In Vitro Analysis of Interactions Between Staphylococcus aureus and Pseudomonas aeruginosa During Biofilm Formation
Abstract
1. Introduction
2. Results
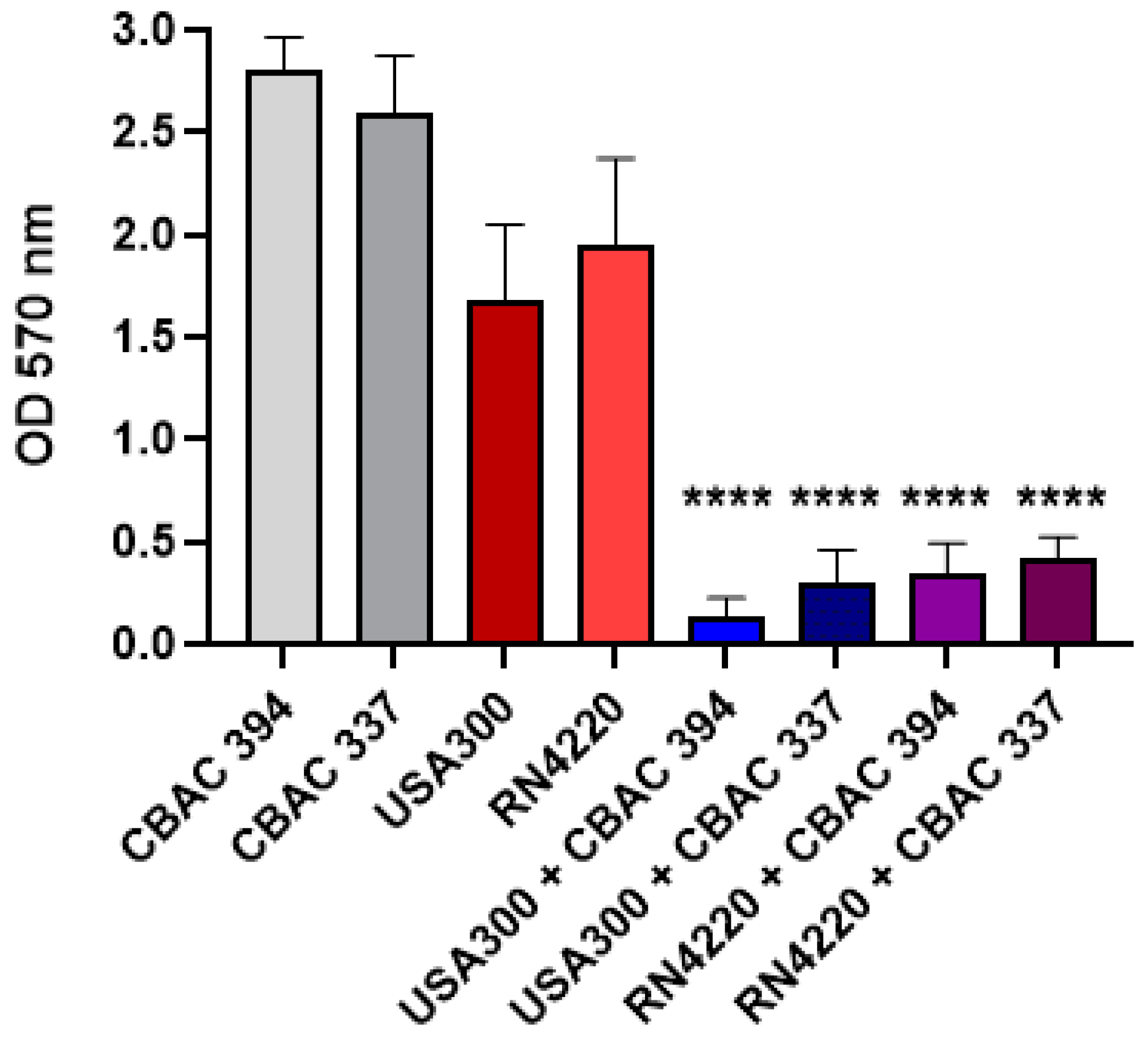
2.1. Polymicrobial Biofilm Strength
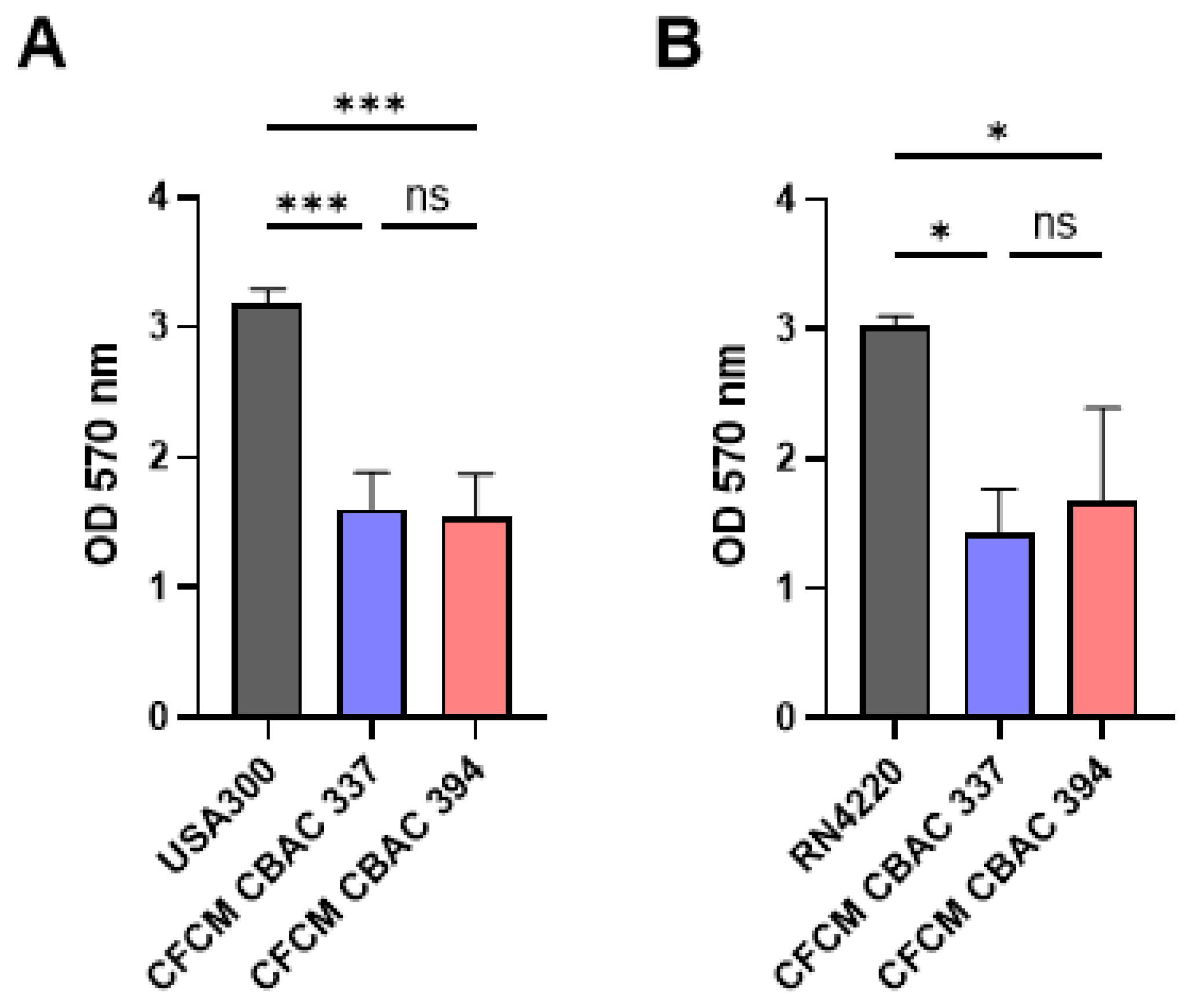
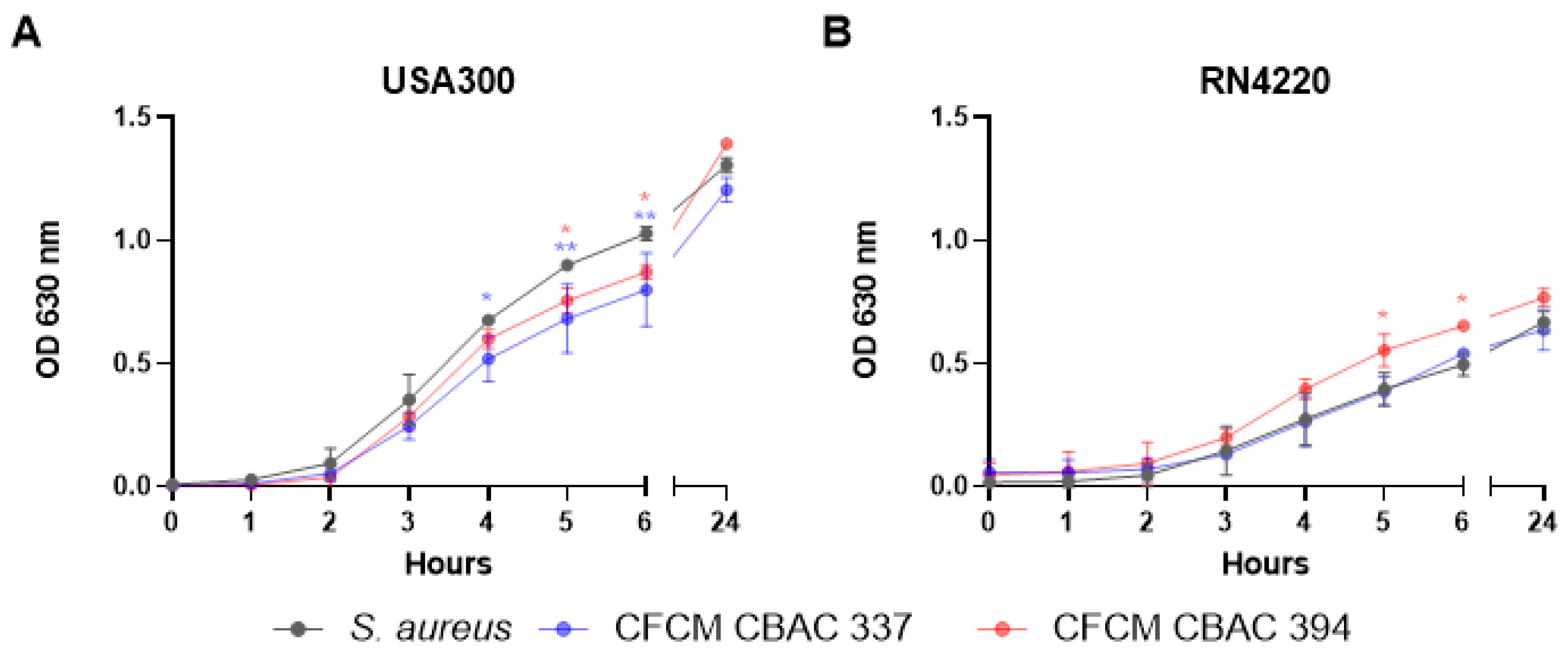
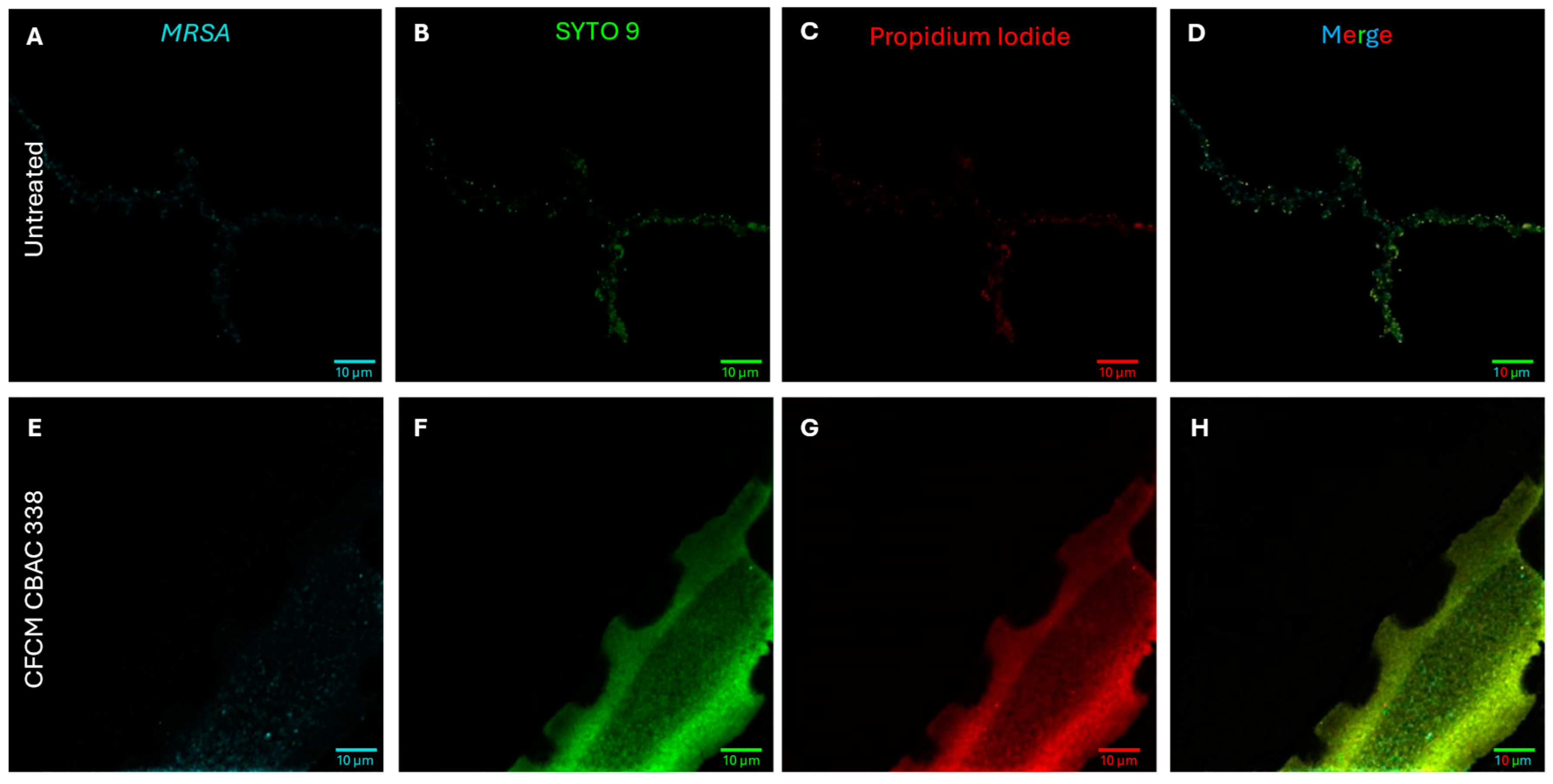
2.2. Inhibition of S. aureus Biofilm Formation and Planktonic Growth with CFCM
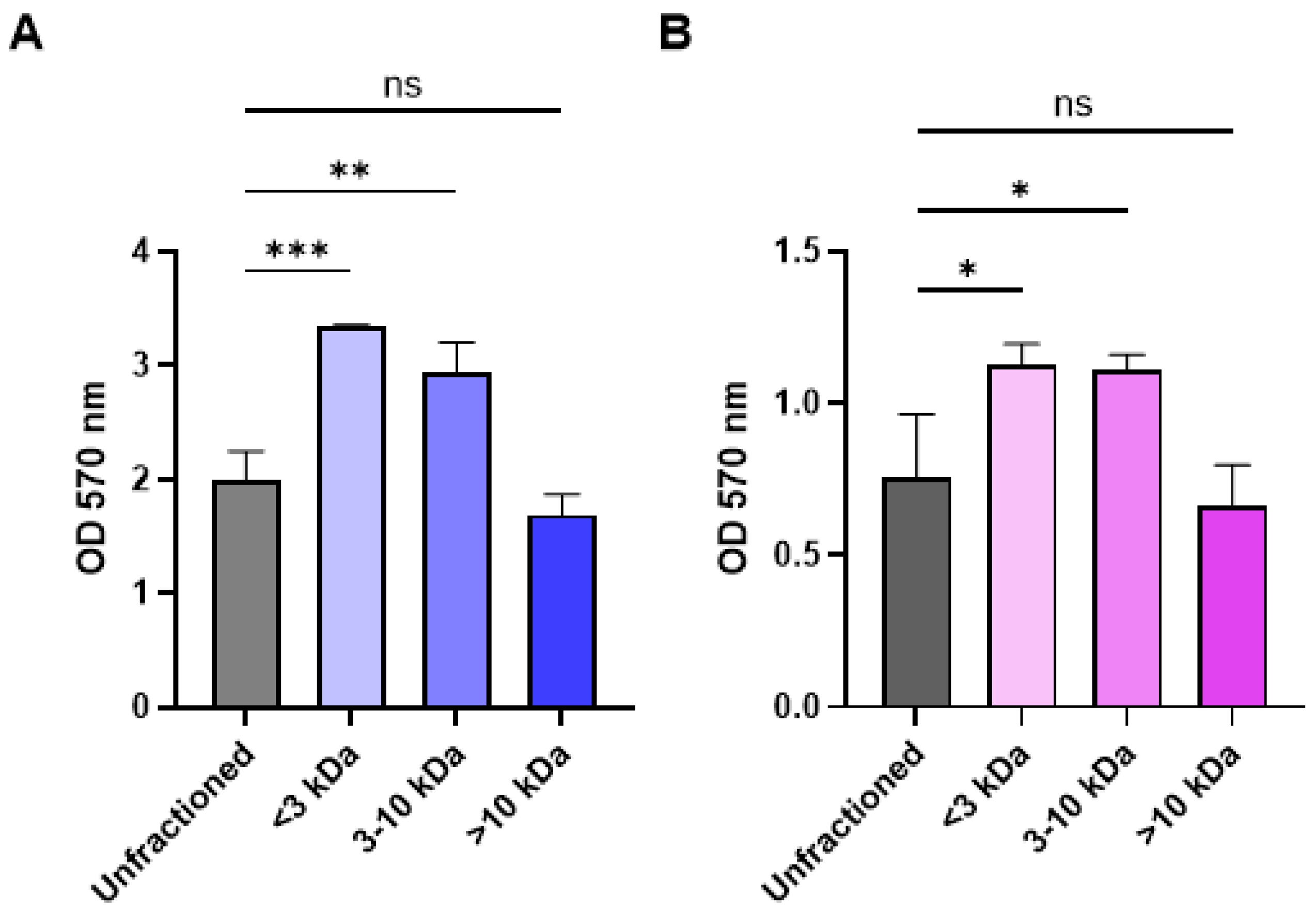
2.3. Fractionation of CFCM Activity by Molecular Weight and Impact on Growth
2.4. Growth in Polymicrobial Biofilm Increases Expression of Virulence Factors and Excreted Proteins
2.5. P. aeruginosa Excreted Proteins Eliminate S. aureus Biofilms
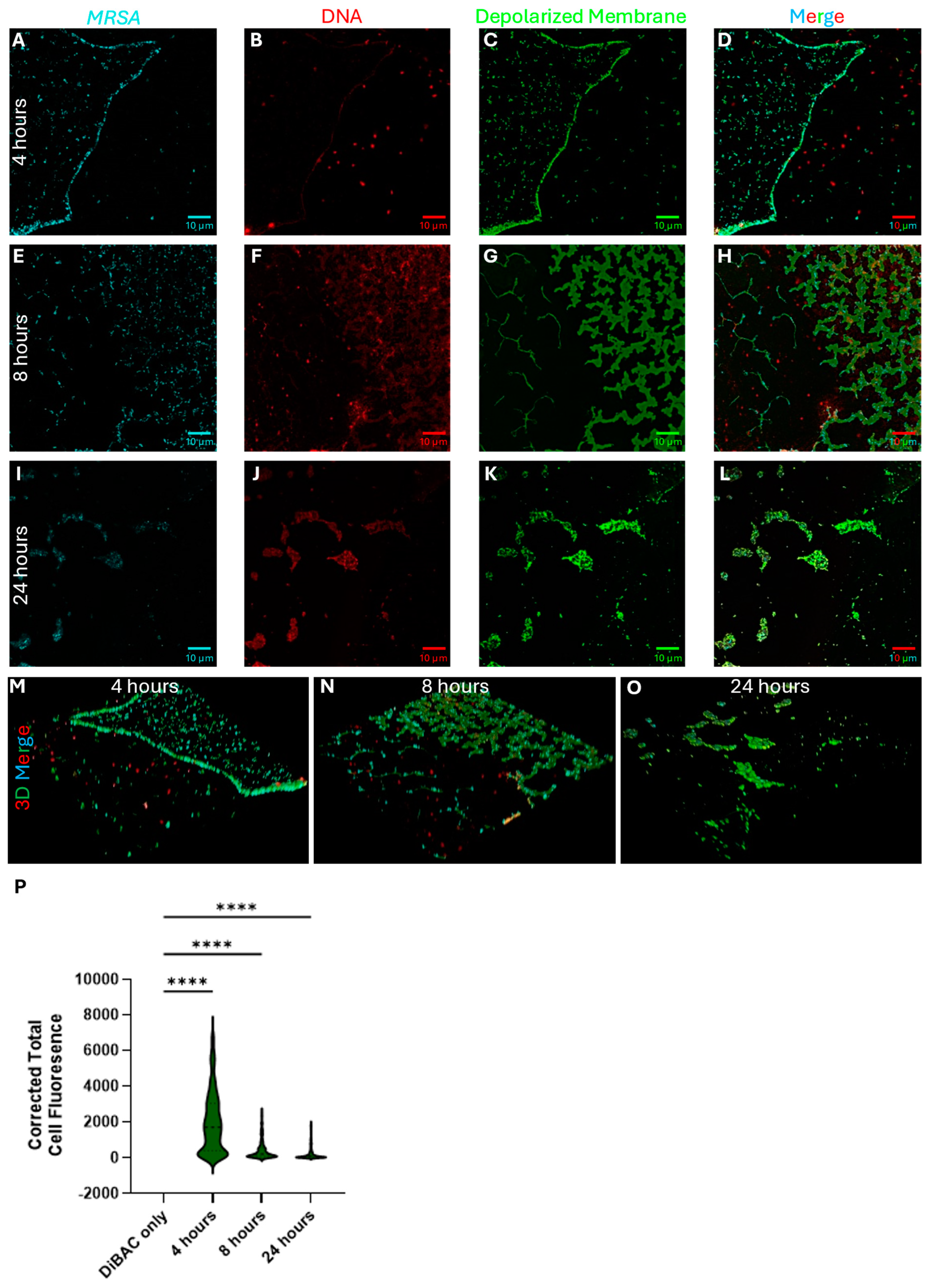
2.5.1. P. aeruginosa Kills S. aureus During Polymicrobial Growth Through Membrane Depolarization
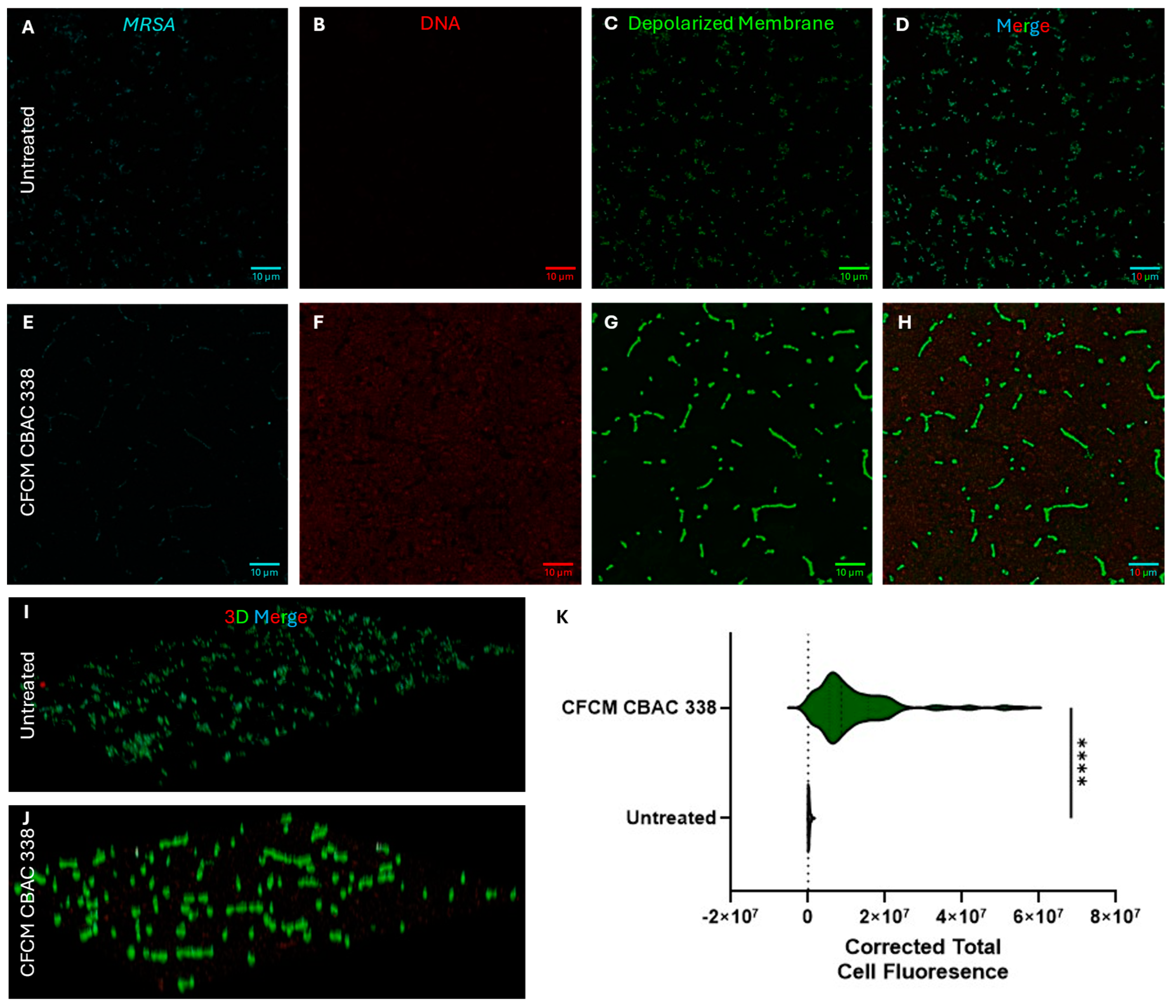
2.5.2. P. aeruginosa CFCM Depolarize S. aureus Membranes, Leading to Cell Death
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Polymicrobial Biofilm Assay
4.3. Preparation of Cell-Free Conditioned Media (CFCM)
4.4. Determination of CFCM Activity on S. aureus Biofilm Formation
4.5. Determination of CFCM Activity on S. aureus Planktonic Growth
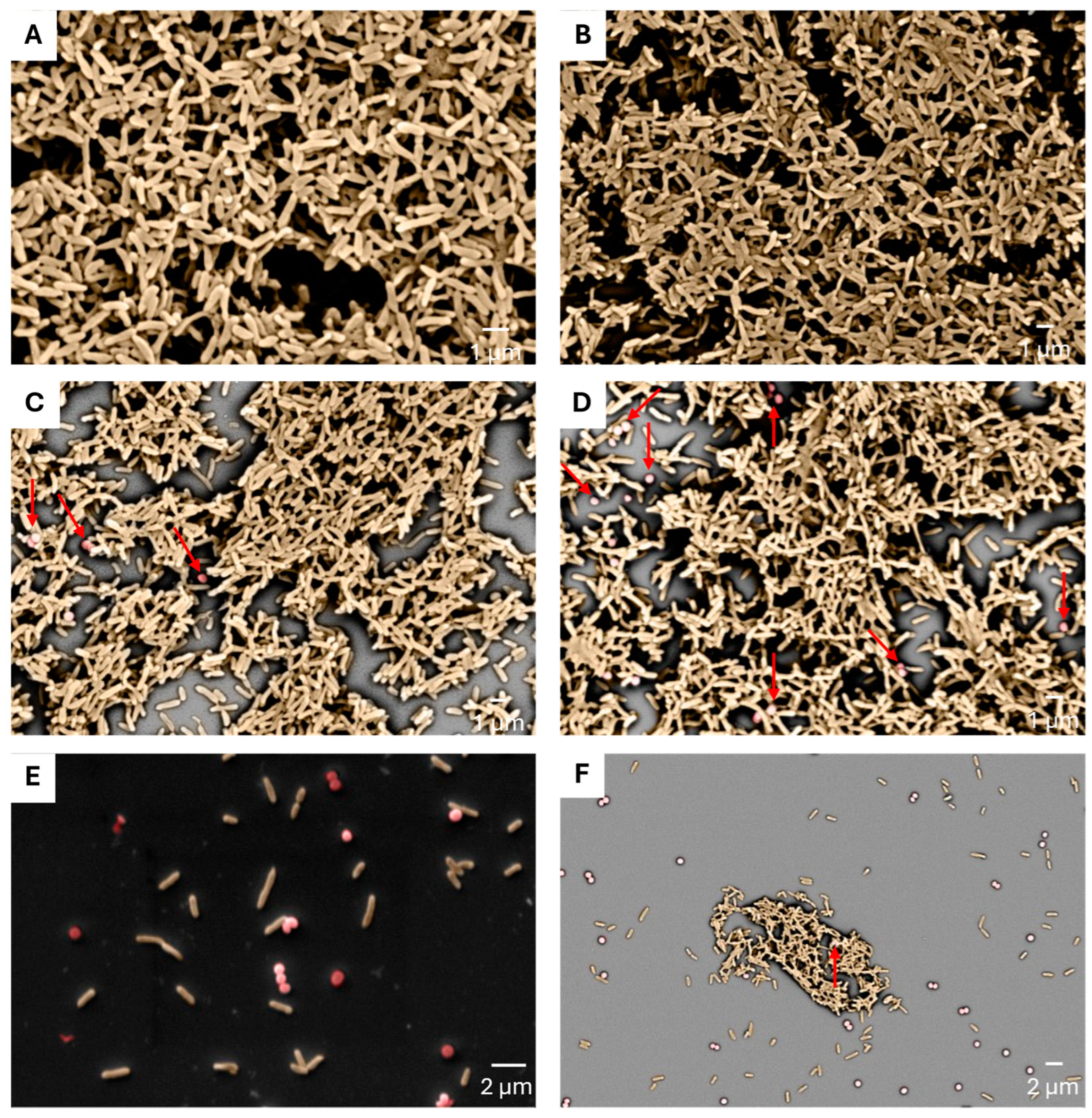
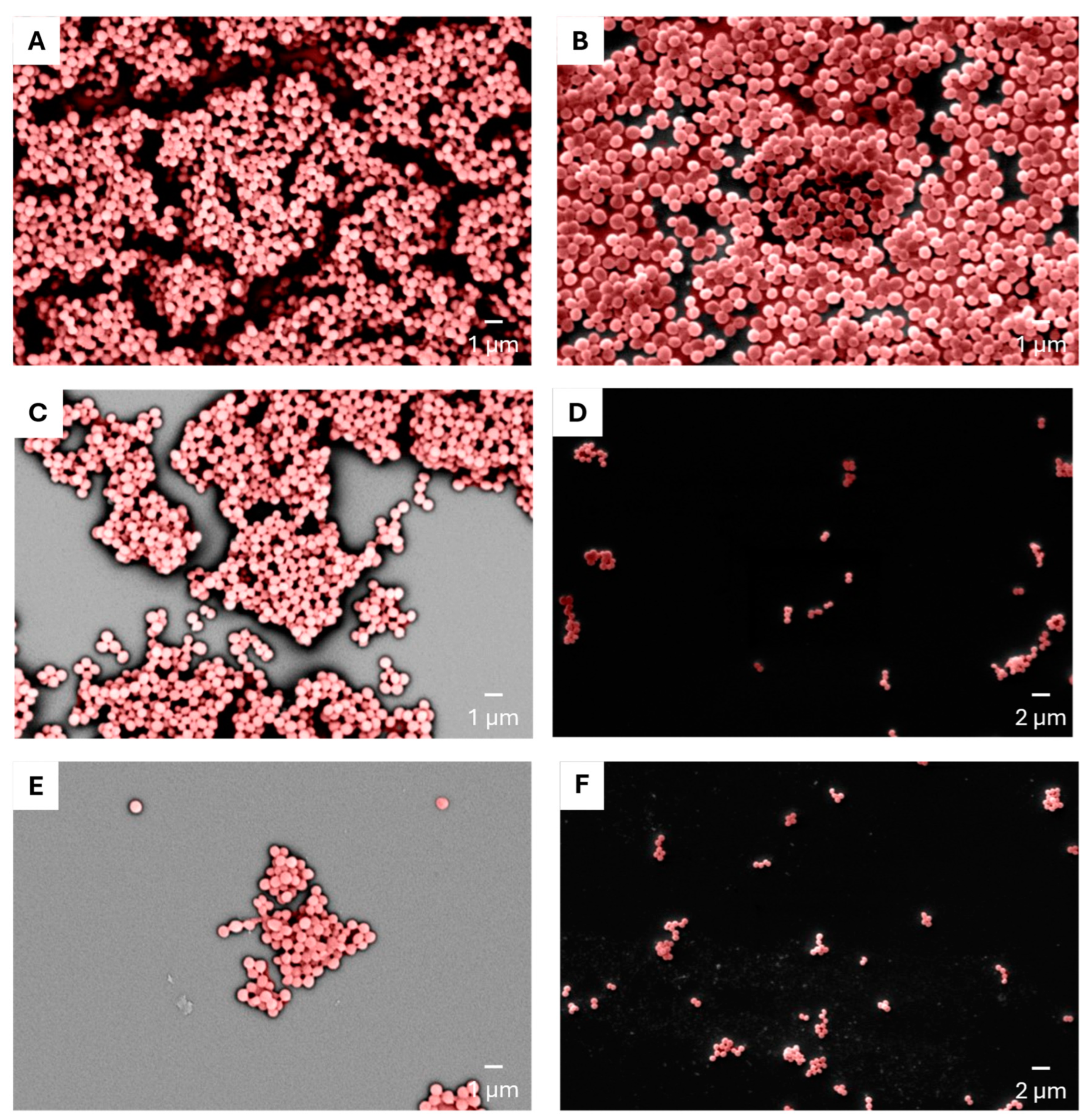
4.6. Determination of CFCM Activity on S. aureus Biofilm by Scanning Electron Microscopy (SEM)
4.7. Determination of Total Colony-Forming Units Obtained from Biofilms
4.8. Gene Expression Analysis
4.9. Exoprotein Analysis
4.10. Laser Scanning Confocal Microscopy
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ESKAPE | Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter species |
| MDR | Multi-drug resistance |
| HQNO | 2-n-heptyl-4-hydroxyquinoline-N-oxide |
| QS | Quorum sensing |
| SEM | Scanning electron microscopy |
| CFU | Colony forming units |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| MSSA | Methicillin-susceptible Staphylococcus aureus |
| CFCM | Cell-free conditioned media |
| eDNA | Extracellular DNA |
| PI | Propidium iodide |
References
- Foster, T. Staphylococcus. In Medical Microbiology; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston TX, USA, 1996. [Google Scholar]
- Laborda, P.; Sanz-García, F.; Hernando-Amado, S.; Martínez, J.L. Pseudomonas aeruginosa: An Antibiotic Resilient Pathogen with Environmental Origin. Curr. Opin. Microbiol. 2021, 64, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef] [PubMed]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Ma, D.; Mandell, J.B.; Donegan, N.P.; Cheung, A.L.; Ma, W.; Rothenberger, S.; Shanks, R.M.Q.; Richardson, A.R.; Urish, K.L. The Toxin-Antitoxin MazEF Drives Staphylococcus aureus Biofilm Formation, Antibiotic Tolerance, and Chronic Infection. mBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Biswas, L.; Götz, F. Molecular Mechanisms of Staphylococcus and Pseudomonas Interactions in Cystic Fibrosis. Front. Cell Infect. Microbiol. 2022, 11, 824042. [Google Scholar] [CrossRef]
- Camus, L.; Briaud, P.; Vandenesch, F.; Doléans-Jordheim, A.; Moreau, K. Mixed Populations and Co-Infection: Pseudomonas aeruginosa and Staphylococcus aureus. Adv. Exp. Med. Biol. 2022, 1386, 397–424. [Google Scholar] [CrossRef]
- Costerton, J.W.; Marrie, T.J.; Cheng, K.-J. Phenomena of Bacterial Adhesion. In Bacterial Adhesion; Springer: New York, NY, USA, 1985; pp. 3–43. [Google Scholar] [CrossRef]
- Su, Y.; Ding, T. Targeting Microbial Quorum Sensing: The next Frontier to Hinder Bacterial Driven Gastrointestinal Infections. Gut Microbes 2023, 15, 2252780. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and Promise of Bacterial Quorum Sensing Research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef]
- Murray, J.L.; Connell, J.L.; Stacy, A.; Turner, K.H.; Whiteley, M. Mechanisms of Synergy in Polymicrobial Infections. J. Microbiol. 2014, 52, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Gambino, E.; Maione, A.; Guida, M.; Albarano, L.; Carraturo, F.; Galdiero, E.; Di Onofrio, V. Evaluation of the Pathogenic-Mixed Biofilm Formation of Pseudomonas aeruginosa/Staphylococcus aureus and Treatment with Limonene on Three Different Materials by a Dynamic Model. Int. J. Environ. Res. Public Health 2022, 19, 3741. [Google Scholar] [CrossRef]
- Yung, D.B.Y.; Sircombe, K.J.; Pletzer, D. Friends or Enemies? The Complicated Relationship between Pseudomonas aeruginosa and Staphylococcus aureus. Mol. Microbiol. 2021, 116, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jenul, C.; Keim, K.C.; Jens, J.N.; Zeiler, M.J.; Schilcher, K.; Schurr, M.J.; Melander, C.; Phelan, V.V.; Horswill, A.R. Pyochelin Biotransformation by Staphylococcus aureus Shapes Bacterial Competition with Pseudomonas aeruginosa in Polymicrobial Infections. Cell Rep. 2023, 42, 112540. [Google Scholar] [CrossRef]
- Zhao, A.; Sun, J.; Liu, Y. Understanding Bacterial Biofilms: From Definition to Treatment Strategies. Front. Cell Infect. Microbiol. 2023, 13, 1137947. [Google Scholar] [CrossRef] [PubMed]
- Ghequire, M.G.K.; De Mot, R. Ribosomally Encoded Antibacterial Proteins and Peptides from Pseudomonas. FEMS Microbiol. Rev. 2014, 38, 523–568. [Google Scholar] [CrossRef]
- Ghssein, G.; Ezzeddine, Z. A Review of Pseudomonas aeruginosa Metallophores: Pyoverdine, Pyochelin and Pseudopaline. Biology 2022, 11, 1711. [Google Scholar] [CrossRef]
- Jeong, G.-J.; Khan, F.; Tabassum, N.; Jo, D.-M.; Jung, W.-K.; Kim, Y.-M. Roles of Pseudomonas aeruginosa Siderophores in Interaction with Prokaryotic and Eukaryotic Organisms. Res. Microbiol. 2024, 175, 104211. [Google Scholar] [CrossRef]
- Purschke, F.G.; Hiller, E.; Trick, I.; Rupp, S. Flexible Survival Strategies of Pseudomonas Aeruginosa in Biofilms Result in Increased Fitness Compared with Candida Albicans. Mol. Cell. Proteom. 2012, 11, 1652–1669. [Google Scholar] [CrossRef]
- Thierbach, S.; Wienhold, M.; Fetzner, S.; Hennecke, U. Synthesis and Biological Activity of Methylated Derivatives of the Pseudomonas Metabolites HHQ, HQNO and PQS. Beilstein J. Org. Chem. 2019, 15, 187. [Google Scholar] [CrossRef]
- Hazan, R.; Que, Y.A.; Maura, D.; Strobel, B.; Majcherczyk, P.A.; Hopper, L.R.; Wilbur, D.J.; Hreha, T.N.; Barquera, B.; Rahme, L.G. Auto Poisoning of the Respiratory Chain by a Quorum Sensing Regulated Molecule Favors Biofilm Formation and Antibiotic Tolerance. Curr. Biol. 2016, 26, 195. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Rao, S.; Bansal, A.; Dang, S.; Gupta, S.; Gabrani, R. Pseudomonas aeruginosa Biofilm: Potential Therapeutic Targets. Biologicals 2014, 42, 1–7. [Google Scholar] [CrossRef]
- Idrees, M.; Sawant, S.; Karodia, N.; Rahman, A. Staphylococcus aureus Biofilm: Morphology, Genetics, Pathogenesis and Treatment Strategies. Int. J. Environ. Res. Public Health 2021, 18, 7602. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, P.S.; Veena, K.; Senthil, R.; Iswamy, K.; Ponmalar, E.M.; Mariappan, V.; Girija, A.S.S.; Vadivelu, J.; Nagarajan, S.; Challabathula, D.; et al. Biofilm-Associated Agr and Sar Quorum Sensing Systems of Staphylococcus aureus Are Inhibited by 3-Hydroxybenzoic Acid Derived from Illicium verum. ACS Omega 2022, 7, 14653–14665. [Google Scholar] [CrossRef] [PubMed]
- Yarwood, J.M.; Schlievert, P.M. Quorum Sensing in Staphylococcus Infections. J. Clin. Investig. 2003, 112, 1620–1625. [Google Scholar] [CrossRef]
- Gao, P.; Wei, Y.; Hou, S.; Lai, P.-M.; Liu, H.; Tai, S.S.C.; Tang, V.Y.M.; Prakash, P.H.; Sze, K.-H.; Chen, J.H.K.; et al. SaeR as a Novel Target for Antivirulence Therapy against Staphylococcus aureus. Emerg. Microbes Infect. 2023, 12, 2254415. [Google Scholar] [CrossRef]
- Moormeier, D.E.; Bose, J.L.; Horswill, A.R.; Bayles, K.W. Temporal and Stochastic Control of Staphylococcus aureus Biofilm Development. mBio 2014, 5, e01341-14. [Google Scholar] [CrossRef]
- Wang, G. Human Antimicrobial Peptides and Proteins. Pharmaceuticals 2014, 7, 545–594. [Google Scholar] [CrossRef]
- Mountcastle, S.E.; Vyas, N.; Villapun, V.M.; Cox, S.C.; Jabbari, S.; Sammons, R.L.; Shelton, R.M.; Walmsley, A.D.; Kuehne, S.A. Biofilm Viability Checker: An Open-Source Tool for Automated Biofilm Viability Analysis from Confocal Microscopy Images. NPJ Biofilms Microbiomes 2021, 7, 44. [Google Scholar] [CrossRef]
- O’Toole, R.F. The Interface between COVID-19 and Bacterial Healthcare-Associated Infections. Clin. Microbiol. Infect. 2021, 27, 1772–1776. [Google Scholar] [CrossRef]
- Folic, M.M.; Djordjevic, Z.; Folic, N.; Radojevic, M.Z.; Jankovic, S.M. Epidemiology and Risk Factors for Healthcare-Associated Infections Caused by Pseudomonas aeruginosa. J. Chemother. 2021, 33, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Biswas, R.; Jangra, B.; Ashok, G.; Ravichandiran, V.; Mohan, U. Current Strategies for Combating Biofilm-Forming Pathogens in Clinical Healthcare-Associated Infections. Indian J. Microbiol. 2024, 64, 781–796. [Google Scholar] [CrossRef]
- Orazi, G.; O’Toole, G.A. Pseudomonas aeruginosa Alters Staphylococcus aureus Sensitivity to Vancomycin in a Biofilm Model of Cystic Fibrosis Infection. mBio 2017, 8, e00873-17. [Google Scholar] [CrossRef]
- Hotterbeekx, A.; Kumar-Singh, S.; Goossens, H.; Malhotra-Kumar, S. In Vivo and In Vitro Interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front. Cell Infect. Microbiol. 2017, 7, 106. [Google Scholar] [CrossRef]
- Kessler, E.; Safrin, M.; Olson, J.C.; Ohman, D.E. Secreted LasA of Pseudomonas aeruginosa Is a Staphylolytic Protease. J. Biol. Chem. 1993, 268, 7503–7508. [Google Scholar] [CrossRef] [PubMed]
- Herzberg, C.; van Hasselt, J.G.C. Pharmacodynamics of Interspecies Interactions in Polymicrobial Infections. NPJ Biofilms Microbiomes 2025, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Koumans, C.I.M.; Tandar, S.T.; Liakopoulos, A.; van Hasselt, J.G.C. Interspecies Interactions Alter the Antibiotic Sensitivity of Pseudomonas aeruginosa. Microbiol. Spectr. 2024, 12, e0201224. [Google Scholar] [CrossRef]
- Glatthardt, T.; Curityba de Mello Campos, J.; Cardoso Chamon, R.; Freitas de Sá Coimbra, T.; de Almeida Rocha, G.; Alves Figueira de Melo, M.; Estevam Parente, T.; Araujo Lobo, L.; Caetano Martha Antunes, L.; Regina Netto dos Santos, K.; et al. Small Molecules Produced by Commensal Staphylococcus Epidermidis Disrupt Formation of Biofilms by Staphylococcus aureus. Appl. Environ. Microbiol. 2020, 86, e02539-19. [Google Scholar] [CrossRef]
- Lima, R.D.; dos Reis, G.A.; da Silva Reviello, J.; Glatthardt, T.; da Silva Coimbra, L.; Lima, C.O.G.X.; Antunes, L.C.M.; Ferreira, R.B.R. Antibiofilm Activity of Cutibacterium Acnes Cell-Free Conditioned Media against Staphylococcus spp. Braz. J. Microbiol. 2021, 52, 2373–2383. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scaffo, J.; Lima, R.D.; Dobrotka, C.; Ribeiro, T.A.N.; Pereira, R.F.A.; Sachs, D.; Ferreira, R.B.R.; Aguiar-Alves, F. In Vitro Analysis of Interactions Between Staphylococcus aureus and Pseudomonas aeruginosa During Biofilm Formation. Antibiotics 2025, 14, 504. https://doi.org/10.3390/antibiotics14050504
Scaffo J, Lima RD, Dobrotka C, Ribeiro TAN, Pereira RFA, Sachs D, Ferreira RBR, Aguiar-Alves F. In Vitro Analysis of Interactions Between Staphylococcus aureus and Pseudomonas aeruginosa During Biofilm Formation. Antibiotics. 2025; 14(5):504. https://doi.org/10.3390/antibiotics14050504
Chicago/Turabian StyleScaffo, Julia, Rayssa Durães Lima, Cameron Dobrotka, Tainara A. N. Ribeiro, Renata F. A. Pereira, Daniela Sachs, Rosana B. R. Ferreira, and Fabio Aguiar-Alves. 2025. "In Vitro Analysis of Interactions Between Staphylococcus aureus and Pseudomonas aeruginosa During Biofilm Formation" Antibiotics 14, no. 5: 504. https://doi.org/10.3390/antibiotics14050504
APA StyleScaffo, J., Lima, R. D., Dobrotka, C., Ribeiro, T. A. N., Pereira, R. F. A., Sachs, D., Ferreira, R. B. R., & Aguiar-Alves, F. (2025). In Vitro Analysis of Interactions Between Staphylococcus aureus and Pseudomonas aeruginosa During Biofilm Formation. Antibiotics, 14(5), 504. https://doi.org/10.3390/antibiotics14050504







