The Prevalence of Antimicrobial Resistance Genes in the Environments of Small Ruminant Farms from Central Portugal
Abstract
1. Introduction
2. Material and Methods
2.1. Sampling
2.2. DNA Extraction
2.3. Molecular Detection of AMR Genes
2.4. Statistical Analysis
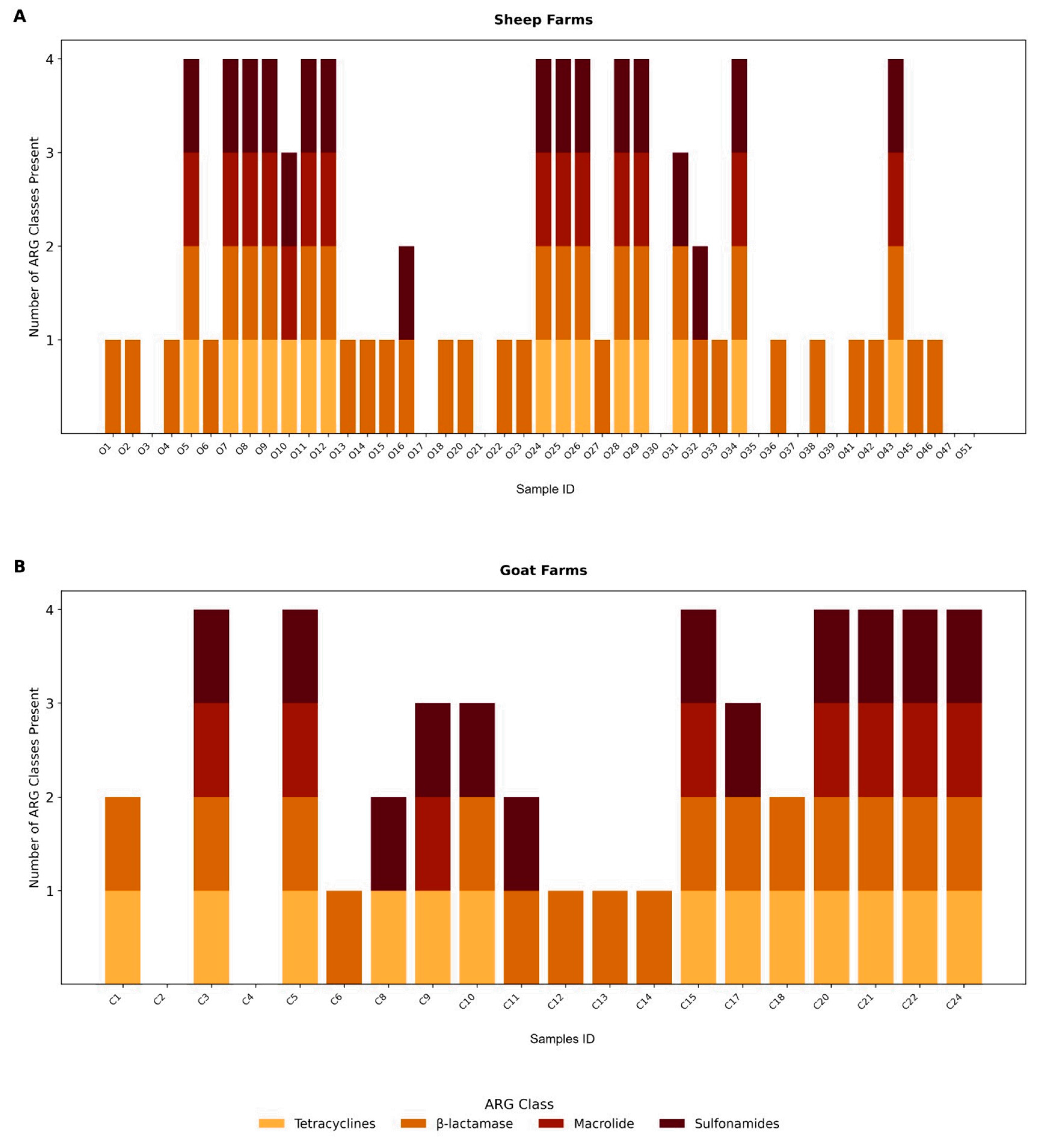
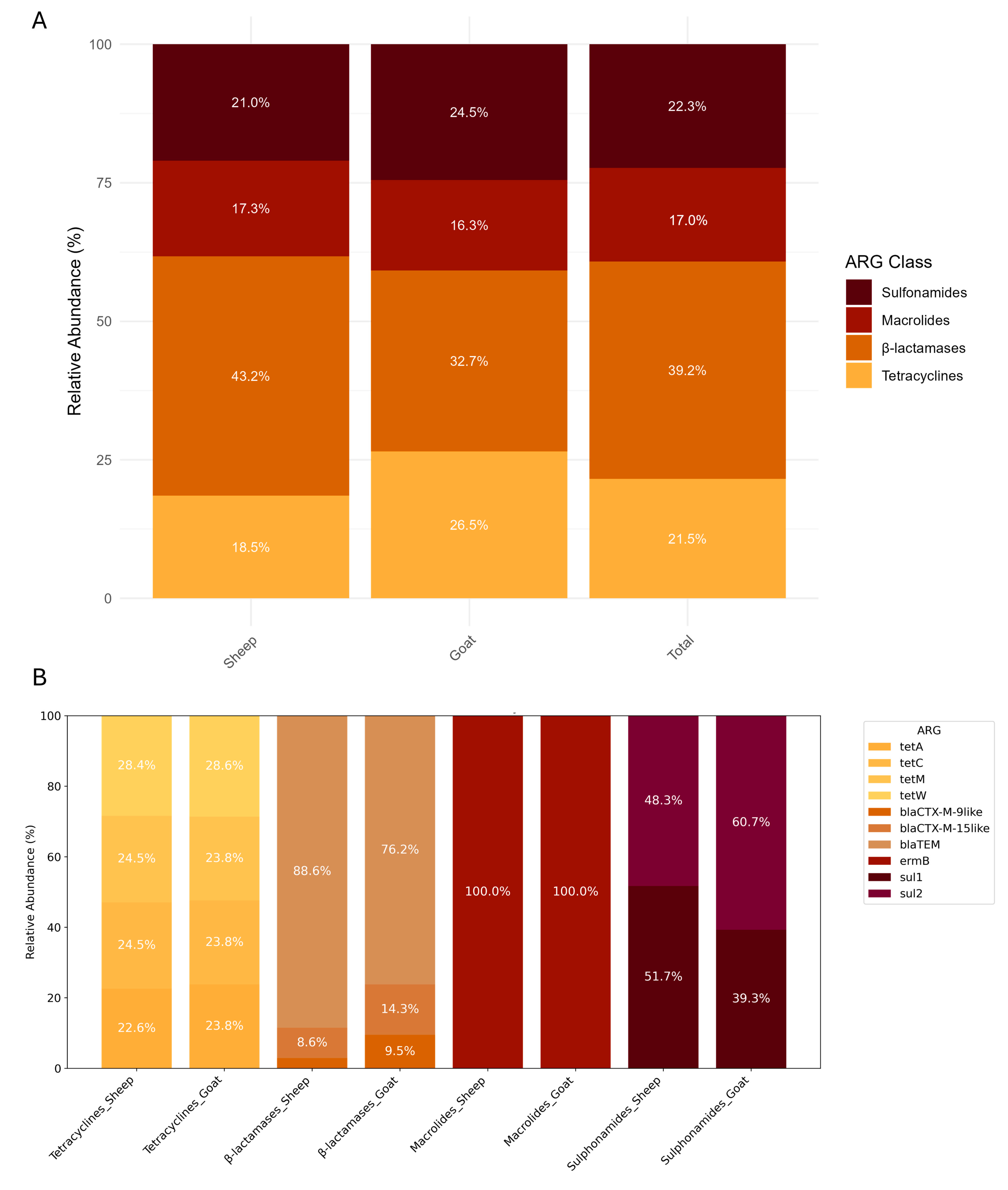
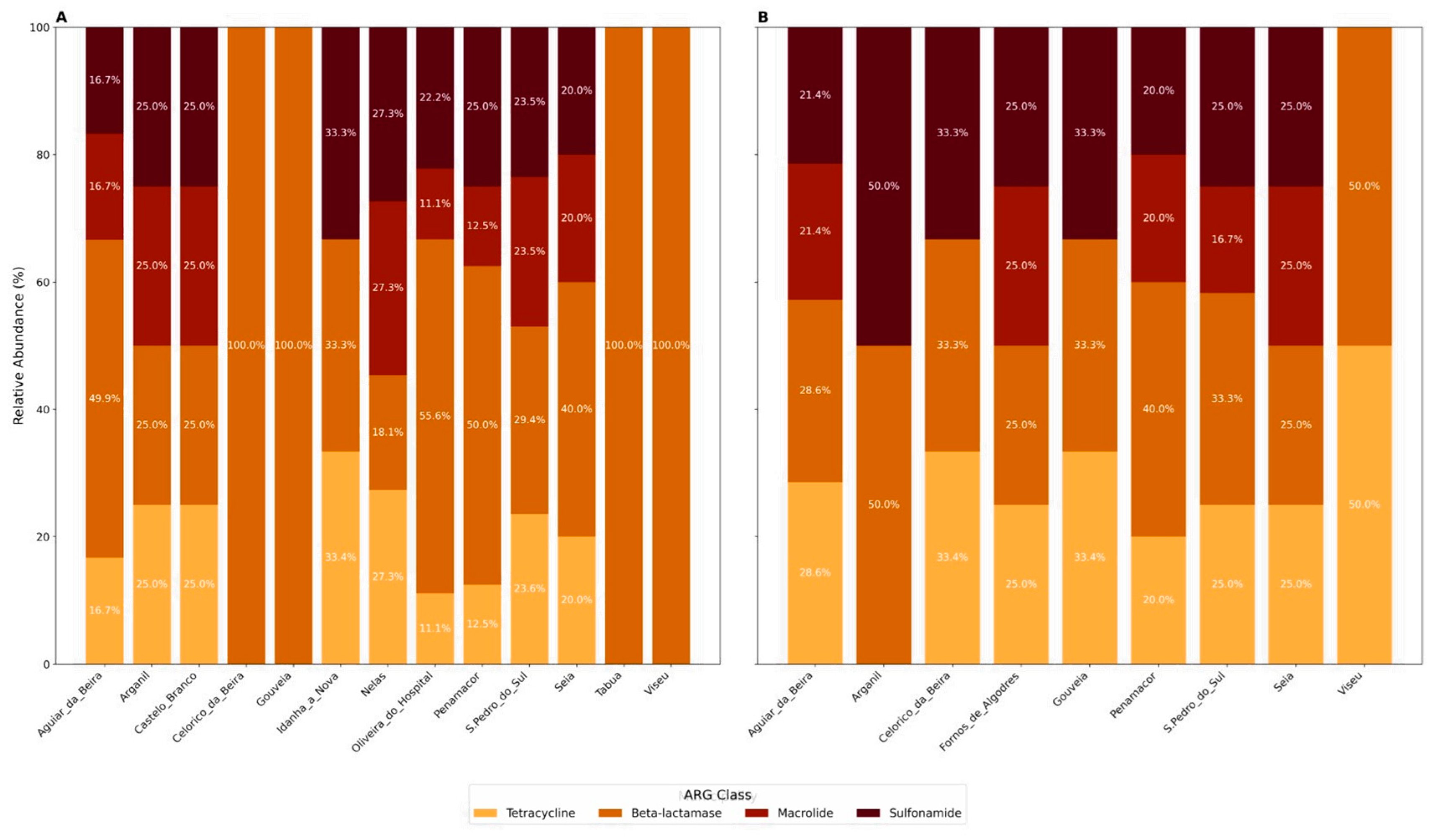
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO Regional Office for Europe/European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe 2022–2020 Data; WHO: Geneva, Switzerland, 2022; ISBN 9789294985521. [Google Scholar]
- Silva, V.; Monteiro, A.; Pereira, J.E.; Maltez, L.; Igrejas, G.; Poeta, P. MRSA in Humans, Pets and Livestock in Portugal: Where We Came from and Where We Are Going. Pathogens 2022, 11, 1110. [Google Scholar] [CrossRef] [PubMed]
- Flynn, C.E.; Guarner, J. Emerging Antimicrobial Resistance. Mod. Pathol. 2023, 36, 100249. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, N.; Joji, R.M.; Shahid, M. Evolution and Implementation of One Health to Control the Dissemination of Antibiotic-Resistant Bacteria and Resistance Genes: A Review. Front. Cell. Infect. Microbiol. 2023, 12, 1065796. [Google Scholar] [CrossRef] [PubMed]
- Van, T.T.H.; Yidana, Z.; Smooker, P.M.; Coloe, P.J. Antibiotic Use in Food Animals Worldwide, with a Focus on Africa: Pluses and Minuses. J. Glob. Antimicrob. Resist. 2020, 20, 170–177. [Google Scholar] [CrossRef]
- Rushton, J. Anti-Microbial Use in Animals: How to Assess the Trade-Offs. Zoonoses Public Health 2015, 62, 10–21. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic Resistance Genes from Livestock Waste: Occurrence, Dissemination, and Treatment. npj Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- Lei, L.; Chen, N.; Chen, Z.; Zhao, Y.; Lin, H.; Li, X.; Hu, W.; Zhang, H.; Shi, J.; Luo, Y. Dissemination of Antibiotic Resistance Genes from Aboveground Sources to Groundwater in Livestock Farms. Water Res. 2024, 256, 121584. [Google Scholar] [CrossRef]
- Ramos, S.; Igrejas, G.; Capelo-Martinez, J.L.; Poeta, P. Antibiotic Resistance and Mechanisms Implicated in Fecal Enterococci Recovered from Pigs, Cattle and Sheep in a Portuguese Slaughterhouse. Ann. Microbiol. 2012, 62, 1485–1494. [Google Scholar] [CrossRef]
- Castro, J.; Castro, M.; Gómez-Sal, A. Changes on the Climatic Edge: Adaptation of and Challenges to Pastoralism in Montesinho (Northern Portugal). Mt. Res. Dev. 2021, 41, R29–R37. [Google Scholar] [CrossRef]
- Amador, P.; Fernandes, R.; Prudêncio, C.; Duarte, I. Prevalence of Antibiotic Resistance Genes in Multidrug-Resistant Enterobacteriaceae on Portuguese Livestock Manure. Antibiotics 2019, 8, 23. [Google Scholar] [CrossRef]
- Antunes, M.I.; Lima, M.S.; Stilwell, G.; Romeiras, M.I.; Fragoso, L.; Madeira de Carvalho, L.M. Anthelmintic Efficacy in Sheep and Goats under Different Management and Deworming Systems in the Region of Lisbon and Tagus Valley, Portugal. Pathogens 2022, 11, 1457. [Google Scholar] [CrossRef]
- Chen, B.; Hao, L.; Guo, X.; Wang, N.; Ye, B. Prevalence of Antibiotic Resistance Genes of Wastewater and Surface Water in Livestock Farms of Jiangsu Province, China. Environ. Sci. Pollut. Res. 2015, 22, 13950–13959. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, M.; Mei, S.; Daras, I.M.; van Doorn, G.S.; Falcao Salles, J.; de Vos, M.G.J. An Eco-Evolutionary Perspective on Antimicrobial Resistance in the Context of One Health. iScience 2025, 28, 111534. [Google Scholar] [CrossRef] [PubMed]
- Kelbrick, M.; Hesse, E.; O’Brien, S. Cultivating Antimicrobial Resistance: How Intensive Agriculture Ploughs the Way for Antibiotic Resistance. Microbiology 2023, 169, 001384. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. The Antibiotic Resistome: The Nexus of Chemical and Genetic Diversity. Nat. Rev. Microbiol. 2007, 5, 175–186. [Google Scholar] [CrossRef]
- D’Costa, V.M.; McGrann, K.M.; Hughes, D.W.; Wright, G.D. Sampling the Antibiotic Resistome. Science 2006, 311, 374–377. [Google Scholar] [CrossRef]
- Elshobary, M.E.; Badawy, N.K.; Ashraf, Y.; Zatioun, A.A.; Masriya, H.H.; Ammar, M.M.; Mohamed, N.A.; Mourad, S.; Assy, A.M. Combating Antibiotic Resistance: Mechanisms, Multidrug-Resistant Pathogens, and Novel Therapeutic Approaches: An Updated Review. Pharmaceuticals 2025, 18, 402. [Google Scholar] [CrossRef]
- Denissen, J.; Reyneke, B.; Waso-Reyneke, M.; Havenga, B.; Barnard, T.; Khan, S.; Khan, W. Prevalence of ESKAPE Pathogens in the Environment: Antibiotic Resistance Status, Community-Acquired Infection and Risk to Human Health. Int. J. Hyg. Environ. Health 2022, 244, 114006. [Google Scholar] [CrossRef]
- Souza, H.C.A.d.; Panzenhagen, P.; dos Santos, A.M.P.; Portes, A.B.; Fidelis, J.; Conte-Junior, C.A. Unravelling the Advances of CRISPR-Cas9 as a Precise Antimicrobial Therapy: A Systematic Review. J. Glob. Antimicrob. Resist. 2025, 42, 51–60. [Google Scholar] [CrossRef]
- Samreen; Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental Antimicrobial Resistance and Its Drivers: A Potential Threat to Public Health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar] [CrossRef]
- Tao, S.; Chen, H.; Li, N.; Wang, T.; Liang, W. The Spread of Antibiotic Resistance Genes In Vivo Model. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 3348695. [Google Scholar] [CrossRef] [PubMed]
- Deekshit, V.K.; Srikumar, S. ‘To Be, or Not to Be’—The Dilemma of ‘Silent’ Antimicrobial Resistance Genes in Bacteria. J. Appl. Microbiol. 2022, 133, 2902–2914. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.A.; Dantas, G. The Shared Antibiotic Resistome of Soil Bacteria and Human Pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Cha, C.J. Antibiotic Resistome from the One-Health Perspective: Understanding and Controlling Antimicrobial Resistance Transmission. Exp. Mol. Med. 2021, 53, 301–309. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.-N.; et al. Tackling Antibiotic Resistance: The Environmental Framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef]
- Valvi, D.; Walker, D.I.; Inge, T.; Bartell, S.M.; Jenkins, T.; Helmrath, M.; Ziegler, T.R.; La Merrill, M.A.; Eckel, S.P.; Conti, D.; et al. Environmental Chemical Burden in Metabolic Tissues and Systemic Biological Pathways in Adolescent Bariatric Surgery Patients: A Pilot Untargeted Metabolomic Approach. Environ. Int. 2020, 143, 105957. [Google Scholar] [CrossRef]
- Tamminen, M.; Karkman, A.; Lõhmus, A.; Muziasari, W.I.; Takasu, H.; Wada, S.; Suzuki, S.; Virta, M. Tetracycline Resistance Genes Persist at Aquaculture Farms in the Absence of Selection Pressure. Environ. Sci. Technol. 2011, 45, 386–391. [Google Scholar] [CrossRef]
- Ismail, Z.B.; Abutarbush, S.M. Molecular Characterization of Antimicrobial Resistance and Virulence Genes of Escherichia coli Isolates from Bovine Mastitis. Vet. World 2020, 13, 1588–1593. [Google Scholar] [CrossRef]
- Hiraoka (Furuya), Y.; Abo, H.; Matsuda, M.; Harada, S.; Kumakawa, M.; Shirakawa, T.; Ozawa, M.; Kawanishi, M.; Sekiguchi, H.; Shimazaki, Y. Genomic Characterization of Third-Generation Cephalosporin-Resistant Escherichia coli Strains Isolated from Diseased Dogs and Cats: Report from Japanese Veterinary Antimicrobial Resistance Monitoring. Vet. Microbiol. 2024, 298, 110220. [Google Scholar] [CrossRef]
- Deng, X.; Wang, S.; Hou, P.; Sun, N.; Yang, Y.; Zeng, Q.; Wang, J.; Wang, C.; Lv, X.; Zhang, W.; et al. Fecal Carriage and Molecular Characterization of Carbapenem-Resistant Enterobacteriaceae from Hospitalized Children in a Tertiary Hospital of Shandong, China. Front. Microbiol. 2025, 16, 1542207. [Google Scholar] [CrossRef]
- Cramer, G.; Solano, L.; Johnson, R. Evaluation of Tetracycline in Milk Following Extra-Label Administration of Topical Tetracycline for Digital Dermatitis in Dairy Cattle. J. Dairy. Sci. 2019, 102, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Quinteira, S.; Dantas, R.; Pinho, L.; Campos, C.; Freitas, A.R.; Brito, N.V.; Miranda, C. Dairy Cattle and the Iconic Autochthonous Cattle in Northern Portugal Are Reservoirs of Multidrug-Resistant Escherichia coli. Antibiotics 2024, 13, 1208. [Google Scholar] [CrossRef] [PubMed]
- Bradford, P.A. Extended-Spectrum β-Lactamases in the 21st Century: Characterization, Epidemiology, and Detection of This Important Resistance Threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, J.; Li, Z.; Guo, S.; Li, K.; Xu, P.; Ok, Y.S.; Jones, D.L.; Zou, J. Antibiotics and Antibiotic Resistance Genes in Agricultural Soils: A Systematic Analysis. Crit. Rev. Environ. Sci. Technol. 2023, 53, 847–864. [Google Scholar] [CrossRef]
- Chopra, I.; Roberts, M. Tetracycline Antibiotics: Mode of Action, Applications, Molecular Biology, and Epidemiology of Bacterial Resistance. Microbiol. Mol. Biol. Rev. 2001, 65, 232–260. [Google Scholar] [CrossRef]
- Roberts, M.C. Tetracycline Resistance Determinants: Mechanisms of Action, Regulation of Expression, Genetic Mobility, and Distribution. FEMS Microbiol. Rev. 1996, 19, 1–24. [Google Scholar] [CrossRef]
- Vázquez-Laslop, N.; Mankin, A.S. How Macrolide Antibiotics Work. Trends Biochem. Sci. 2018, 43, 668–684. [Google Scholar] [CrossRef]
- Gaynor, M.; Mankin, A.S. Macrolide Antibiotics: Binding Site, Mechanism of Action, Resistance. Front. Med. Chem. 2012, 21–35, 949–961. [Google Scholar] [CrossRef][Green Version]
- Venkatesan, M.; Fruci, M.; Verellen, L.A.; Skarina, T.; Mesa, N.; Flick, R.; Pham, C.; Mahadevan, R.; Stogios, P.J.; Savchenko, A. Molecular Mechanism of Plasmid-Borne Resistance to Sulfonamide Antibiotics. Nat. Commun. 2023, 14, 4031. [Google Scholar] [CrossRef]
- Kang, J.; Liu, Y.; Chen, X.; Xu, F.; Wang, H.; Xiong, W.; Li, X. Metagenomic Insights into the Antibiotic Resistomes of Typical Chinese Dairy Farm Environments. Front. Microbiol. 2022, 13, 990272. [Google Scholar] [CrossRef]
- Li, B.; Li, X.; Wang, B.; Yan, T. A Metagenomic Approach for Characterizing Antibiotic Resistance Genes in Specific Bacterial Populations: Demonstration with Escherichia coli in Cattle Manure. Appl. Environ. Microbiol. 2022, 88, e0255421. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Yang, J.; Zeng, J.; Xiao, D.; Tong, C.; Zeng, Z. Metagenomic Analysis of Antimicrobial Resistance in Ducks, Workers, and the Environment in Duck Farms, Southern China. Ecotoxicol. Environ. Saf. 2023, 262, 115191. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Li, Y.; Yang, Y.; Shen, Z.; Cai, C.; Wang, Y.; Walsh, T.R.; Shen, J.; Wu, Y.; Wang, S. High Prevalence and Persistence of Carbapenem and Colistin Resistance in Livestock Farm Environments in China. J. Hazard. Mater. 2021, 406, 124298. [Google Scholar] [CrossRef] [PubMed]
- Ndegwa, E.; Almehmadi, H.; Kim, C.; Kaseloo, P.; Ako, A.A. Longitudinal Shedding Patterns and Characterization of Antibiotic Resistant E. coli in Pastured Goats Using a Cohort Study. Antibiotics 2019, 8, 136. [Google Scholar] [CrossRef]
- OECD (Organisation for Economic Co-operation and Development). Stemming the Superbug Tide Policy Brief; OECD: Paris, France, 2018; ISBN 9789264307582. [Google Scholar]
- IHME; University of Washington. Measuring Infectious Causes and Resistance Outcomes for Burden Estimation—Institute for Health Metrics and Evaluation. Available online: https://vizhub.healthdata.org/microbe/?settings=eyIxIjoia2V5X2ZpbmRpbmdzIiwiMiI6ImJhciIsIjMiOiJhbXIiLCI0IjoyMiwiNSI6MSwiNiI6MywiNyI6MywiOCI6OTEsIjkiOjEsIjEyIjozLCIxMyI6MSwiMTQiOjEsIjE1IjoxLCIxNiI6MSwiMTciOjMsIjE4IjoyMDIxLCIxOSI6ZmFsc2UsIjIwIjpmYWxzZSwiMjI (accessed on 15 May 2025).
- He, L.Y.; Ying, G.G.; Liu, Y.S.; Su, H.C.; Chen, J.; Liu, S.S.; Zhao, J.L. Discharge of Swine Wastes Risks Water Quality and Food Safety: Antibiotics and Antibiotic Resistance Genes from Swine Sources to the Receiving Environments. Environ. Int. 2016, 92–93, 210–219. [Google Scholar] [CrossRef]
- Zhu, T.; Chen, T.; Cao, Z.; Zhong, S.; Wen, X.; Mi, J.; Ma, B.; Zou, Y.; Zhang, N.; Liao, X.; et al. Antibiotic Resistance Genes in Layer Farms and Their Correlation with Environmental Samples. Poult. Sci. 2021, 100, 101485. [Google Scholar] [CrossRef]
- Zalewska, M.; Błażejewska, A.; Czapko, A.; Popowska, M. Antibiotics and Antibiotic Resistance Genes in Animal Manure—Consequences of Its Application in Agriculture. Front. Microbiol. 2021, 12, 610656. [Google Scholar] [CrossRef]
- Jung, H.R.; Lee, Y.J.; Hong, S.; Yoon, S.; Lim, S.K.; Lee, Y.J. Current Status of β-Lactam Antibiotic Use and Characterization of β-Lactam-Resistant Escherichia coli from Commercial Farms by Integrated Broiler Chicken Operations in Korea. Poult. Sci. 2023, 102, 103091. [Google Scholar] [CrossRef]
- Turner, J.; Muraoka, A.; Bedenbaugh, M.; Childress, B.; Pernot, L.; Wiencek, M.; Peterson, Y.K. The Chemical Relationship Among Beta-Lactam Antibiotics and Potential Impacts on Reactivity and Decomposition. Front. Microbiol. 2022, 13, 807955. [Google Scholar] [CrossRef]
- Laconi, A.; Mughini-Gras, L.; Tolosi, R.; Grilli, G.; Trocino, A.; Carraro, L.; Di Cesare, F.; Cagnardi, P.; Piccirillo, A. Microbial Community Composition and Antimicrobial Resistance in Agricultural Soils Fertilized with Livestock Manure from Conventional Farming in Northern Italy. Sci. Total Environ. 2021, 760, 143404. [Google Scholar] [CrossRef]
- Hille, K.; Felski, M.; Ruddat, I.; Woydt, J.; Schmid, A.; Friese, A.; Fischer, J.; Sharp, H.; Valentin, L.; Michael, G.B.; et al. Association of Farm-Related Factors with Characteristics Profiles of Extended-Spectrum β-Lactamase-/Plasmid-Mediated AmpC β-Lactamase-Producing Escherichia coli Isolates from German Livestock Farms. Vet. Microbiol. 2018, 223, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Jin, S.; Guo, X.; Tong, H.; Ren, N.; You, S. Distribution and Transmission of β-Lactamase Resistance Genes in Meal-to-Milk Chain on Dairy Farm. Environ. Pollut. 2023, 331, 121831. [Google Scholar] [CrossRef] [PubMed]
- Ovung, A.; Bhattacharyya, J. Sulfonamide Drugs: Structure, Antibacterial Property, Toxicity, and Biophysical Interactions. Biophys. Rev. 2021, 13, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, T.; Lin, X.; Huang, J.; Zeng, J.; Cai, Q.; Zhang, Y.; Rong, J.; Yu, W.; Qiu, J.; et al. Isolation, Identification, and Optimization of Conditions for the Degradation of Four Sulfonamide Antibiotics and Their Metabolic Pathways in Pseudomonas Stutzeri Strain DLY-21. Heliyon 2024, 10, e29123. [Google Scholar] [CrossRef]
- Mulchandani, R.; Wang, Y.; Gilbert, M.; Van Boeckel, T.P. Global Trends in Antimicrobial Use in Food-Producing Animals: 2020 to 2030. PLoS Glob. Public Health 2023, 3, e0001305. [Google Scholar] [CrossRef]
- Wang, Z.; Li, Y.; Wang, J.; Li, S. Tetracycline Antibiotics in Agricultural Soil: Dissipation Kinetics, Transformation Pathways, and Structure-Related Toxicity. Sci. Total Environ. 2024, 949, 175126. [Google Scholar] [CrossRef]
- Yoshizawa, N.; Usui, M.; Fukuda, A.; Asai, T.; Higuchi, H.; Okamoto, E.; Seki, K.; Takada, H.; Tamura, Y. Manure Compost Is a Potential Source of Tetracycline-Resistant Escherichia coli and Tetracycline Resistance Genes in Japanese Farms. Antibiotics 2020, 9, 76. [Google Scholar] [CrossRef]
- Cheng, W.; Chen, H.; Su, C.; Yan, S. Abundance and Persistence of Antibiotic Resistance Genes in Livestock Farms: A Comprehensive Investigation in Eastern China. Environ. Int. 2013, 61, 1–7. [Google Scholar] [CrossRef]
- Shang, M.; Gao, Y.; Zheng, L.; Ji, L.; Du, J.; Kong, X.; Wang, H.; Shi, F.; Wang, H.; Liu, J.; et al. Vertical Distribution and Drivers of Antibiotic Resistance Genes in Agricultural Soil Irrigated with Livestock Wastewater. Microorganisms 2025, 13, 610. [Google Scholar] [CrossRef]
- Jensen, L.B.; Agersø, Y.; Sengeløv, G. Presence of Erm Genes among Macrolide-Resistant Gram-Positive Bacteria Isolated from Danish Farm Soil. Environ. Int. 2002, 28, 487–491. [Google Scholar] [CrossRef]
- Wang, N.; Guo, X.; Yan, Z.; Wang, W.; Chen, B.; Ge, F.; Ye, B. A Comprehensive Analysis on Spread and Distribution Characteristic of Antibiotic Resistance Genes in Livestock Farms of Southeastern China. PLoS ONE 2016, 11, e0156889. [Google Scholar] [CrossRef] [PubMed]
- Lund, D.; Kieffer, N.; Parras-Moltó, M.; Ebmeyer, S.; Berglund, F.; Johnning, A.; Joakim Larsson, D.G.; Kristiansson, E. Large-Scale Characterization of the Macrolide Resistome Reveals High Diversity and Several New Pathogen-Associated Genes. Microb. Genom. 2022, 8, 000770. [Google Scholar] [CrossRef]
- Min, J.Y.; Jang, Y.J. Macrolide Therapy in Respiratory Viral Infections. Mediat. Inflamm. 2012, 2012, 649570. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, H.E.; Jernberg, C.; Andersson, A.F.; Sjölund-Karlsson, M.; Jansson, J.K.; Engstrand, L. Short-Term Antibiotic Treatment Has Differing Long-Term Impacts on the Human Throat and Gut Microbiome. PLoS ONE 2010, 5, e9836. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Y.; Zhao, C.; Ma, J.; Guo, J. Antibiotic Resistance Gene Pollution in Poultry Farming Environments and Approaches for Mitigation: A System Review. Poult. Sci. 2025, 104, 104858. [Google Scholar] [CrossRef]
- Wen, R.; Yang, M.; Xu, Q.; Xu, W.; Zhou, Q.; Ma, B.; Lin, X.; Lei, C.; Wang, H. Assessing the Pig Microbial Health Impacts of Smallholder Farming. Ecotoxicol. Environ. Saf. 2024, 286, 117204. [Google Scholar] [CrossRef]
- Luiken, R.E.C.; Heederik, D.J.J.; Scherpenisse, P.; Van Gompel, L.; van Heijnsbergen, E.; Greve, G.D.; Jongerius-Gortemaker, B.G.M.; Tersteeg-Zijderveld, M.H.G.; Fischer, J.; Juraschek, K.; et al. Determinants for Antimicrobial Resistance Genes in Farm Dust on 333 Poultry and Pig Farms in Nine European Countries. Environ. Res. 2022, 208, 112715. [Google Scholar] [CrossRef]
- Allen, H.K.; Levine, U.Y.; Looft, T.; Bandrick, M.; Casey, T.A. Treatment, Promotion, Commotion: Antibiotic Alternatives in Food-Producing Animals. Trends Microbiol. 2013, 21, 114–119. [Google Scholar] [CrossRef]
- Cheng, G.; Hao, H.; Xie, S.; Wang, X.; Dai, M.; Huang, L.; Yuan, Z. Antibiotic Alternatives: The Substitution of Antibiotics in Animal Husbandry? Front. Microbiol. 2014, 5, 217. [Google Scholar] [CrossRef]
- Rizzo, A.; Piccinno, M.; Lillo, E.; Carbonari, A.; Jirillo, F.; Sciorsci, R.L. Antimicrobial Resistance and Current Alternatives in Veterinary Practice: A Review. Curr. Pharm. Des. 2023, 29, 312–322. [Google Scholar]
- Pandey, S.; Doo, H.; Keum, G.B.; Kim, E.S.; Kwak, J.; Ryu, S.; Choi, Y.; Kang, J.; Kim, S.; Lee, N.R.; et al. Antibiotic Resistance in Livestock, Environment and Humans: One Health Perspective. J. Anim. Sci. Technol. 2024, 62, 266–278. [Google Scholar] [CrossRef]
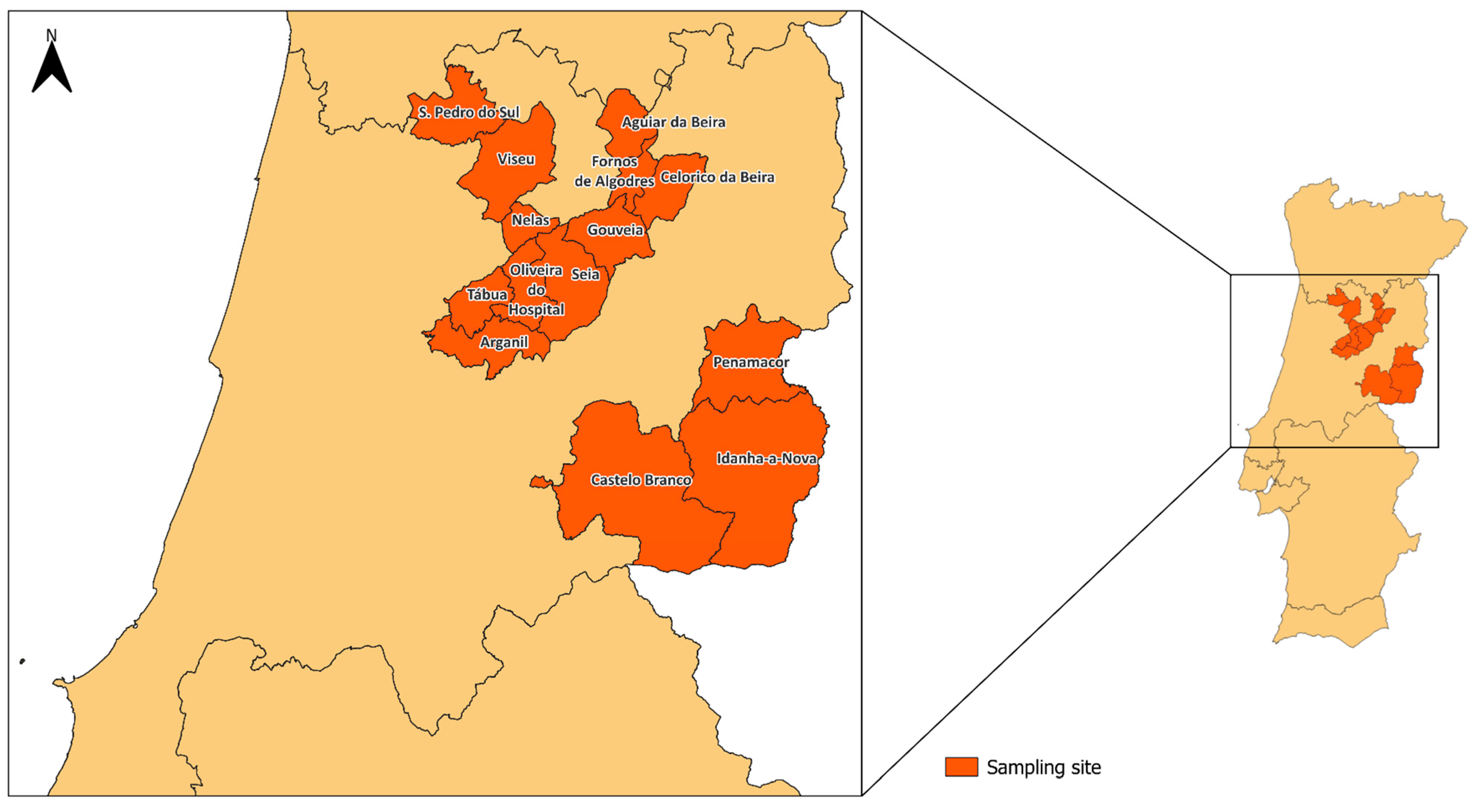
| Municipality | Goat Farms (n) | Sheep Farms (n) | Total |
|---|---|---|---|
| Aguiar da Beira | 4 | 4 | 8 |
| Arganil | 2 | 1 | 3 |
| Castelo Branco | - | 1 | 1 |
| Celorico da Beira | 1 | 5 | 6 |
| Fornos de Algodres | 1 | - | 1 |
| Gouveia | 2 | 6 | 8 |
| Idanha-a-Nova | - | 1 | 1 |
| Nelas | - | 3 | 3 |
| Oliveira do Hospital | - | 5 | 5 |
| Penamacor | 2 | 7 | 9 |
| S. Pedro do Sul | 6 | 5 | 11 |
| Seia | 1 | 5 | 6 |
| Tábua | - | 1 | 1 |
| Viseu | 1 | 1 | 2 |
| Total | 20 | 45 | 65 |
| Class | Gene | Sequence (5′ > 3′) | Annealing Temperature (°C) | Product Size (bp) | Reference |
|---|---|---|---|---|---|
| Tetracyclines | tetA | F:GCTACATCCTGCTTGCCTTC R:CATAGATCGCCGTGAAGAGG | 60 | 210 | [13] |
| tetC | F:TGCAACTCGTAGGACAGGTG R:ACCAGTGACGAAGGCTTGAG | 60 | 139 | ||
| tetM | F:ACAGAAAGCTTATTATATAAC R:GGCGTGTCTATGATGTTCAC | 51 | 171 | ||
| tetW | F:GAGAGCCTGCTATATGCCAGC R:GGGCGTATCCACAATGTTAAC | 60 | 168 | ||
| Sulfonamides | sul1 | F:TGTCGAACCTTCAAAAGCTG R:TGGACCCAGATCCTTTACAG | 60 | 113 | [13] |
| sul2 | F:ATCTGCCAAACTCGTCGTTA R:CAATGTGATCCATGATGTCG | 60 | 89 | ||
| Macrolides | ermB | F: AGGGTTGCTCTTGCACACTC R: CTGTGGTATGGCGGGTAAGT | 58 | 119 | [13] |
| β-lactamase (ESBL) | blaCTX-M-15like | F: GCTGGTGACATGGATGAAAG R: TAGGTTGAGGCTGGGTGAAG | 60 | 87 | [44] |
| blaCTX-M-9like | F: GTTGGTGACGTGGCTCAAAG R: GTTGCGGCTGGGTAAAATAG | 60 | 89 | ||
| blaTEM | F: TCTGACAACGATCGGAGGAC R: TGCCGGGAAGCTAGAGTAAG | 60 | 86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bento, J.T.; Gomes-Gonçalves, S.; Cruz, R.; Esteves, F.; Baptista, A.L.; Aires Pereira, M.; Caseiro, P.; Carreira, P.; Figueira, L.; Mesquita, J.R.; et al. The Prevalence of Antimicrobial Resistance Genes in the Environments of Small Ruminant Farms from Central Portugal. Antibiotics 2025, 14, 576. https://doi.org/10.3390/antibiotics14060576
Bento JT, Gomes-Gonçalves S, Cruz R, Esteves F, Baptista AL, Aires Pereira M, Caseiro P, Carreira P, Figueira L, Mesquita JR, et al. The Prevalence of Antimicrobial Resistance Genes in the Environments of Small Ruminant Farms from Central Portugal. Antibiotics. 2025; 14(6):576. https://doi.org/10.3390/antibiotics14060576
Chicago/Turabian StyleBento, Jaqueline T., Sara Gomes-Gonçalves, Rita Cruz, Fernando Esteves, Alexandra Lameira Baptista, Maria Aires Pereira, Pedro Caseiro, Pedro Carreira, Luís Figueira, João R. Mesquita, and et al. 2025. "The Prevalence of Antimicrobial Resistance Genes in the Environments of Small Ruminant Farms from Central Portugal" Antibiotics 14, no. 6: 576. https://doi.org/10.3390/antibiotics14060576
APA StyleBento, J. T., Gomes-Gonçalves, S., Cruz, R., Esteves, F., Baptista, A. L., Aires Pereira, M., Caseiro, P., Carreira, P., Figueira, L., Mesquita, J. R., Bordalo, A. A., & Machado, A. (2025). The Prevalence of Antimicrobial Resistance Genes in the Environments of Small Ruminant Farms from Central Portugal. Antibiotics, 14(6), 576. https://doi.org/10.3390/antibiotics14060576









