The Co-Existence of mcr-1.1 and mcr-3.5 in Escherichia coli Isolated from Clinical Samples in Thailand
Abstract
1. Introduction
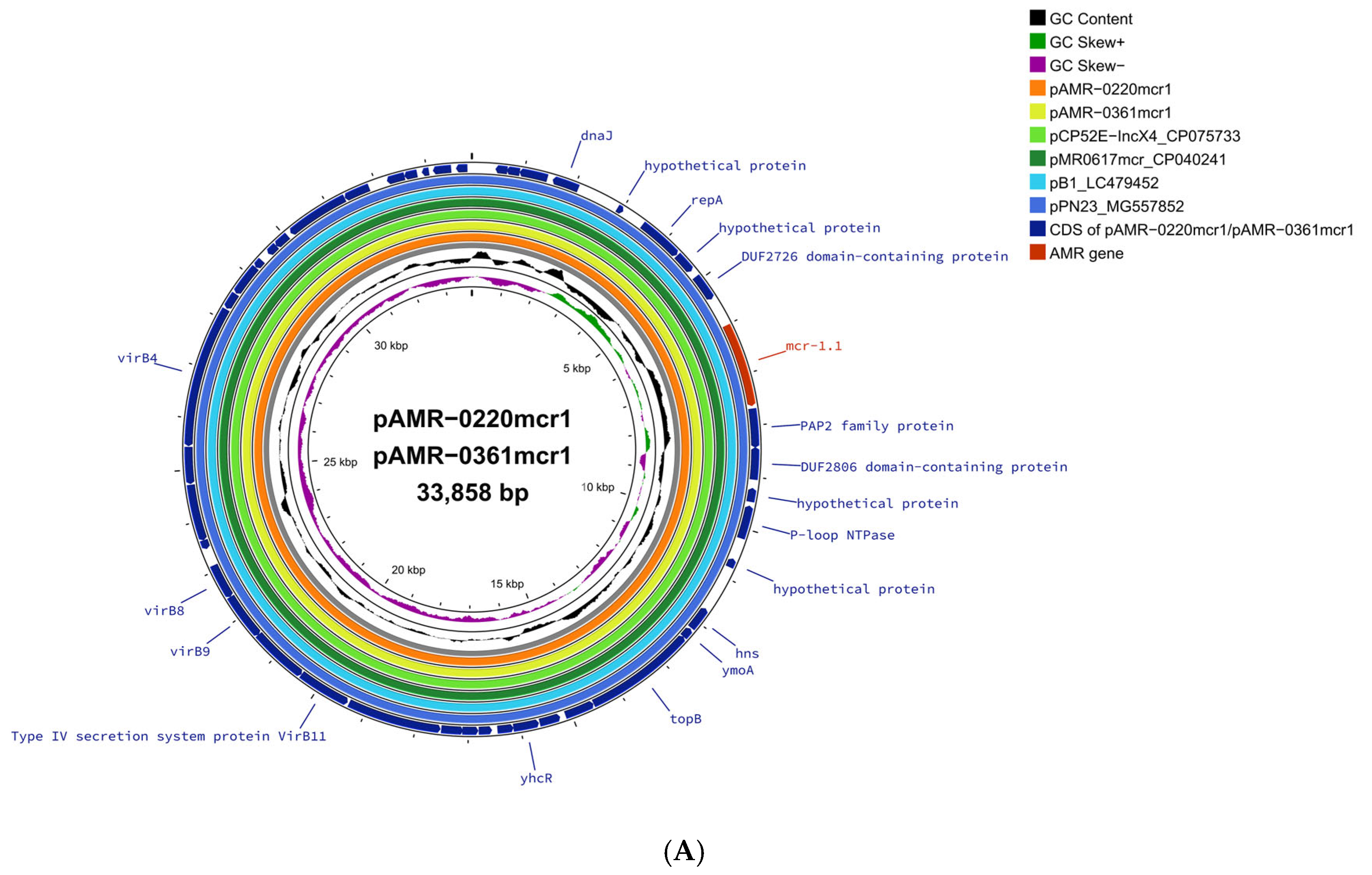
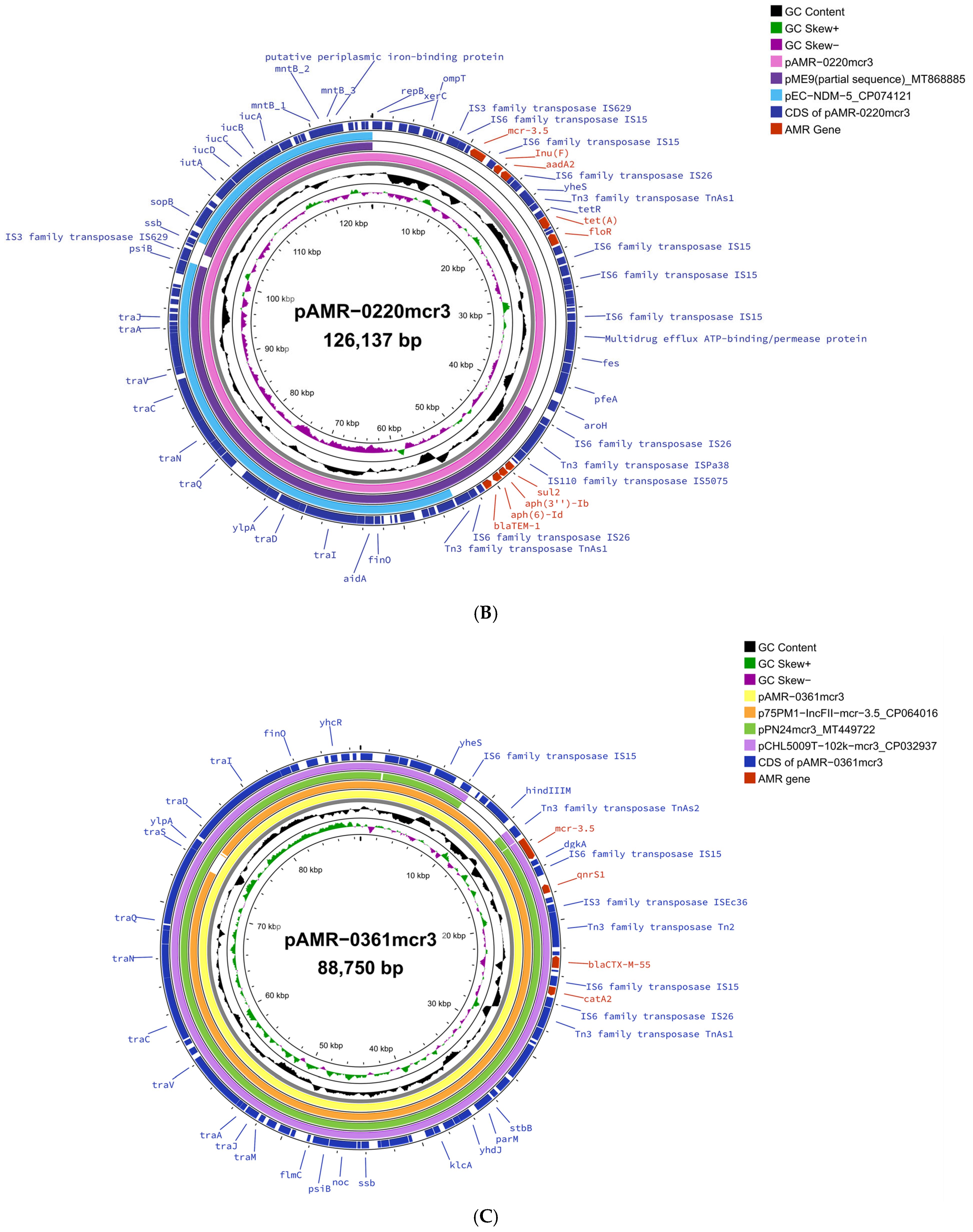
2. Results and Discussion
3. Material and Methods
3.1. Bacterial Strains and Antibiotic Susceptibility Testing
3.2. Detection of Extended Spectrum Beta-Lactamase (ESBL), Carbapenemase, and Colistin Resistance Genes
3.3. Short- and Long-Read Whole-Genome Sequencing
3.4. Analysis of Whole-Genome Sequencing Data
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- El-Sayed Ahmed, M.A.E.; Zhong, L.L.; Shen, C.; Yang, Y.; Doi, Y.; Tian, G.B. Colistin and its role in the Era of antibiotic resistance: An extended review (2000–2019). Emerg. Microbes Infect. 2020, 9, 868–885. [Google Scholar] [CrossRef]
- Zellweger, R.M.; Carrique-Mas, J.; Limmathurotsakul, D.; Day, N.P.J.; Thwaites, G.E.; Baker, S.; Southeast Asia Antimicrobial Resistance Network. A current perspective on antimicrobial resistance in Southeast Asia. J. Antimicrob. Chemother. 2017, 72, 2963–2972. [Google Scholar] [CrossRef] [PubMed]
- HPSR-AMR. Highlights Thailand One Health Report 2022: Antimicrobial Consumption and Antimicrobial Resistance. 2025. Available online: https://www.thaiamrwatch.net/materials/92 (accessed on 30 April 2025).
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Hussein, N.H.; AL-Kadmy, I.M.S.; Taha, B.M.; Hussein, J.D. Mobilized colistin resistance (mcr) genes from 1 to 10: A comprehensive review. Mol. Biol. Rep. 2021, 48, 2897–2907. [Google Scholar] [CrossRef]
- Olaitan, A.O.; Chabou, S.; Okdah, L.; Morand, S.; Rolain, J.M. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 2016, 16, 147. [Google Scholar] [CrossRef]
- Phuadraksa, T.; Wichit, S.; Arikit, S.; Songtawee, N.; Yainoy, S. Co-occurrence of mcr-2 and mcr-3 genes on chromosome of multidrug-resistant Escherichia coli isolated from healthy individuals in Thailand. Int. J. Antimicrob. Agents 2022, 60, 106662. [Google Scholar] [CrossRef] [PubMed]
- Khanawapee, A.; Kerdsin, A.; Chopjitt, P.; Boueroy, P.; Hatrongjit, R.; Akeda, Y.; Tomono, K.; Nuanualsuwan, S.; Hamada, S. Distribution and Molecular Characterization of Escherichia coli Harboring mcr Genes Isolated from Slaughtered Pigs in Thailand. Microb. Drug Resist. 2021, 27, 971–979. [Google Scholar] [CrossRef]
- Paveenkittiporn, W.; Kamjumphol, W.; Ungcharoen, R.; Kerdsin, A. Whole-Genome Sequencing of Clinically Isolated Carbapenem-Resistant Enterobacterales Harboring mcr Genes in Thailand, 2016–2019. Front. Microbiol. 2020, 11, 586368. [Google Scholar] [CrossRef]
- Liu, L.; Feng, Y.; Zhang, X.; McNally, A.; Zong, Z. New Variant of mcr-3 in an Extensively Drug-Resistant Escherichia coli Clinical Isolate Carrying mcr-1 and blaNDM-5. Antimicrob. Agents Chemother. 2017, 61, e01757-17. [Google Scholar] [CrossRef]
- Creighton, J.; Anderson, T.; Howard, J.; Dyet, K.; Ren, X.; Freeman, J. Co-occurrence of mcr-1 and mcr-3 genes in a single Escherichia coli in New Zealand. J. Antimicrob. Chemother. 2019, 74, 3113–3116. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Zhang, P.; Yang, X.; Wang, Z.; Fanning, S.; Wang, J.; Du, P.; Bai, L. Identification of a novel hybrid plasmid coproducing MCR-1 and MCR-3 variant from an Escherichia coli strain. J. Antimicrob. Chemother. 2019, 74, 1517–1520. [Google Scholar] [CrossRef]
- Hernandez, M.; Iglesias, M.R.; Rodriguez-Lazaro, D.; Gallardo, A.; Quijada, N.; Miguela-Villoldo, P.; Campos, M.J.; Piriz, S.; Lopez-Orozco, G.; de Frutos, C.; et al. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Euro Surveill. 2017, 22, 30586. [Google Scholar] [CrossRef] [PubMed]
- Hamame, A.; Davoust, B.; Rolain, J.M.; Diene, S.M. Genomic characterisation of an mcr-1 and mcr-3-producing Escherichia coli strain isolated from pigs in France. J. Glob. Antimicrob. Resist. 2022, 28, 174–179. [Google Scholar] [CrossRef]
- Yu, Y.; Andrey, D.O.; Yang, R.S.; Sands, K.; Tansawai, U.; Li, M.; Portal, E.; Gales, A.C.; Niumsup, P.R.; Sun, J.; et al. A Klebsiella pneumoniae strain co-harbouring mcr-1 and mcr-3 from a human in Thailand. J. Antimicrob. Chemother. 2020, 75, 2372–2374. [Google Scholar] [CrossRef] [PubMed]
- Leangapichart, T.; Stosic, M.S.; Hickman, R.A.; Lunha, K.; Jiwakanon, J.; Angkititrakul, S.; Magnusson, U.; Van Boeckel, T.P.; Jarhult, J.D.; Sunde, M. Exploring the epidemiology of mcr genes, genetic context and plasmids in Enterobacteriaceae originating from pigs and humans on farms in Thailand. J. Antimicrob. Chemother. 2023, 78, 1395–1405. [Google Scholar] [CrossRef]
- Roer, L.; Overballe-Petersen, S.; Hansen, F.; Schonning, K.; Wang, M.; Roder, B.L.; Hansen, D.S.; Justesen, U.S.; Andersen, L.P.; Fulgsang-Damgaard, D.; et al. Escherichia coli Sequence Type 410 Is Causing New International High-Risk Clones. mSphere 2018, 3, e00337-18. [Google Scholar] [CrossRef]
- Dadashi, M.; Sameni, F.; Bostanshirin, N.; Yaslianifard, S.; Khosravi-Dehaghi, N.; Nasiri, M.J.; Goudarzi, M.; Hashemi, A.; Hajikhani, B. Global prevalence and molecular epidemiology of mcr-mediated colistin resistance in Escherichia coli clinical isolates: A systematic review. J. Glob. Antimicrob. Resist. 2022, 29, 444–461. [Google Scholar] [CrossRef]
- Ruekit, S.; Srijan, A.; Serichantalergs, O.; Margulieux, K.R.; Mc Gann, P.; Mills, E.G.; Stribling, W.C.; Pimsawat, T.; Kormanee, R.; Nakornchai, S.; et al. Molecular characterization of multidrug-resistant ESKAPEE pathogens from clinical samples in Chonburi, Thailand (2017–2018). BMC Infect. Dis. 2022, 22, 695. [Google Scholar] [CrossRef]
- Srijan, A.; Margulieux, K.R.; Ruekit, S.; Snesrud, E.; Maybank, R.; Serichantalergs, O.; Kormanee, R.; Sukhchat, P.; Sriyabhaya, J.; Hinkle, M.; et al. Genomic Characterization of Nonclonal mcr-1-Positive Multidrug-Resistant Klebsiella pneumoniae from Clinical Samples in Thailand. Microb. Drug Resist. 2018, 24, 403–410. [Google Scholar] [CrossRef]
- Khine, N.O.; Wongsurawat, T.; Jenjaroenpun, P.; Hampson, D.J.; Prapasarakul, N. Comparative genomic analysis of Colistin resistant Escherichia coli isolated from pigs, a human and wastewater on colistin withdrawn pig farm. Sci. Rep. 2023, 13, 5124. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, W.; Tanaka, H.; Taniguchi, Y.; Iimura, M.; Soga, E.; Kubo, R.; Matsuo, N.; Kawamura, K.; Arakawa, Y.; Nagano, Y.; et al. Acquisition of mcr-1 and Cocarriage of Virulence Genes in Avian Pathogenic Escherichia coli Isolates from Municipal Wastewater Influents in Japan. Appl. Environ. Microbiol. 2019, 85, e01661-19. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, M.; Spiller, O.B.; Andrey, D.O.; Hinchliffe, P.; Li, H.; MacLean, C.; Niumsup, P.; Powell, L.; Pritchard, M.; et al. Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat. Commun. 2017, 8, 2054. [Google Scholar] [CrossRef] [PubMed]
- Boueroy, P.; Wongsurawat, T.; Jenjaroenpun, P.; Chopjitt, P.; Hatrongjit, R.; Jittapalapong, S.; Kerdsin, A. Plasmidome in mcr-1 harboring carbapenem-resistant enterobacterales isolates from human in Thailand. Sci. Rep. 2022, 12, 19051. [Google Scholar] [CrossRef]
- Matamoros, S.; van Hattem, J.M.; Arcilla, M.S.; Willemse, N.; Melles, D.C.; Penders, J.; Vinh, T.N.; Thi Hoa, N.; Bootsma, M.C.J.; van Genderen, P.J.; et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 2017, 7, 15364. [Google Scholar] [CrossRef]
- Huang, J.; Zhu, J.; Gong, D.; Wu, L.; Zhu, Y.; Hu, L. Whole genome sequence of EC16, a bla(NDM-5)-, bla(CTX-M-55)-, and fosA3-coproducing Escherichia coli ST167 clinical isolate from China. J. Glob. Antimicrob. Resist. 2022, 29, 296–298. [Google Scholar] [CrossRef]
- Shi, J.P.; Zhu, H.; Liu, C.; Xie, H.; Li, C.C.; Cao, X.L.; Shen, H. Epidemiological and genomic characteristics of global mcr-positive Escherichia coli isolates. Front. Microbiol. 2023, 13, 1105401. [Google Scholar] [CrossRef]
- Tansawai, U.; Yu, Y.; Kiddee, A.; Assawatheptawee, K.; Sands, K.; Hassan, B.; Walsh, T.R.; Niumsup, P.R. Emergence of mcr-3-mediated IncP and IncFII plasmids in Thailand. J. Glob. Antimicrob. Resist. 2021, 24, 446–447. [Google Scholar] [CrossRef]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef]
- Boonyasiri, A.; Brinkac, L.M.; Jauneikaite, E.; White, R.C.; Greco, C.; Seenama, C.; Tangkoskul, T.; Nguyen, K.; Fouts, D.E.; Thamlikitkul, V. Characteristics and genomic epidemiology of colistin-resistant Enterobacterales from farmers, swine, and hospitalized patients in Thailand, 2014–2017. BMC Infect. Dis. 2023, 23, 556. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017. [Google Scholar]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef] [PubMed]
| AMR-0201 | AMR-0220 * | AMR-0251 | AMR-0354 | AMR-0361 * | AMR-0429 | ||
|---|---|---|---|---|---|---|---|
| Antimicrobial Class | Antimicrobial Agent | Minimum Inhibitory Concentration (μg/mL) | |||||
| Carbapenem | Ertapenem | ≤0.25 (S) | ≤0.25 (S) | ≤0.25 (S) | ≤0.25 (S) | ≤0.25 (S) | ≤0.25 (S) |
| Imipenem | ≤1 (S) | ≤1 (S) | ≤1 (S) | ≤1 (S) | ≤1 (S) | ≤1 (S) | |
| Meropenem | ≤1 (S) | ≤1 (S) | ≤1 (S) | ≤1 (S) | ≤1 (S) | ≤1 (S) | |
| Cephem | Cefepime 4 | >16 (R) | >16 (R) | >16 (R) | >16 (R) | 8 (R) | 16 (R) |
| Ceftazidime 3 | >16 (R) | >16 (R) | >16 (R) | >16 (R) | 8 (R) | 16 (R) | |
| Cephalothin 1 | >16 (R) | >16 (R) | >16 (R) | >16 (R) | >16 (R) | >16 (R) | |
| Ceftriaxone 3 | >16 (R) | >16 (R) | >16 (R) | >16 (R) | >16 (R) | >16 (R) | |
| Cefuroxime 2 | >16 (R) | >16 (R) | >16 (R) | >16 (R) | >16 (R) | >16 (R) | |
| Cefoxitin 2 | >16 (R) | 8 (S) | 8 (S) | 8 (S) | ≤4 (S) | ≤4 (S) | |
| β-lactam/β-lactam combination agent | Ampicillin | >16 (R) | >16 (R) | >16 (R) | >16 (R) | >16 (R) | >16 (R) |
| Amoxicillin-Clavulanate | >16/8 (R) | 8/4 (R) | ≤4/2 (R) | 8/4 (R) | >16/8 (R) | >16/8 (R) | |
| Piperacillin-Tazobactam | ≤4/4 (S) | ≤4/4 (S) | ≤4/4 (S) | ≤4/4 (S) | ≤4/4 (S) | ≤4/4 (S) | |
| 5-Fluoroquinolone | Ciprofloxacin | >2 (R) | >2 (R) | >2 (R) | ≤0.5 (S) | >2 (R) | 1 (S) |
| Norfloxacin | >8 (R) | >8 (R) | >8 (R) | ≤2 (S) | >8 (R) | ≤2 (S) | |
| Folate Antagonist | Trimethoprim | >8 (R) | ≤1 (S) | >8 (R) | ≤1 (S) | >8 (R) | >8 (R) |
| Trimethoprim-Sulfamethoxazole | >2/38 (R) | ≤0.5/9.5 (S) | >2/38 (R) | ≤0.5/9.5 (S) | >2/38 (R) | >2/38 (R) | |
| Aminoglycoside | Gentamicin | >8 (R) | >8 (R) | >8 (R) | ≤2 (S) | >8 (R) | >8 (R) |
| Tobramycin | 8 (I) | 8 (I) | 8 (I) | ≤2 (S) | 8 (I) | 8 (I) | |
| Nitrofurantoin Polymyxin | Nitrofurantoin | ≤16 (S) | ≤16 (S) | 32 (S) | ≤16 (S) | 64 (I) | 32 (S) |
| Colistin a | 4 (R) | 4 (R) | 4 (R) | 4 (R) | 4 (R) | 4 (R) | |
| Aminoglycoside | Beta-Lactam | Polymyxin | Phenicol | Quinolone | Lincosamide | Macrolide | Tetracycline | Trimethoprim | Sulfonamide | Rifampin | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample ID | MLST | Replicon Type | aac(3)-IIa | aac(3)-IId | aadA1 | aadA2 | aadA22 | aph(3)-Ia | aph(3)-Ib | aph(6)-Id | blaCMY-2 | blaCTX-M-15 | blaCTX-M-55 | blaTEM-1 | mcr-1.1 | mcr-3.4 | mcr-3.5 | catA2 | cmlA1 | floR | qnrS1 | qnrS13 | lnu(F) | mdf(A) | mph(A) | erm(B) | tet(A) | tet(B) | tet(M) | dfrA12 | dfrA14 | dfrA17 | sul1 | sul2 | sul3 | arr-2 |
| AMR-0201 | 410 | IncFIB, IncI1, IncX4 | ||||||||||||||||||||||||||||||||||
| AMR-0220 * | 410 | IncFIB, IncI1, IncX4, IncY | ||||||||||||||||||||||||||||||||||
| AMR-0251 | 10 | IncHI2, IncY | ||||||||||||||||||||||||||||||||||
| AMR-0354 | 38 | IncFII, IncI1,IncI2 | ||||||||||||||||||||||||||||||||||
| AMR-0361 * | 46 | IncFIB, IncFII, IncN, IncX4 | ||||||||||||||||||||||||||||||||||
| AMR-0429 | 617 | incFII, IncI1, IncQ1, IncX1, IncFI, IncY | ||||||||||||||||||||||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nobthai, P.; Ruekit, S.; Peerapongpaisarn, D.; Sukhchat, P.; Swierczewski, B.E.; Ruamsap, N.; Lertsethtakarn, P. The Co-Existence of mcr-1.1 and mcr-3.5 in Escherichia coli Isolated from Clinical Samples in Thailand. Antibiotics 2025, 14, 596. https://doi.org/10.3390/antibiotics14060596
Nobthai P, Ruekit S, Peerapongpaisarn D, Sukhchat P, Swierczewski BE, Ruamsap N, Lertsethtakarn P. The Co-Existence of mcr-1.1 and mcr-3.5 in Escherichia coli Isolated from Clinical Samples in Thailand. Antibiotics. 2025; 14(6):596. https://doi.org/10.3390/antibiotics14060596
Chicago/Turabian StyleNobthai, Panida, Sirigade Ruekit, Dutsadee Peerapongpaisarn, Prawet Sukhchat, Brett E. Swierczewski, Nattaya Ruamsap, and Paphavee Lertsethtakarn. 2025. "The Co-Existence of mcr-1.1 and mcr-3.5 in Escherichia coli Isolated from Clinical Samples in Thailand" Antibiotics 14, no. 6: 596. https://doi.org/10.3390/antibiotics14060596
APA StyleNobthai, P., Ruekit, S., Peerapongpaisarn, D., Sukhchat, P., Swierczewski, B. E., Ruamsap, N., & Lertsethtakarn, P. (2025). The Co-Existence of mcr-1.1 and mcr-3.5 in Escherichia coli Isolated from Clinical Samples in Thailand. Antibiotics, 14(6), 596. https://doi.org/10.3390/antibiotics14060596






