 36 | 12 | 13 | S | S |
| Phenicol | Chloramphenicol | 53 | 1 | 7 | S | S |
| Fluoroquinolone | Ciprofloxacin | 59 | 0 | 2 | S | S |
| Aminoglycosides | Kanamycin | 52 | 1 | 8 | S | S |
| Gentamicin | 60 | 1 | 0 | S | S | |
| Tetracycline | Doxycycline | 49 | 0 | 12 | S | S |
| Antifolate | Trimethoprim | 41 | 16 | 4 | S | S |
| MDR Pattern | No. of Drug Class | No. of Isolates |
|---|---|---|
| PEN-ERY-DOX | 3 | 1 (9.09%) |
| PEN-AMP-KAN-DOX | 3 | 6 (54.55%) |
| PEN-AMP-DOX-TRI | 3 | 2 (18.18%) |
| PEN-AMP-CIP-KAN-TRI | 4 | 1 (9.09%) |
| PEN-AMP-MET-ERY-CHL-CIP-KAN-DOX-TRI | 7 | 1 (9.09%) |
| Total | - | 11 (100%) |
| Isolates | Antibiotic-Resistant Genes | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta-Lactam | Trimethoprim | Aminoglycoside | Chloramphenicol | Tetracycline | |||||
| blaZ | mecA | aac | dfrK | aph2 | aph3 | cat1 | tetK | tetM | |
| S. aureus | |||||||||
| WU1-1 | + | - | - | - | - | + | - | - | - |
| WU7-2 | + | - | - | - | - | + | - | - | - |
| WU13-1 | + | - | - | - | - | + | - | - | - |
| WU20-1 | + | - | - | - | - | - | - | - | + |
| WU52-2 | + | - | - | - | - | + | - | - | - |
| S. pseudintermedius | |||||||||
| WU47-1 | + | - | - | - | - | - | - | - | + |
| WU48-1 | + | + | + | + | - | + | - | - | + |
| WU55-1 | - | - | - | - | - | - | - | - | + |
| S. sciuri | |||||||||
| WU10-2 | - | - | - | - | - | + | - | - | - |
| WU29-1 | + | - | - | - | - | - | - | - | - |
| WU30-2 | - | - | - | - | - | - | - | - | - |
| WU34-1 | - | - | - | - | - | - | - | - | - |
| WU35-1 | - | - | - | - | - | - | - | - | - |
| WU35-2 | + | - | - | - | - | - | - | - | - |
| WU37-2 | - | - | - | - | - | - | - | - | - |
| WU53-1 | + | - | - | - | - | + | - | - | - |
| S. epidermidis | |||||||||
| WU24-1 | + | - | - | - | - | - | - | - | - |
| WU15-1 | + | + | - | - | - | - | - | + | - |
| No. of isolates | 12 | 2 | 1 | 1 | 0 | 7 | 0 | 1 | 4 |
| Percentage (%) | 67 | 11 | 6 | 6 | 0 | 39 | 0 | 6 | 22 |
| Feature | S. aureus WU1-1 | S. pseudintermedius WU48-1 |
|---|---|---|
| 1. Information on NCBI database | ||
| Assembly accession | GCA_037083895 | GCA_037132735 |
| 2. Genome characteristics | ||
| Genome size | 2.8 Mbp | 2.7 Mbp |
| G+C content of genome | 32.5% | 37.5% |
| 3. Genes | ||
| Total gene | 2814 | 2625 |
| Protein coding | 2668 | 2475 |
| Non-coding | 1 | 1 |
| rRNA | 13 | 15 |
| tRNA | 59 | 58 |
| Pseudogene | 70 | 73 |
| Others | 3 | 3 |
| 4. Virulent factor annotation | ||
| Capsule | 16 | 1 |
| Staphyloferrin A and B | 19 | 1 |
| Fibronectin binding protein | 11 | nf |
| Exotoxin | 9 | nf |
| Hemolysin (α, β, γ, and δ) | 7 | nf |
| Type VII secretion system | 6 | nf |
| Coagulation factors | 4 | nf |
| Leukocidin | 3 | 2 |
| Leukotoxin | 2 | nf |
| Other binding proteins | 2 | nf |
| Aureolysin | 1 | nf |
| Autolysin | 1 | nf |
| Exfoliative toxin | 1 | nf |
| Others | 46 | 1 |
| 5. Antimicrobial resistance gene annotation | ||
| Antibiotic efflux | 88 | 75 |
| Antibiotic target alteration | 67 | 64 |
| Antibiotic target protection | 7 | 9 |
| Antibiotic inactivation | 6 | 16 |
| Antibiotic target replacement | 5 | 6 |
| 6. Resistance of common antibiotic group | ||
| Macrolide group | 27 | 25 |
| Tetracycline group | 21 | 25 |
| Fluoroquinolone group | 18 | 14 |
| Beta-lactam group | 14 | 19 |
| Aminoglycoside group | 7 | 13 |
| Sulphonamide group | 1 | 1 |
| 7. Genes related to methicillin resistance | ||
| mecA | nf | Found in genome |
| mecC | Found in genome | nf |
| mecR1 | Found in genome | Found in genome |
| mecI | Found in genome | nf |
| Resistant Genes | Target Gene | Primer Sequence | Size (bp) | Ref. |
|---|---|---|---|---|
| Beta-lactam | blaZ | F: 5′-TGACCACTTTTATCAGCAACC-3′ | 240 | [77] |
| R: 5′-GCCATTTCAACACCTTCTTTC-3′ | ||||
| mecA | F: 5′-AAAATCGATGGTAAAGGTTGGC-3′ | 533 | [78] | |
| R: 5′-AGTTCTGCAGTACCGGATTTGC-3′ | ||||
| Trimethoprim | dfrK | F: 5′-GCTGCGATGGATAAGAACAG-3′ | 214 | [79] |
| R: 5′-GGACGATTTCACAACCATTAAAGC-3′ | ||||
| Aminoglycoside | aph2 | F: 5′-GAAGTACGCAGAAGAGA-3′ | 491 | [80] |
| R: 5′-ACATGGCAAGCTCTAGGA-3′ | ||||
| aph3 | F: 5′-AAATACCGCTGCGTA-3′ | 242 | [80] | |
| R: 5′-CATACTCTTCCGAGCAA-3′ | ||||
| Chloramphenicol | cat1 | F: 5′-GCGAACGAAAAACAATTGCA-3′ | 748 | [81] |
| R: 5′-TGAAGCTGTAAGGCAACTGG-3′ | ||||
| Tetracycline | tetK | F: 5′-GTAGCGACAATAGGTAATAGT-3′ | 360 | [82] |
| R: 5′-GTAGTGACAATAAACCTCCTA-3′ | ||||
| tetM | F: 5′-AGTGGAGCGATTACAGAA-3′ | 158 | [83] | |
| R: 5′-CATATGTCCTGGCGTGTCTA-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saengsawang, P.; Tanonkaew, R.; Kimseng, R.; Nissapatorn, V.; Wintachai, P.; Rodríguez-Ortega, M.J.; Mitsuwan, W. Whole Genome Sequence Analysis of Multidrug-Resistant Staphylococcus aureus and Staphylococcus pseudintermedius Isolated from Superficial Pyoderma in Dogs and Cats. Antibiotics 2025, 14, 643. https://doi.org/10.3390/antibiotics14070643
Saengsawang P, Tanonkaew R, Kimseng R, Nissapatorn V, Wintachai P, Rodríguez-Ortega MJ, Mitsuwan W. Whole Genome Sequence Analysis of Multidrug-Resistant Staphylococcus aureus and Staphylococcus pseudintermedius Isolated from Superficial Pyoderma in Dogs and Cats. Antibiotics. 2025; 14(7):643. https://doi.org/10.3390/antibiotics14070643
Chicago/Turabian StyleSaengsawang, Phirabhat, Ruedeechanok Tanonkaew, Rungruedee Kimseng, Veeranoot Nissapatorn, Phitchayapak Wintachai, Manuel J. Rodríguez-Ortega, and Watcharapong Mitsuwan. 2025. "Whole Genome Sequence Analysis of Multidrug-Resistant Staphylococcus aureus and Staphylococcus pseudintermedius Isolated from Superficial Pyoderma in Dogs and Cats" Antibiotics 14, no. 7: 643. https://doi.org/10.3390/antibiotics14070643
APA StyleSaengsawang, P., Tanonkaew, R., Kimseng, R., Nissapatorn, V., Wintachai, P., Rodríguez-Ortega, M. J., & Mitsuwan, W. (2025). Whole Genome Sequence Analysis of Multidrug-Resistant Staphylococcus aureus and Staphylococcus pseudintermedius Isolated from Superficial Pyoderma in Dogs and Cats. Antibiotics, 14(7), 643. https://doi.org/10.3390/antibiotics14070643










