First Report of Stenotrophomonas maltophilia from Canine Dermatological Infections: Unravelling Its Antimicrobial Resistance, Biofilm Formation, and Virulence Traits
Abstract
1. Introduction
2. Results
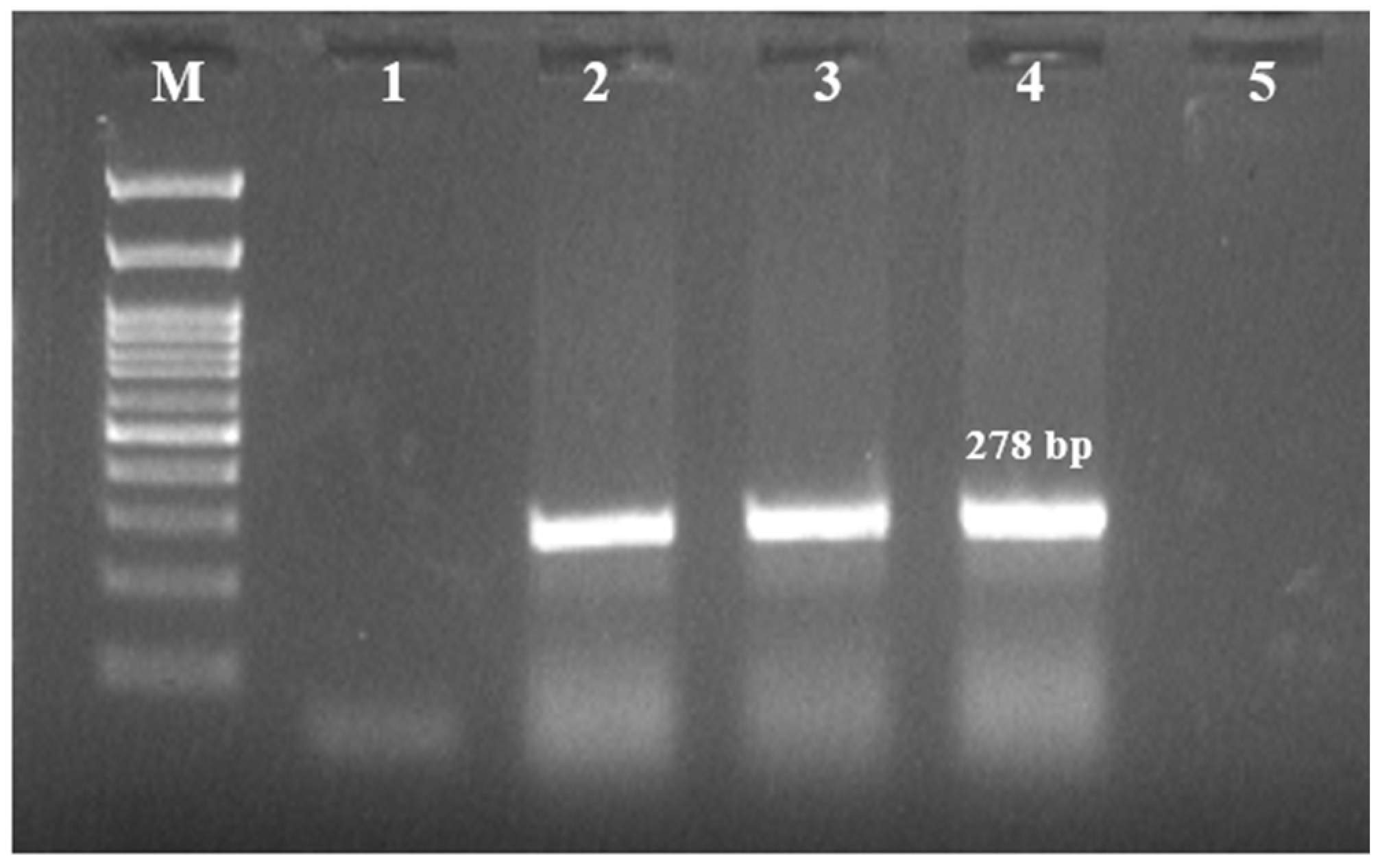
2.1. Prevalence of S. maltophilia
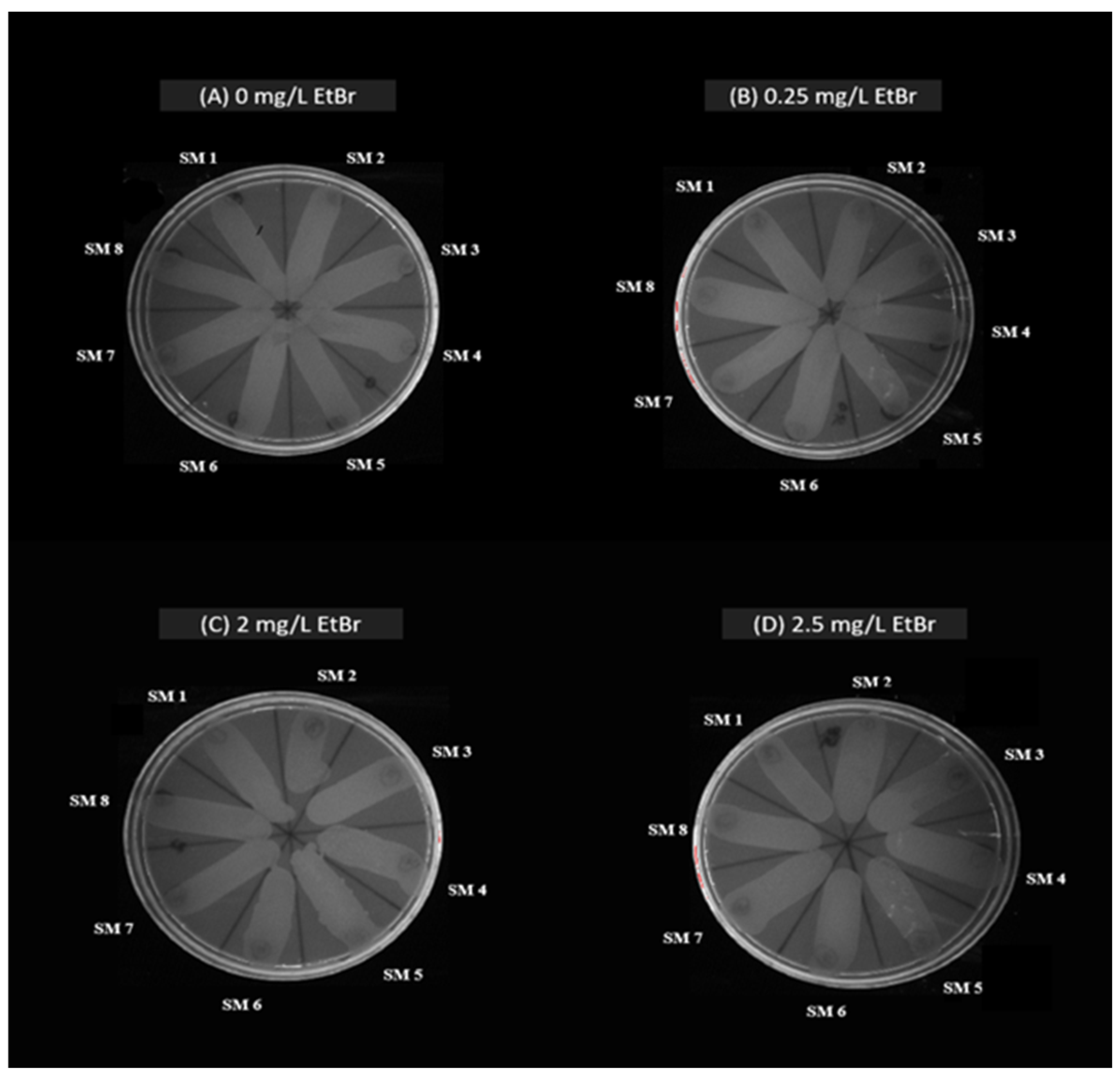
2.2. Characterization of Antimicrobial Resistance in S. maltophilia
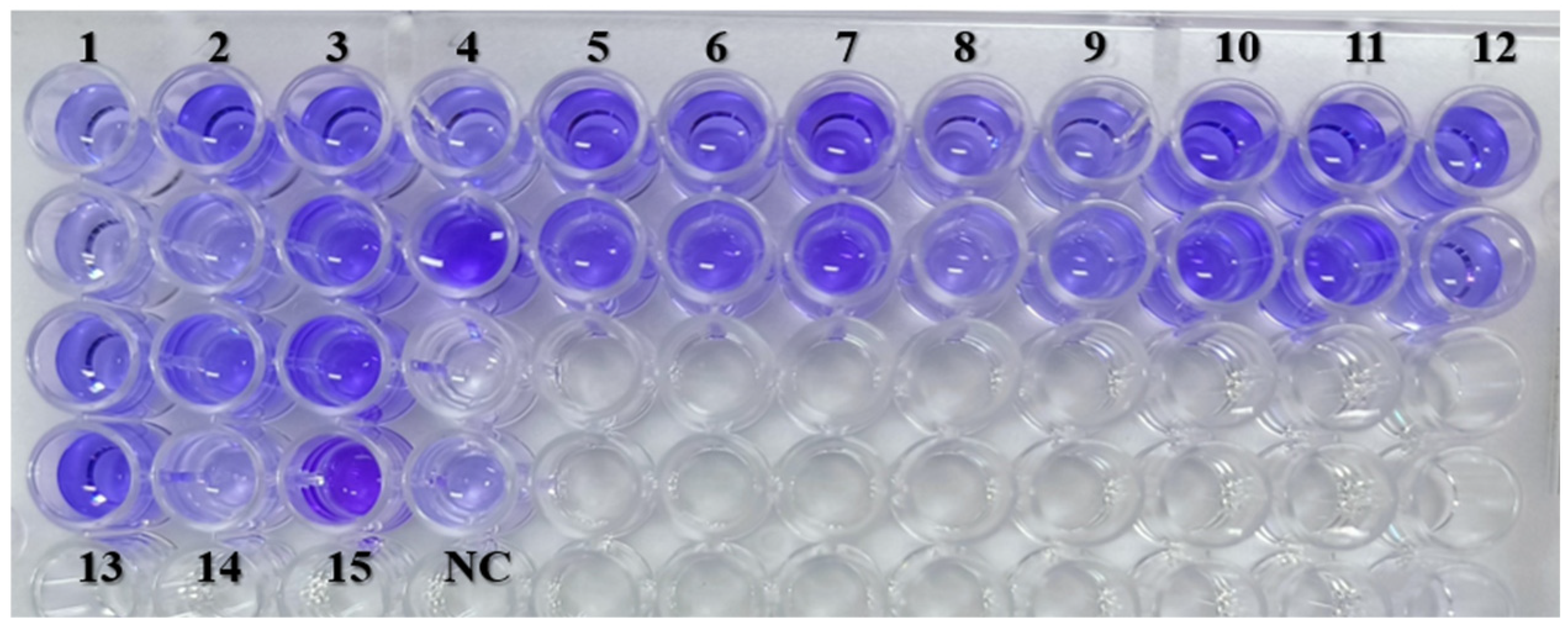
2.3. Virulence Characterization of S. maltophilia
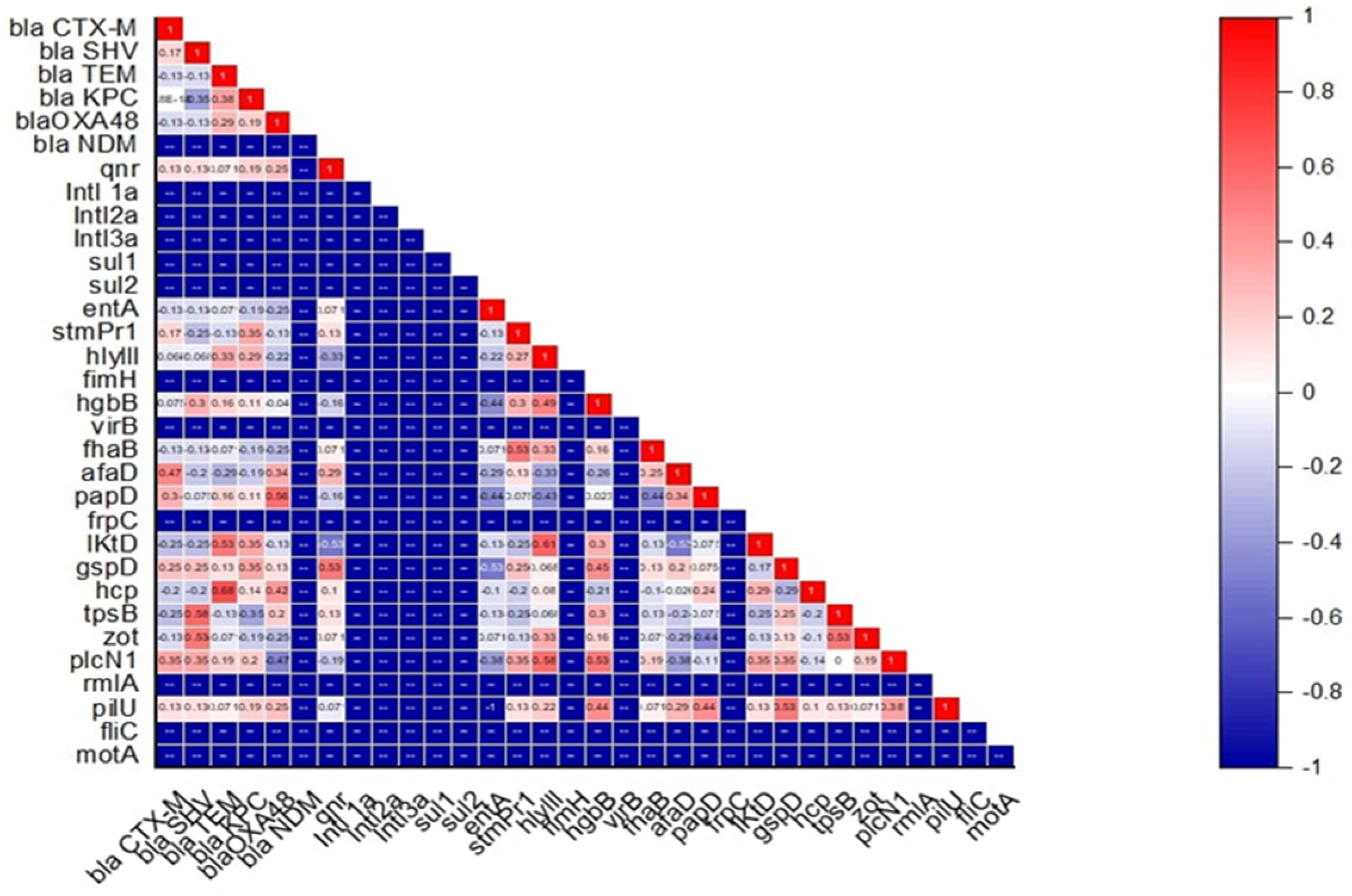
2.4. Association Between Antimicrobial Resistance, Virulence, Motility Pattern, and Biofilm-Forming Ability in S. maltophilia
3. Discussion
4. Materials and Methods
4.1. Isolation and Characterization of S. maltophilia
4.1.1. Sample Collection
4.1.2. Bacterial Isolation, Identification, and Molecular Confirmation
4.1.3. Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry (MALDI-TOF MS) Identification
| Gene | Primer Sequence | Cyclic Condition | No. of Cycles | Amplicon Size (bp) | Reference | |
|---|---|---|---|---|---|---|
| Step | Temperature and Time | |||||
| Species specific PCR | ||||||
| 23S rRNA | F-GCTGGATTGGTTCTAGGAAAACGC R-ACGCAGTCACTCCTTGCG | Initial denaturation | 94 °C–5 min | 278 | [26] | |
| Denaturation | 94 °C–45 s | 30 | ||||
| Annealing | 68 °C–45 s | |||||
| Extension | 72 °C–45 s | |||||
| Final extension | 72 °C–10 min | |||||
| Antimicrobial resistance genes | ||||||
| blaSIM | F-GTACAAGGGATTCGGCATCG R-TGGCCTGTTCCCATGTGAG | Initial denaturation | 95 °C–4 min | 569 | [27] | |
| Denaturation | 95 °C–45 s | 30 | ||||
| Annealing | 58 °C–60 s | |||||
| blaVIM | F-GTTTGGTCGCATATCGCAAC R-AATGCGCAGCACCAGGATAG | 382 | ||||
| Extension | 72 °C–40 s | |||||
| Final Extension | 72 °C–5 min | |||||
| blaTEM | F-TCCGCTCATGAGACAATAACC R-TTGGTCTGACAGTTACCAATGC | Initial denaturation | 94 °C–5 min | 931 | [28] | |
| Denaturation | 94 °C–30 s | 35 | ||||
| Annealing | 60 °C–15 s | |||||
| blaCTX-M | F-ATGTGCAGYACCAGTAARGTKATGGC R-TGGGTRAARTARGTSACCAGAAYCAGCGG | Extension | 72 °C–30 s | 593 | ||
| Final Extension | 72 °C–5 min | |||||
| blaSHV | F-AGCCGCTTGAGCAAATTAAAC R-ATCCCGCAGATAAATCACCAC | Initial denaturation | 95 °C–5 min | 713 | [29] | |
| Denaturation | 94 °C–45 s | 35 | ||||
| Annealing | 53 °C–45 s | |||||
| Extension | 72 °C–1 min | |||||
| Final Extension | 72 °C–10 min | |||||
| blaKPC | F-CGTCTAGTTCTGCTGTCTTG R-CTTGTCATCCTTGTTAGGCG | Initial denaturation | 94 °C–10 min | 798 | [30] | |
| Denaturation | 94 °C–30 s | 30 | ||||
| Annealing | 52 °C–40 s | |||||
| Extension | 72 °C–50 s | |||||
| Final Extension | 72 °C–5 min | |||||
| blaNDM | F-TCGCATAAAACGCCTCTG R-GAAACTGTCGCACCTCAT | Initial denaturation | 95 °C–6 min | 1001 | [31] | |
| Denaturation | 95 °C–45 s | 32 | ||||
| Annealing | 55 °C–45 s | |||||
| Extension | 72 °C–60 s | |||||
| Final Extension | 72 °C–7 min | |||||
| qnr | F-ACACAGAACGGCTGGACTGC R-TTCAACGACGTGGAGCTGT | Initial denaturation | 95 °C–5 min | 817 | [32] | |
| Denaturation | 95 °C–60 s | 30 | ||||
| Annealing | 55 °C–60 s | |||||
| Extension | 68 °C–60 s | |||||
| Final Extension | 68 °C–5 min | |||||
| Sul1 | F-TAGCGAGGGCTTTACTAAGC R-ATTCAGAATGCCGAACACCG | Initial denaturation | 95 °C–5 min | 437 | [33] | |
| Denaturation | 95 °C–1 min | 35 | ||||
| Annealing | 55 °C–60 s | |||||
| Sul2 | F-CCTGTTTCGTCCGACACAGA R-GAAGCGCAGCCGCAATTCAT | 956 | ||||
| Extension | 72 °C–1 min | |||||
| Final Extension | 72 °C–10 min | |||||
| blaOXA-48 | F-GCGTGGTTAAGGATGAACAC R-CATCAAGTTCAACCCAACCG | Initial denaturation | 95 °C–5 min | 438 | [34] | |
| Denaturation | 95 °C–45 s | 35 | ||||
| Annealing | 60 °C–45 s | |||||
| Extension | 72 °C–1 min | |||||
| Final Extension | 72 °C–8 min | |||||
| Virulence genes | ||||||
| entA | F: CGTTCGCACTCGACGTGAC R: CGAACTGACGGTAACGATCACG | Initial denaturation | 94 °C–5 min | 251 | [35] | |
| Denaturation | 94 °C–30 s | 34 | ||||
| Annealing | 60 °C–30 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| stmPr1 | F: TGAAAGCAAATGCGCCGTTG R: GTGATGGCGTCGGTGATGTC | Initial denaturation | 94 °C–5 min | 852 | ||
| Denaturation | 94 °C–30 s | 34 | ||||
| Annealing | 60 °C–30 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| hlyIII |
F: CGTCCATTGCTTCGATCCGTG R: GACGAAGTGGCAGACGCTG | Initial denaturation | 94 °C–5 min | 607 | ||
| Denaturation | 94 °C–30 s | 34 | ||||
| Annealing | 60 °C–30 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| fimH |
F: GATCCGCCTGAACTGCCAG R: CTGGCAGTTCAGGCGGATC | Initial denaturation | 94 °C–5 min | 576 | ||
| Denaturation | 94 °C–30 s | 34 | ||||
| Annealing | 60 °C–30 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| hgbB | F: GGACATCCAGAACATGGGTGC R: GGATCGATCGTGTACGGACC | Initial denaturation | 94 °C–5 min | 1239 | ||
| Denaturation | 94 °C–30 s | 34 | ||||
| Annealing | 60 °C–30 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| virB | F: GCATCATGCAGAACGAGCTG R: GACGGCTCGTACTTCTGCAC | Initial denaturation | 95 °C–5 min | 1075 | ||
| Denaturation | 95 °C–45 s | 35 | ||||
| Annealing | 60 °C–45 s | |||||
| Extension | 72 °C–1 min | |||||
| Final Extension | 72 °C–8 min | |||||
| frpC | F: CCAGTTCAACCTGTCGATGCTG R: CACCGAACAGGTTGTCCCAG | Initial denaturation | 95 °C–5 min | 653 | ||
| Denaturation | 95 °C–45 s | 35 | ||||
| Annealing | 60 °C–45 s | |||||
| Extension | 72 °C–1 min | |||||
| Final Extension | 72 °C–8 min | |||||
| afaD | F: GAAGCGCCTGACTGCCTTTTG R: GATCACGTTGTAAGGCCGCC | Initial denaturation | 95 °C–5 min | 328 | ||
| Denaturation | 95 °C–45 s | 35 | ||||
| Annealing | 60 °C–45 s | |||||
| Extension | 72 °C–1 min | |||||
| Final Extension | 72 °C–8 min | |||||
| fhaB | F: GTATCGCACAACCGCTTCCAG R: CGTCGTTGATGACCTTCTGCAC | Initial denaturation | 95 °C–5 min | 1744 | ||
| Denaturation | 95 °C–45 s | 35 | ||||
| Annealing | 60 °C–45 s | |||||
| Extension | 72 °C–1 min | |||||
| Final Extension | 72 °C–8 min | |||||
| papD | F: CACGCGAGTGATCTATCCGG R: GTGATGAAGCGCACCTGGTC | Initial denaturation | 95 °C–5 min | 579 | ||
| Denaturation | 95 °C–45 s | 35 | ||||
| Annealing | 60 °C–45 s | |||||
| Extension | 72 °C–1 min | |||||
| Final Extension | 72 °C–8 min | |||||
| gspD | F: GTCGACACCGATATCGGTGG R: GGTAGACCACATGCAGGTTGC | Initial denaturation | 94 °C–5 min | 694 | ||
| Denaturation | 94 °C–30 s | 32 | ||||
| Annealing | 60 °C–15 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| hcp | F: GACGGCAACGCGATCAATTAC R: GTTCTTGGTTGCACTCCACTG | Initial denaturation | 94 °C–5 min | 201 | ||
| Denaturation | 94 °C–30 s | 32 | ||||
| Annealing | 60 °C–15 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| zot | F: GCGTCAGTACACCGATGGTTG R: GCAGGCAGTGTCCAGCATG | Initial denaturation | 94 °C–5 min | 431 | ||
| Denaturation | 94 °C–30 s | 32 | ||||
| Annealing | 60 °C–15 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| plcN1 | F: GTGACCGATATCGGCCGAC R: CTGGAAGTGGCGGTGGAAG | Initial denaturation | 94 °C–5 min | 1779 | ||
| Denaturation | 94 °C–30 s | 34 | ||||
| Annealing | 62 °C–30 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| pilU | F: CGACCACCATCGATTTCACTTCG R: GACAGGTCCATCAGCAGCTG | Initial denaturation | 94 °C–5 min | 778 | ||
| Denaturation | 94 °C–30 s | 34 | ||||
| Annealing | 60 °C–30 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| fliC | F: CGATCTCCGAGCGCTTCG R: GAACAGCTGGCTGGAGAACG | Initial denaturation | 94 °C–5 min | 296 | ||
| Denaturation | 94 °C–30 s | 34 | ||||
| Annealing | 60 °C–30 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| rmlA | F: CTCAGCGTGCTGATGCTGG R: GATGAAGTTGGAGGCTTCCAGC | Initial denaturation | 94 °C–5 min | 600 | ||
| Denaturation | 94 °C–30 s | 34 | ||||
| Annealing | 60 °C–30 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| tpsB | F: GTGGACATCGTGATGAAGCGC R: CTTGCCGATGAAGTGACGGTG | Initial denaturation | 94 °C–5 min | 822 | ||
| Denaturation | 94 °C–30 s | 34 | ||||
| Annealing | 54 °C–30 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| motA | F: CGTTGGATTCCTGGTCGTCATC R: GAGCCCATGGTGATGACGATG | Initial denaturation | 94 °C–5 min | 558 | ||
| Denaturation | 94 °C–30 s | 34 | ||||
| Annealing | 54 °C–30 s | |||||
| Extension | 72 °C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
| lktD | F: GCACATCCGTGATGCAGTCG R: CGAGATTCTCGTCCTGCATGG | Initial denaturation | 94 °C–5 min | 1235 | ||
| Denaturation | 94 °C–30 s | 34 | ||||
| Annealing | 54 °C–30 s | |||||
| Extension | 72° C–30 s | |||||
| Final Extension | 72 °C–5 min | |||||
4.2. Screening of the Isolates for the Presence of Antimicrobial Resistance
4.2.1. Antimicrobial Susceptibility Testing (AST)
4.2.2. Assessment of Bacterial Efflux Pump Activity in S. maltophilia
4.2.3. Antimicrobial Resistance Genes (ARGs)
4.3. Characterization of the Virulence Properties of S. maltophilia
4.3.1. Evaluation of Biofilm-Forming Ability
4.3.2. Motility Assay
4.3.3. Molecular Characterization of Virulence Genes
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| ARGs | Antimicrobial resistant genes |
| AST | Antimicrobial susceptibility testing |
| AT | Aztreonam |
| CLSI | Clinical and Laboratory Standards Institute |
| CPD | Cefpodoxime |
| CTR | Ceftriaxone |
| Etbr | Ethidium bromide |
| IPM | Imipenem |
| lktD | Leukotoxin D |
| LPS | Lipopolysaccharides |
| MALDI-TOF MS | Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry |
| MDR | Multidrug resistant |
| NC | Negative control |
| NFGNB | Non-fermenting gram-negative bacilli |
| OD | Optical density |
| PCR | Polymerase Chain Reaction |
| PIT | Piperacillin-tazobactam |
| S. maltophilia | Stenotrophomonas maltophilia |
| T2SS | Type II secretion system |
| T4SS | Type IV secretion system |
| TCC | Ticarcillin-clavulanate |
| TMP/SMX | Trimethoprim/sulfamethoxazole |
| zot | Zonula occludens toxin |
References
- Brooke, J.S. Advances in the microbiology of Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 2021, 34, e0003019. [Google Scholar] [CrossRef] [PubMed]
- Gibb, J.; Wong, D.W. Antimicrobial treatment strategies for Stenotrophomonas maltophilia: A focus on novel therapies. Antibiotics 2021, 10, 1226. [Google Scholar] [CrossRef]
- Mikhailovich, V.; Heydarov, R.; Zimenkov, D.; Chebotar, I. Stenotrophomonas maltophilia virulence: A current view. Front. Microbiol. 2024, 15, 1385631. [Google Scholar] [CrossRef]
- Bhaumik, R.; Aungkur, N.Z.; Anderson, G.G. A guide to Stenotrophomonas maltophilia virulence capabilities, as we currently understand them. Front. Cell. Infect. Microbiol. 2023, 13, 1322853. [Google Scholar] [CrossRef]
- Albini, S.; Abril, C.; Franchini, M.; Hüssy, D.; Filioussis, G. Stenotrophomonas maltophilia isolated from the airways of animals with chronic respiratory disease. Schweiz. Arch. Tierheilkd. 2009, 151, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Kralova-Kovarikova, S.; Husnik, R.; Honzak, D.; Kohout, P.; Fictum, P. Stenotrophomonas maltophilia urinary tract infections in three dogs: A case report. Vet. Med. 2012, 57, 380–383. [Google Scholar] [CrossRef]
- Thenissery, A.; Chandran, R.; Ravindran, R.; Ravishankar, C.; Piriyath, J.H. Isolation and identification of emergent multidrug resistant Stenotrophomonas maltophilia from skin ulcers of Sarcoptes infested pigs. Vet. Rec. Case Rep. 2022, 10, e312. [Google Scholar] [CrossRef]
- Jayol, A.; Corlouer, C.; Haenni, M.; Darty, M.; Maillard, K.; Desroches, M.; Lamy, B.; Jumas-Bilak, E.; Madec, J.Y.; Decousser, J.W. Are animals a source of Stenotrophomonas maltophilia in human infections? Contributions of a nationwide molecular study. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1039–1045. [Google Scholar] [CrossRef]
- Sheu, C.C.; Chang, Y.T.; Lin, S.Y.; Chen, Y.H.; Hsueh, P.R. Infections caused by carbapenem-resistant Enterobacteriaceae: An update on therapeutic options. Front. Microbiol. 2019, 10, 80. [Google Scholar] [CrossRef]
- Mojica, M.F.; Humphries, R.; Lipuma, J.J.; Mathers, A.J.; Rao, G.G.; Shelburne, S.A.; Fouts, D.E.; Van Duin, D.; Bonomo, R.A. Clinical challenges treating Stenotrophomonas maltophilia infections: An update. JAC Antimicrob. Resist. 2022, 4, dlac040. [Google Scholar] [CrossRef]
- Sánchez, M.B.; Hernández, A.; Rodríguez-Martínez, J.M.; Martínez-Martínez, L.; Martínez, J.L. Predictive analysis of transmissible quinolone resistance indicates Stenotrophomonas maltophilia as a potential source of a novel family of Qnr determinants. BMC Microbiol. 2018, 8, 148. [Google Scholar] [CrossRef]
- Huang, Y.W.; Hu, R.M.; Yang, T.C. Role of the pcm-mph operon in resistance to several antibiotics in Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 2013, 68, 1987–1993. [Google Scholar] [CrossRef] [PubMed]
- Cha, M.K.; Kang, C.I.; Kim, S.H.; Cho, S.Y.; Ha, Y.E.; Chung, D.R.; Peck, K.R.; Song, J.H. Emergence of fluoroquinolone-resistant Stenotrophomonas maltophilia in blood isolates causing bacteremia: Molecular epidemiology and microbiologic characteristics. Diagn. Microbiol. Infect. Dis. 2016, 85, 210–212. [Google Scholar] [CrossRef] [PubMed]
- Windels, E.M.; Michiels, J.E.; Van den Bergh, B.; Fauvart, M.; Michiels, J. Antibiotics: Combatting tolerance to stop resistance. mBio 2019, 10, e02095-19. [Google Scholar] [CrossRef]
- Adegoke, A.A.; Stenström, T.A.; Okoh, A.I. Stenotrophomonas maltophilia as an emerging ubiquitous pathogen: Looking beyond contemporary antibiotic therapy. Front. Microbiol. 2017, 8, 2276. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; McCusker, M.P.; Viveiros, M.; Couto, I.; Fanning, S.; Pagès, J.M.; Amaral, L. A simple method for assessment of MDR bacteria for over-expressed efflux pumps. Open Microbiol. J. 2013, 7, 72–82. [Google Scholar] [CrossRef]
- Pompilio, A.; Piccolomini, R.; Picciani, C.; D’Antonio, D.; Savini, V.; Di Bonaventura, G. Factors associated with adherence to and biofilm formation on polystyrene by Stenotrophomonas maltophilia: The role of cell surface hydrophobicity and motility. FEMS Microbiol. Lett. 2008, 287, 41–47. [Google Scholar] [CrossRef]
- Bostanghadiri, N.; Ghalavand, Z.; Fallah, F.; Yadegar, A.; Ardebili, A.; Tarashi, S.; Pournajaf, A.; Mardaneh, J.; Shams, S.; Hashemi, A. Characterization of phenotypic and genotypic diversity of Stenotrophomonas maltophilia strains isolated from selected hospitals in Iran. Front. Microbiol. 2019, 10, 1191. [Google Scholar] [CrossRef]
- Bostanghadiri, N.; Ardebili, A.; Ghalavand, Z.; Teymouri, S.; Mirzarazi, M.; Goudarzi, M.; Ghasemi, E.; Hashemi, A. Antibiotic resistance, biofilm formation, and biofilm-associated genes among Stenotrophomonas maltophilia clinical isolates. BMC Res. Notes 2021, 14, 151. [Google Scholar] [CrossRef]
- Amoli, R.I.; Nowroozi, J.; Sabokbar, A.; Rajabniya, R. Isolation of Stenotrophomonas maltophilia from clinical samples: An investigation of patterns motility and production of melanin pigment. Asian Pac. J. Trop. Biomed. 2017, 7, 826–830. [Google Scholar] [CrossRef]
- Yinsai, O.; Deeudom, M.; Duangsonk, K. Genotypic diversity, antibiotic resistance, and virulence phenotypes of Stenotrophomonas maltophilia clinical isolates from a Thai University Hospital Setting. Antibiotics 2023, 12, 410. [Google Scholar] [CrossRef] [PubMed]
- Nas, M.Y.; White, R.C.; DuMont, A.L.; Lopez, A.E.; Cianciotto, N.P. Stenotrophomonas maltophilia encodes a VirB/VirD4 type IV secretion system that modulates apoptosis in human cells and promotes competition against heterologous bacteria, including Pseudomonas aeruginosa. Infect. Immun. 2019, 87, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Azimi, A.; Aslanimehr, M.; Yaseri, M.; Shadkam, M.; Douraghi, M. Distribution of smf-1, rmlA, spgM and rpfF genes among Stenotrophomonas maltophilia isolates in relation to biofilm-forming capacity. J. Glob. Antimicrob. Resist. 2020, 23, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Youenou, B.; Favre-Bonté, S.; Bodilis, J.; Brothier, E.; Dubost, A.; Muller, D.; Nazaret, S. Comparative genomics of environmental and clinical Stenotrophomonas maltophilia strains with different antibiotic resistance profiles. Genome Biol. Evol. 2015, 7, 2484–2505. [Google Scholar] [CrossRef]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef]
- Gallo, S.W.; Ramos, P.L.; Ferreira, C.A.S.; Oliveira, S.D. A specific polymerase chain reaction method to identify Stenotrophomonas maltophilia. Mem. Inst. Oswaldo Cruz. 2013, 108, 390–391. [Google Scholar] [CrossRef]
- Saleh, R.O.; Hussen, B.M.; Mubarak, S.M.H.; Mostafavi, S.K.S. High diversity of virulent and multidrug-resistant Stenotrophomonas maltophilia in Iraq. Gene Rep. 2021, 23, 101124. [Google Scholar] [CrossRef]
- Sakthikarthikeyan, S.; Sivakumar, M.; Manikandan, R.; Senthilkumar, P.; Sureshkumar, V.; Malmarugan, S. Prevalence and Molecular Characterization of Multidrug-resistant ESBL-producing E. coli in Commercial Poultry. Indian J. Anim. Res. 2024, B-5166, 1–6. [Google Scholar] [CrossRef]
- Ibrahim, M.E.; Algak, T.B.; Abbas, M.; Elamin, B.K. Emergence of blaTEM, blaCTX-M, blaSHV and blaOXA genes in multidrug-resistant Enterobacteriaceae and Acinetobacter baumannii in Saudi Arabia. Exp. Ther. Med. 2021, 22, 1450. [Google Scholar] [CrossRef]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef]
- Rizi, K.S.; Jamehdar, S.A.; Sasan, M.S.; Ghazvini, K.; Aryan, E.; Safdari, H.; Farsiani, H. Detection of extended-spectrum beta-lactamases and carbapenemases in clinical isolates of Stenotrophomonas maltophilia in the northeast of Iran. Gene Rep. 2023, 34, 101857. [Google Scholar] [CrossRef]
- Malekan, M.; Tabaraie, B.; Akhoundtabar, L.; Afrough, P.; Behrouzi, A. Distribution of class I integron and smqnr resistance gene among Stenotrophomonas maltophilia isolated from clinical samples in Iran. Avicenna J. Med. Biotechnol. 2017, 9, 138–141. [Google Scholar] [PubMed]
- Hu, L.F.; Chen, G.S.; Kong, Q.X.; Gao, L.P.; Chen, X.; Ye, Y.; Li, J.B. Increase in the prevalence of resistance determinants to trimethoprim/sulfamethoxazole in clinical Stenotrophomonas maltophilia isolates in China. PLoS ONE 2016, 11, e0157693. [Google Scholar] [CrossRef] [PubMed]
- Doyle, D.; Peirano, G.; Lascols, C.; Lloyd, T.; Church, D.L.; Pitout, J.D.D. Laboratory detection of Enterobacteriaceae that produce carbapenemases. J. Clin. Microbiol. 2012, 50, 3877–3880. [Google Scholar] [CrossRef]
- Cruz-Córdova, A.; Mancilla-Rojano, J.; Luna-Pineda, V.M.; Escalona-Venegas, G.; Cázares-Domínguez, V.; Ormsby, C.; Franco-Hernández, I.; Zavala-Vega, S.; Hernández, M.A.; Medina-Pelcastre, M.; et al. Molecular epidemiology, antibiotic resistance and virulence traits of Stenotrophomonas maltophilia strains associated with an outbreak in a Mexican tertiary care hospital. Front. Cell. Infect. Microbiol. 2020, 10, 50. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 34th ed.; Clinical and Laboratory Standards Institute: Berwyn, PA, USA, 2024; pp. 88–89. [Google Scholar]
- Di Bonaventura, G.; Spedicato, I.; Antonio, D.; Robuffo, I.; Piccolomini, R. Biofilm formation by Stenotrophomonas maltophilia: Modulation by quinolones, trimethoprim-sulfamethoxazole, and ceftazidime. Antimicrob. Agents Chemother. 2004, 48, 151–160. [Google Scholar] [CrossRef]
- Pompilio, A.; Pomponio, S.; Crocetta, V.; Gherardi, G.; Verginelli, F.; Fiscarelli, E.; Dicuonzo, G.; Savini, V.; Antonio, D.; Di Bonaventura, G. Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: Genome diversity, biofilm formation, and virulence. BMC Microbiol. 2011, 11, 159. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty heatmaps. R Package Version 2012, 1, 726. [Google Scholar]



| Isolates | Date of Collection | Type of Sample | Gram’s Staining | Catalase Test | Oxidase Test | Glucose Fermentation Test | 23S rRNA | MALDI-TOF SCORE |
|---|---|---|---|---|---|---|---|---|
| SM 1 | 12.01.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.25 |
| SM 2 | 18.01.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2 |
| SM 3 | 21.01.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.06 |
| SM 4 | 02.02.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.17 |
| SM 5 | 07.02.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2 |
| SM 6 | 09.02.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.2 |
| SM 7 | 17.02.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.26 |
| SM 8 | 17.02.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.15 |
| SM 9 | 19.02.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.23 |
| SM 10 | 26.02.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.27 |
| SM 11 | 11.03.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.26 |
| SM 12 | 11.03.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.24 |
| SM 13 | 13.03.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.26 |
| SM 14 | 16.03.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.23 |
| SM 15 | 16.03.24 | Skin swab | Negative | Positive | Negative | Negative | Positive | 2.17 |
| Strain Number | Phenotypic Resistance Pattern | AMR Genes | Virulence Genes | Biofilm Activity | Efflux Activity |
|---|---|---|---|---|---|
| SM 1 | IPM, CTR, CPD, AT | blaKPC, blaNDM, qnr | virB, motA, rmlA, fliC, pilU, gspD, hgbB, plcN1, hlyIII, lktD | STRONG | ACTIVE |
| SM 2 | CPD, AT | qnr | virB, motA, rmlA, fliC, entA | MODERATE | ACTIVE |
| SM 3 | IPM, PIT, TCC, CTR, CPD, AT | blaKPC, blaOXA-48, qnr | virB, motA, rmlA, fliC, pilU, gspD, papD, afaD | MODERATE | ACTIVE |
| SM 4 | IPM, PIT, TCC, CTR, CPD, AT | blaSHV, blaOXA-48,qnr | virB, motA, rmlA, fliC, pilU, gspD, papD, hgbB plcN1, tpsB | STRONG | ACTIVE |
| SM 5 | IPM, PIT, CTR, CPD, AT | blaKPC, qnr | virB, motA, rmlA, fliC, pilU, gspD, papD, hgbB, plcN1, stmPr1 | MODERATE | ACTIVE |
| SM 6 | IPM, CTR, CPD, AT | blaTEM, blaKPC blaNDM, qnr | virB, motA, rmlA, fliC, pilU, gspD, papD, hgbB, plcN1, hlyIII, lktD, hcp | MODERATE | ACTIVE |
| SM 7 | IPM, PIT, CTR, CPD, AT | blaNDM | virB, motA, rmlA, fliC, pilU, papD, hgbB, plcN1, hlyIII, lktD | STRONG | ACTIVE |
| SM 8 | IPM, PIT, CTR, CPD, AT | qnr | virB, motA, rmlA, fliC, pilU, gspD, hgbB, plcN1, afaD, hlyIII, fhaB, stmPr1 | STRONG | ACTIVE |
| SM 9 | IPM, PIT, TCC, CTR, CPD, AT | blaCTX-M, blaKPC, blaOXA-48, blaNDM, qnr | virB, motA, rmlA, fliC, pilU gspD, papD, hgbB plcN1, afaD, hlyIII, stmPr1 | STRONG | ACTIVE |
| SM 10 | IPM, PIT, CTR, CPD, AT | blaCTX-M, qnr | virB, motA, rmlA, fliC, pilU, gspD, papD, plcN1, afaD | MODERATE | ACTIVE |
| SM 11 | IPM, TCC, CTR, CPD, AT | blaOXA-48, qnr | virB, motA, rmlA, fliC, pilU, gspD, papD, hgbB, afaD | STRONG | ACTIVE |
| SM 12 | IPM, PIT, CTR, CPD, AT | blaOXA-48, qnr | virB, motA, rmlA, fliC, pilU, papD, afaD, hcp | MODERATE | ACTIVE |
| SM 13 | IPM, PIT, TCC, CTR, CPD, AT | blaSHV, blaNDM, qnr | virB, motA, rmlA, fliC, pilU, gspD, hgbB, plcN1, hlyIII, tpsB, zot | STRONG | ACTIVE |
| SM 14 | IPM, PIT, CTR, CPD, AT | blaCTX-M, blaSHV, qnr | virB, motA, rmlA, fliC, pilU, gspD, papD, hgbB, plcN1, afaD | STRONG | ACTIVE |
| SM 15 | IPM, TCC, CTR, CPD, AT | qnr | virB, motA, rmlA, fliC, pilU, gspD, papD, hgbB, afaD tpsB | STRONG | ACTIVE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajeev, R.; Kannan, P.; Sundaram, S.; Mohan, S.B.; Radjendirane, S.; Harnathbhai, C.J.; Subbaiyan, A.; Naveenkumar, V.; Mohanadasse, N.Q.; Savariraj, W.R.; et al. First Report of Stenotrophomonas maltophilia from Canine Dermatological Infections: Unravelling Its Antimicrobial Resistance, Biofilm Formation, and Virulence Traits. Antibiotics 2025, 14, 639. https://doi.org/10.3390/antibiotics14070639
Rajeev R, Kannan P, Sundaram S, Mohan SB, Radjendirane S, Harnathbhai CJ, Subbaiyan A, Naveenkumar V, Mohanadasse NQ, Savariraj WR, et al. First Report of Stenotrophomonas maltophilia from Canine Dermatological Infections: Unravelling Its Antimicrobial Resistance, Biofilm Formation, and Virulence Traits. Antibiotics. 2025; 14(7):639. https://doi.org/10.3390/antibiotics14070639
Chicago/Turabian StyleRajeev, Ria, Porteen Kannan, Sureshkannan Sundaram, Sandhya Bhavani Mohan, Sivachandiran Radjendirane, Chaudhary Jeetendrakumar Harnathbhai, Anbazhagan Subbaiyan, Viswanathan Naveenkumar, Nithya Quintoil Mohanadasse, Wilfred Ruban Savariraj, and et al. 2025. "First Report of Stenotrophomonas maltophilia from Canine Dermatological Infections: Unravelling Its Antimicrobial Resistance, Biofilm Formation, and Virulence Traits" Antibiotics 14, no. 7: 639. https://doi.org/10.3390/antibiotics14070639
APA StyleRajeev, R., Kannan, P., Sundaram, S., Mohan, S. B., Radjendirane, S., Harnathbhai, C. J., Subbaiyan, A., Naveenkumar, V., Mohanadasse, N. Q., Savariraj, W. R., Cull, C. A., & Amachawadi, R. G. (2025). First Report of Stenotrophomonas maltophilia from Canine Dermatological Infections: Unravelling Its Antimicrobial Resistance, Biofilm Formation, and Virulence Traits. Antibiotics, 14(7), 639. https://doi.org/10.3390/antibiotics14070639








