Bacterivorous Ciliate Tetrahymena pyriformis Facilitates vanA Antibiotic Resistance Gene Transfer in Enterococcus faecalis
Abstract
1. Introduction
2. Results
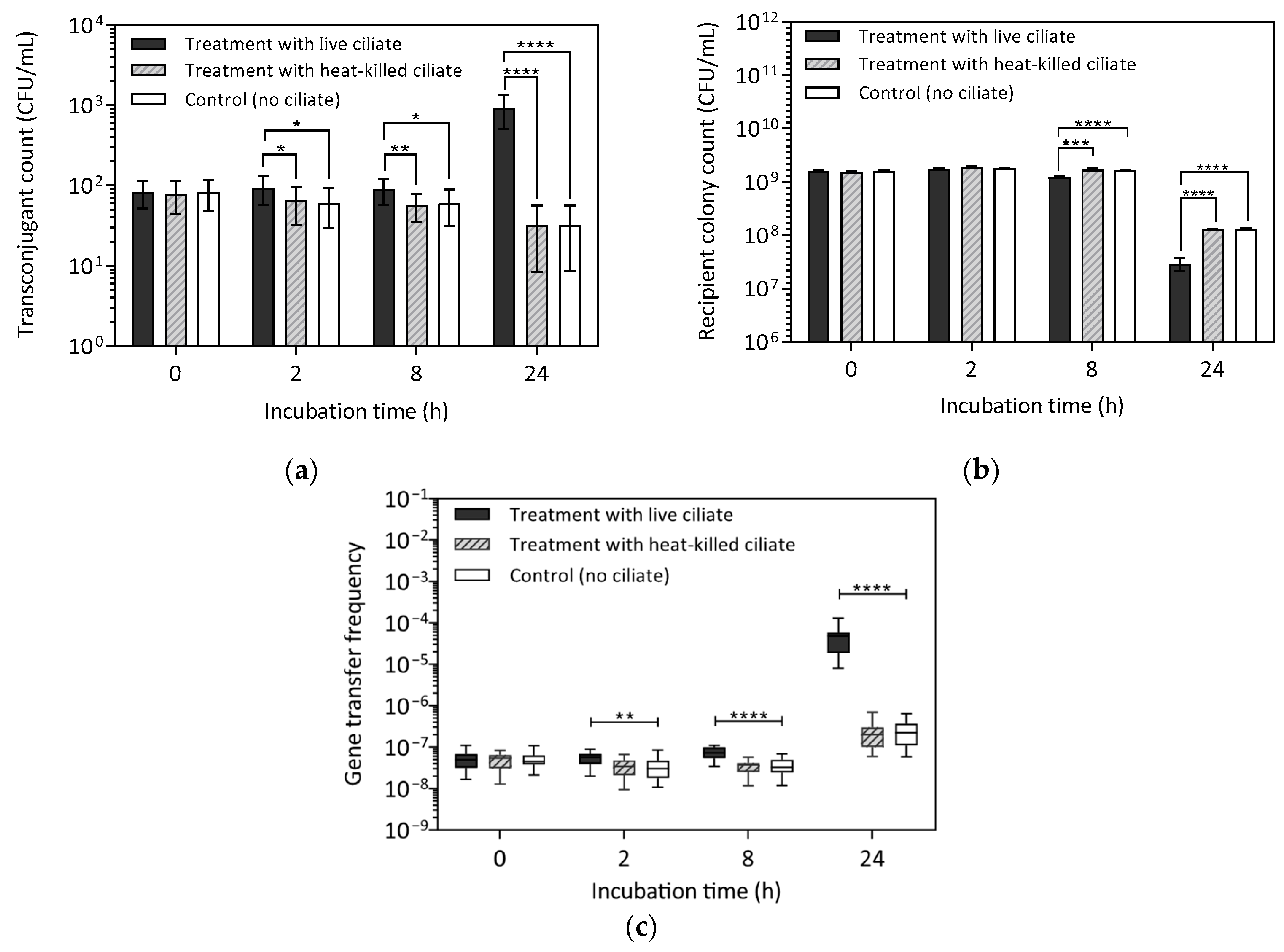
2.1. Effect of Ciliate Viability on Gene Transfer
2.2. Effect of Initial Bacterial Growth Phase on Gene Transfer
2.3. Effect of Phenotypic Differences in Recipients on Gene Transfer
2.4. Effect of Energy Source Availability on Gene Transfer
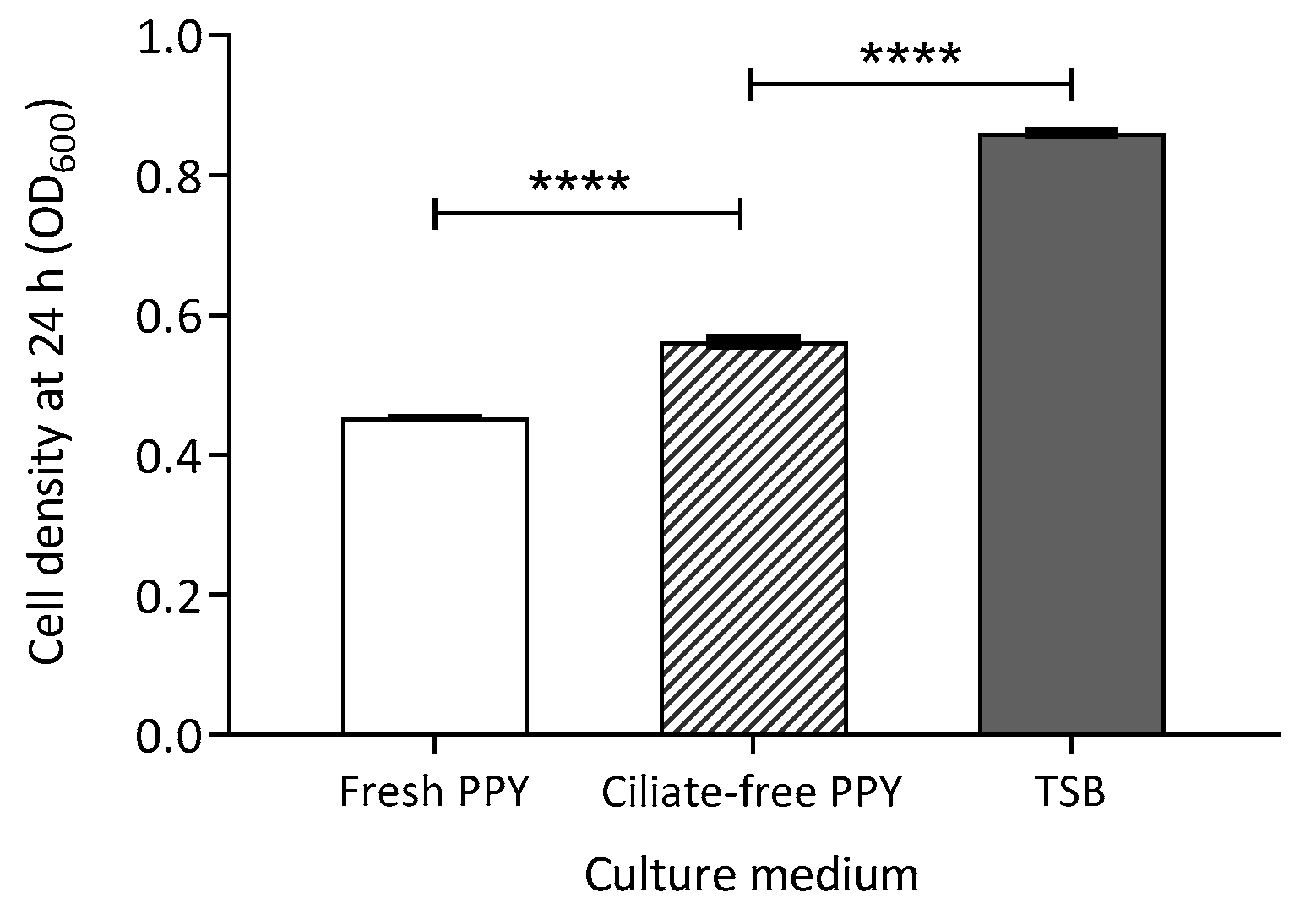
2.4.1. Exposure of Bacteria to Spent Ciliate Culture Medium
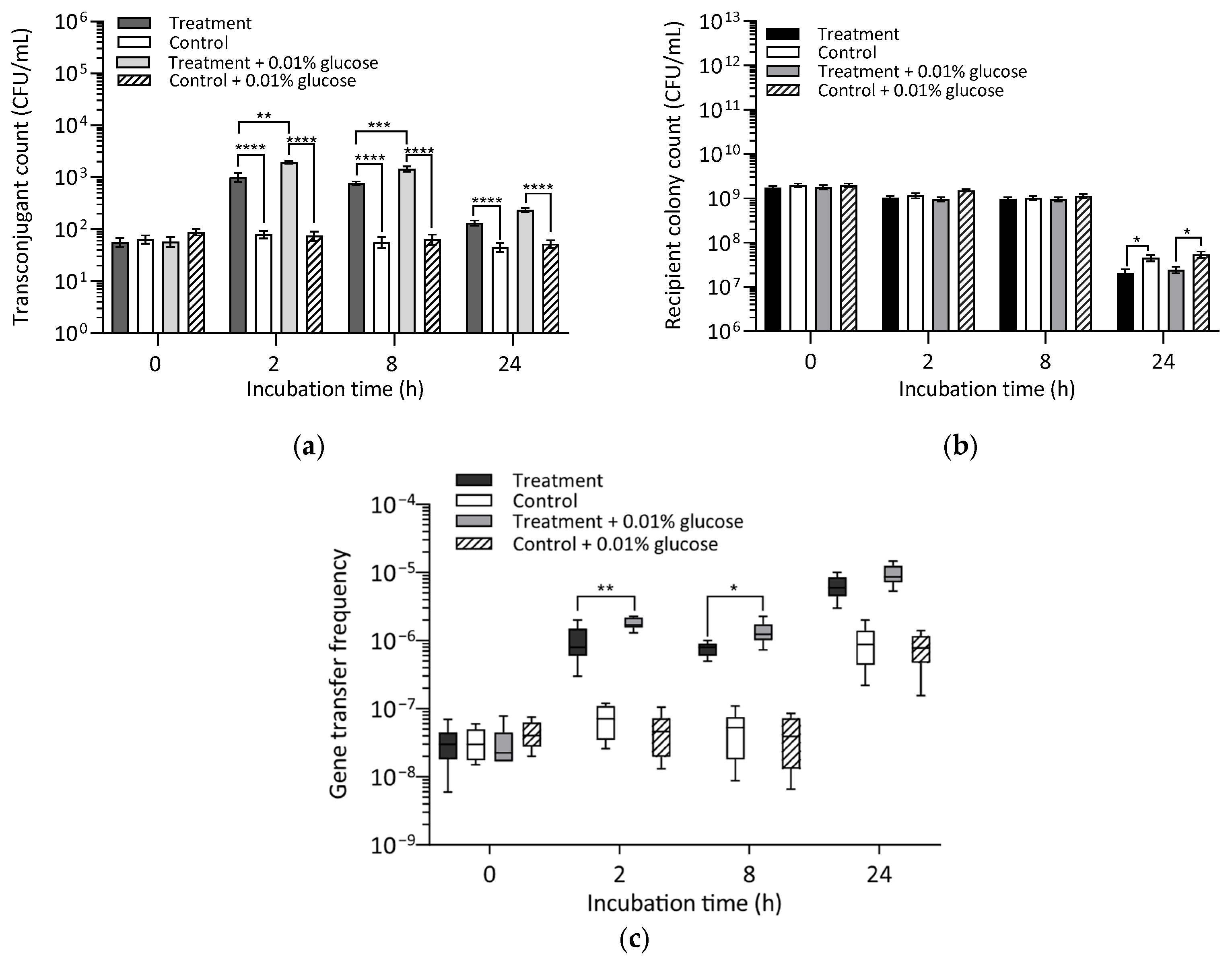
2.4.2. Exposure of Bacteria to Glucose as an Energy Source
3. Discussion
3.1. Effect of Ciliate Viability on Transconjugant Formation
3.2. Effect of Bacterial Growth Phase on Transconjugant Formation
3.3. Effect of Distinct Recipient Phenotypes on Gene Transfer Frequency
3.4. Effect of Energy Source Availability on Transconjugant Formation
3.5. Limitations of Conjugation Studies
3.5.1. Ciliate Grazing and Transconjugant Formation
3.5.2. Use of Selective Media in Transconjugant Detection
3.5.3. Ciliate Waste Products as Potential Energy Sources
3.5.4. Bacterial Growth Phase and Gene Transfer Frequency
3.6. Implications for Wastewater Treatment Plants
4. Materials and Methods
4.1. Protozoa
4.2. Bacteria
Preparation of Fluorescence-Stained Enterococcus faecalis
4.3. Reagents
4.4. Conjugation Studies
4.4.1. Conjugation Assay—Effect of Ciliate Viability
Emergence of Bacterial Cells Within Ciliates
4.4.2. Conjugation Assay—Effect of Phenotypically Distinct Recipients
4.4.3. Conjugation Assay—Effect of Starting Bacterial Growth Phase
4.4.4. Conjugation Assay—Effect of Energy Source Availability
Exposure of Bacteria to Spent Ciliate Culture Medium
Exposure of Bacteria to Glucose as an Energy Source
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| ANOVA | Analysis of variance |
| ARB | Antibiotic-resistant bacteria |
| ARGs | Antibiotic resistance genes |
| BOD | Biological oxygen demand |
| CFU | Colony forming unit |
| DNA | Deoxyribonucleic acid |
| MDR | Multidrug resistant |
| MGEs | Mobile genetic elements |
| PPY | Proteose peptone yeast |
| TC | Transconjugant count |
| TSA | Tryptone soya agar |
| TSB | Tryptone soya broth |
| VBNC | Viable but non-culturable |
| VRE | Vancomycin resistant enterococci |
| WWTPs | Wastewater treatment plants |
References
- Larsson, D.G.J. Pollution from drug manufacturing: Review and perspectives. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130571. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sanchez-Melsio, A.; Borrego, C.M.; Barcelo, D.; Luis Balcazar, J. Occurrence of antibiotics and antibiotic resistance genes in hospital and urban wastewaters and their impact on the receiving river. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dechesne, A.; He, Z.; Madsen, J.S.; Nesme, J.; Soren, J.; Smets, B.F. Estimating the transfer range of plasmids encoding antimicrobial resistance in a wastewater treatment plant microbial community. Environ. Sci. Technol. Lett. 2018, 5, 260–265. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2021, 20, 257–269. [Google Scholar] [CrossRef]
- Wang, Z.; Cai, M.; Du, P.; Li, X. Wastewater surveillance for antibiotics and resistance genes in a river catchment: Spatiotemporal variations and the main drivers. Water Res. 2024, 251, 121090. [Google Scholar] [CrossRef]
- La Rosa, M.C.; Maugeri, A.; Favara, G.; La Mastra, C.; Magnano San Lio, R.; Barchitta, M.; Agodi, A. The Impact of Wastewater on Antimicrobial Resistance: A Scoping Review of Transmission Pathways and Contributing Factors. Antibiotics 2025, 14, 131. [Google Scholar] [CrossRef]
- Mareković, I.; Markanović, M.; Lešin, J.; Ćorić, M. Vancomycin-Resistant Enterococci: Current Understandings of Resistance in Relation to Transmission and Preventive Strategies. Pathogens 2024, 13, 966. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, X.; Ni, N.; Shi, M.; Wang, N. Fate and Proliferation of Vancomycin Resistance Genes in Two Typical Pharmaceutical Wastewater Treatment Plants. Water 2024, 16, 114. [Google Scholar] [CrossRef]
- Suzuki, S.; Pruden, A.; Virta, M.; Zhang, T. Editorial: Antibiotic resistance in aquatic systems, volume II. Front. Microbiol. 2023, 14, 1298681. [Google Scholar] [CrossRef]
- Pärnänen, K.M.M.; Narciso-da-Rocha, C.; Kneis, D.; Berendonk, T.U.; Cacace, D.; Do, T.T.; Elpers, C.; Fatta-Kassinos, D.; Henriques, I.; Jaeger, T.; et al. Antibiotic resistance in European wastewater treatment plants mirrors the prevalence of clinical antibiotic resistance. Sci. Adv. 2019, 5, eaau9124. [Google Scholar] [CrossRef]
- Lopatkin, A.J.; Huang, S.; Smith, R.P.; Srimani, J.K.; Sysoeva, T.A.; Bewick, S.; Karig, D.K.; You, L. Antibiotics as a selective driver for conjugation dynamics. Nat. Microbiol. 2016, 1, 16044. [Google Scholar] [CrossRef] [PubMed]
- Pauli, W.; Jax, K.; Berger, S. Protozoa in wastewater treatment: Function and importance. In Biodegradation and Persistence; Springer: Berlin, Germany, 2001; pp. 203–252. [Google Scholar] [CrossRef]
- Tezuka, Y. Bacterial regeneration of ammonium and phosphorus ratio of organic substrates. Microb. Ecol. 1990, 29, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Madoni, P. Protozoa in wastewater treatment processes: A minireview. Ital. J. Zool. 2011, 78, 3–11. [Google Scholar] [CrossRef]
- Madoni, P. Protozoa as indicators of wastewater treatment frequency. In The Handbook of Water and Wastewater Microbiology; Mara, D., Horan, N., Eds.; Academic Press: London, UK, 2003; pp. 361–371. [Google Scholar] [CrossRef]
- Marti, E.; Variatza, E.; Balcazar, J.L. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014, 22, 36–41. [Google Scholar] [CrossRef]
- Lupo, A.; Coyne, S.; Berendonk, T. Origin and evolution of antibiotic resistance: The common mechanisms of emergence and spread in water bodies. Front. Microbiol. 2012, 3, 115–128. [Google Scholar] [CrossRef]
- Olanrewaju, T.O.; McCarron, M.; Dooley, J.S.G.; Arnscheidt, J. Transfer of antibiotic resistance genes between Enterococcus faecalis strains in filter feeding zooplankton Daphnia magna and Daphnia pulex. Sci. Total Environ. 2019, 659, 1168–1175. [Google Scholar] [CrossRef]
- Suzuki, S.; Sano, D. Effect of protists on horizontal transfer of antimicrobial resistance genes in water environment. J. Water Environ. Technol 2023, 21, 97–107. [Google Scholar] [CrossRef]
- Schlimme, W.; Marchiani, M.; Hanselmann, K.; Jenni, B. Gene transfer between bacteria within digestive vacuoles of protozoa. FEMS Microbiol. Ecol. 1997, 23, 239–247. [Google Scholar] [CrossRef][Green Version]
- Matsuo, J.; Oguri, S.; Nakamura, S.; Hanawa, T.; Fukumoto, T.; Hayashi, Y.; Kawaguchi, K.; Mizutani, Y.; Yao, T.; Akizawa, K.; et al. Ciliates rapidly enhance conjugation frequency between Escherichia coli strains through bacterial accumulation in vesicles. Res. Microbiol. 2010, 161, 711–719. [Google Scholar] [CrossRef]
- Oguri, S.; Matsuo, J.; Hayashi, Y.; Nakamura, S.; Hanawa, T.; Fukumotot, T.; Mizutaini, Y.; Yao, T.; Akizawa, K.; Suzuki, H.; et al. Ciliates promote the transfer of the gene encoding the extended-spectrum β-lactamase CTX-M-27 between Escherichia coli strains. J. Antimicrob. Chemother. 2011, 66, 527–530. [Google Scholar] [CrossRef]
- Balcázar, J.L. Effect of ciliates in transfer of plasmid-mediated quinolone-resistance genes in bacteria. Emerg. Infect. Dis. 2015, 21, 547–548. [Google Scholar] [CrossRef] [PubMed]
- Iriberri, J.; Azúa, I.; Labirua-Iturburu, A.; Artolozaga, I.; Barcina, I. Differential elimination of enteric bacteria by protist in a freshwater system. J. Appl. Bacteriol. 1994, 77, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Baquero, F.; Tedim, A.P.; Coque, T.M. Antibiotic resistance shaping multi-level population biology of bacteria. Front. Microbiol. 2013, 4, 15–23. [Google Scholar] [CrossRef]
- Lindberg, R.; Jarnheimer, P.; Olsen, B.; Johansson, M.; Tysklind, M. Determination of antibiotic substances in hospital sewage water using solid phase extraction and liquid chromatography/mass spectrometry and group analogue internal standards. Chemosphere 2004, 57, 1479–1488. [Google Scholar] [CrossRef]
- Bouki, C.; Venieri, D.; Diamadopoulos, E. Detection and fate of antibiotic resistant bacteria in wastewater treatment plants: A review. Ecotoxicol. Environ. Saf. 2013, 91, 1–9. [Google Scholar] [CrossRef]
- Curds, C.R.; Fey, G.J. The effect of ciliated protozoa on fate of Escherichia coli in the activated sludge process. Water Res. 1969, 3, 853–867. [Google Scholar] [CrossRef]
- Marcinek, H.; Wirth, R.; Muscholl-Silborhorn, A.; Gauer, M. Enterococcus faecalis gene transfer under natural conditions in municipal sewage water treatment plants. Appl. Environ. Microbiol. 1998, 64, 626–632. [Google Scholar] [CrossRef]
- Nilsson, J.R. Structural aspects of digestion of Escherichia coli in Tetrahymena. J. Protozool. 1987, 34, 1–6. [Google Scholar] [CrossRef]
- King, C.H.; Shotts, E.B., Jr.; Wooley, R.E.; Porter, K.G. Survival of coliforms and bacterial pathogens with protozoa during chlorination. Appl. Environ. Microbiol. 1988, 54, 3023–3033. [Google Scholar] [CrossRef]
- Muela, A.; Pocino, M.; Arana, I.; Justo, J.I.; Iriberri, J.; Barcina, I. Effect of growth phase and parental cell survival in river water on plasmid transfer between Escherichia coli strains. Appl. Environ. Microbiol. 1994, 60, 4273–4278. [Google Scholar] [CrossRef]
- Gonzalez, J.M.; Iriberri, J.; Egea, L.; Barcina, B. Differential rates of digestion of bacteria by freshwater and marine phagotrophic protozoa. Appl. Environ. Microbiol. 1990, 56, 1851–1857. [Google Scholar] [CrossRef] [PubMed]
- Hartke, A.; Lemarinier, S.; Pichereau, V.; Auffray, Y. Survival of Enterococcus faecalis in seawater microcosms is limited in the presence of bacterivorous zooflagellates. Curr. Microbiol. 2002, 44, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Hartke, A.; Giard, J.C.; Laplace, J.M.; Auffray, Y. Survival of Enterococcus faecalis in an oligotrophic microcosm: Changes in morphology, development of general stress resistance, and analysis of protein synthesis. Appl. Environ. Microbiol. 1998, 64, 4238–4245. [Google Scholar] [CrossRef] [PubMed]
- Himeoka, Y.; Kaneko, K. Theory for transitions between exponential and stationary phases: Universal laws for lag time. Phys. Rev. X 2017, 7, 021049. [Google Scholar] [CrossRef]
- Tomita, H.; Yasuyoshi, I. Genetic analysis of the Enterococcus vancomycin resistance conjugative plasmid pHTβ: Identification of the region involved in cell aggregation and traB, a key regulator gene for plasmid transfer and cell aggregation. J. Bacteriol. 2008, 190, 7739–7753. [Google Scholar] [CrossRef]
- Iriberri, J.; Ayo, B.; Artolozaga, I.; Barcina, I.; Egea, L. Grazing on allochthonous vs autochthonous bacteria in river water. Lett. Appl. Microbiol. 1994, 18, 12–14. [Google Scholar] [CrossRef]
- Davies, C.M.; Long, J.A.H.; Donald, M.; Ashbolt, N. Survival of fecal microorganisms in marine and freshwater sediments. Appl. Environ. Microbiol. 1995, 61, 1888–1896. [Google Scholar] [CrossRef]
- Sherr, B.F.; Sherr, E.B.; Rassoulazdegan, F. Rates of digestion of bacteria by marine phagotrophic protozoa: Temperature dependence. Appl. Environ. Microbiol. 1998, 54, 1091–1095. [Google Scholar] [CrossRef]
- Nisbet, B. Nutrition and Feeding Strategies in Protozoa; Croom Helm: London, UK, 1984. [Google Scholar]
- Horan, N.J. Biological Wastewater Treatment Systems. Theory and Operation; John Wiley and Sons: Chichester, UK, 1990. [Google Scholar]
- Jürgens, K.; Matz, C. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leuwenhoek 2002, 81, 413–434. [Google Scholar] [CrossRef]
- Pogue, A.J.; Gilbride, K.A. Impact of protozoan grazing on nitrification and the ammonia- and nitrite-oxidizing bacterial communities in activated sludge. Can. J. Microbiol. 2007, 53, 559–571. [Google Scholar] [CrossRef]
- Arregui, L.; Serrano, S.; Maria, L.; Perez-Uz, B.; Guinea, A. Ciliate contributions to bioaggregation; laboratory assays with axenic cultures of Tetrahymena thermophila. Int. Microbiol. 2007, 10, 91–96. [Google Scholar] [PubMed]
- Ding, Z.; Bourven, I.; Guibaud, G.; van Hullebusch, E.D.; Panico, A.; Pirozzi, F.; Esposito, G. Role of extracellular polymeric substances (EPS) production in bioaggregation: Application to wastewater treatment. Appl. Microbiol. Biotechnol. 2015, 99, 9883–9905. [Google Scholar] [CrossRef] [PubMed]
- Ratsak, C.H.; Maarsen, K.A.; Kooijman, S.A.L. Effects of protozoa on carbon mineralization in activated sludge. Water Res. 1996, 30, 1–12. [Google Scholar] [CrossRef]
- Dröge, M.; Pühler, A.; Selbitschka, W. Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol. Gen. Genet. 2000, 263, 471–482. [Google Scholar] [CrossRef]
- Schlüter, A.; Szczepanowski, R.; Kurz, N.; Schneiker, S.; Krahn, I.; Puhler, A. Erythromycin resistance-conferring plasmid pRSB105, isolated from a sewage treatment plant, harbours a new macrolide resistance determinant, an integron-containing Tn402-like element, and a large region of unknown function. Appl. Environ. Microbiol. 2007, 73, 1952–1960. [Google Scholar] [CrossRef]
- Milligan, E.G.; Calarco, J.; Davis, B.C.; Keenum, I.M.; Liguori, K.; Pruden, A.; Harwood, V.J. A systematic review of culture-based methods for monitoring antibiotic-resistant Acinetobacter, Aeromonas, and Pseudomonas as environmentally relevant pathogens in wastewater and surface water. Curr. Environ. Health Rep. 2023, 10, 154–171. [Google Scholar] [CrossRef]
- Łuczkiewicz, A.; Jankowska, K.; Fudala-Ksiazek, S.; Olanczuk-Neyman, K. Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Research 2010, 44, 5089–5097. [Google Scholar] [CrossRef]
- Li, L.; Mendis, N.; Trigui, H.; Oliver, J.D.; Faucher, S.P. The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 2014, 5, 258. [Google Scholar] [CrossRef]
- Taylor, W.D.; Berger, J. Growth responses of cohabiting ciliate protozoa to various prey bacteria. Can. J. Zool. 1976, 54, 1111–1114. [Google Scholar] [CrossRef]
- Grimes, D.J.; Atwell, R.W.; Brayton, P.R.; Palmer, L.M.; Rollins, D.M.; Roszak, D.B.; Singleton, F.L.; Tamplin, M.L.; Colwell, R.R. The fate of enteric pathogenic bacteria in estuarine and marine environments. Microbiol. Sci. 1986, 3, 324–329. [Google Scholar] [CrossRef]
- Pazda, M.; Kumirska, J.; Stepnowski, P.; Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems—A review. Sci. Total Environ. 2019, 697, 134023. [Google Scholar] [CrossRef] [PubMed]
- Batra, A.R.; Cottam, D.; Lepesteur, M.; Dexter, C.; Zuccala, K.; Martino, C.; Khudur, L.; Daniel, V.; Ball, A.S.; Soni, S.K. Development of A Rapid, Low-Cost Portable Detection Assay for Enterococci in Wastewater and Environmental Waters. Microorganisms 2023, 11, 381. [Google Scholar] [CrossRef] [PubMed]
- McConn, B.R.; Kraft, A.L.; Durso, L.M.; Ibekwe, A.M.; Frye, J.G.; Wells, J.E.; Tobey, E.M.; Ritchie, S.; Williams, C.F.; Cook, K.L.; et al. An analysis of culture-based methods used for the detection and isolation of Salmonella spp., Escherichia coli, and Enterococcus spp. from surface water: A systematic review. Sci. Total Environ. 2024, 927, 172190. [Google Scholar] [CrossRef]
- Castañeda-Barba, S.; Top, E.M.; Stalder, T. Plasmids, a molecular cornerstone of antimicrobial resistance in the One Health era. Nat. Rev. Microbiol. 2024, 22, 18–32. [Google Scholar] [CrossRef]
- Conwell, M.; Daniels, V.; Naughton, P.J.; Dooley, J.S.G. Interspecies transfer of vancomycin, erythromycin, and tetracycline resistance among Enterococcus species recovered from agrarian sources. BMC Microbiol. 2017, 17, 19. [Google Scholar] [CrossRef]
- Sterling, A.J.; Snelling, W.J.; Naughton, P.J.; Ternan, N.G.; Dooley, J.S.G. Competent but complex communication: The phenomena of pheromone-responsive plasmids. PLoS Pathog. 2020, 16, e1008310. [Google Scholar] [CrossRef]
- Cairns, J.; Jalasvuori, M.; Ojala, O.; Brockhurst, M.; Hiltunen, T. Conjugation is necessary for a bacterial plasmid to survive under protozoan predation. Biol. Lett. 2016, 12, 20150953. [Google Scholar] [CrossRef]
- Bien, T.L.T.; Thao, N.V.; Kitamura, S.I.; Obayashi, Y.; Suzuki, S. Release and constancy of an antibiotic resistance gene in seawater under grazing stress by ciliates and heterotrophic nanoflagellates. Microbes Environ. 2017, 32, 174–179. [Google Scholar] [CrossRef]
- Lorenz, M.G.; Wackernagel, W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 1994, 58, 563–602. [Google Scholar] [CrossRef]
- Jacquiod, S.; Brejnrod, A.; Morberg, S.M.; Abu, A.W.; Sørensen, S.J.; Riber, L. Deciphering conjugative plasmid permissiveness in wastewater microbiomes. Mol. Ecol. 2017, 26, 3556–3571. [Google Scholar] [CrossRef]
- Bengston-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2018, 42, 68–80. [Google Scholar] [CrossRef]
- Ellabaan, M.M.H.; Munck, C.; Porse, A.; Imamovic, L.; Sommer, M.O.A. Forecasting the dissemination of antibiotic resistance genes across bacterial genomes. Nat Commun. 2021, 12, 2435. [Google Scholar] [CrossRef] [PubMed]
- Douidah, L.; De Zutter, J.; Baré, J.; Houf, K. Towards a typing strategy for Arcobacter species isolated from humans and animals and assessment of the in vitro genomic stability. Foodborne Pathog. Dis. 2014, 11, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Bollmann, A.; Seitz, W.; Schwartz, T. Microbiological characterization of aquatic microbiomes targeting taxonomical marker genes and antibiotic resistance genes of opportunistic bacteria. Sci. Total Environ. 2015, 512–513, 316–325. [Google Scholar] [CrossRef]
- Shrestha, R.G.; Tanaka, Y.; Malla, B.; Tandukar, S.; Bhandari, D.; Inoue, D.; Sei, K.; Sherchand, J.B.; Haramoto, E. Development of a quantitative PCR assay for Arcobacter sp. and its application to environmental water samples. Microbes Environ. 2018, 33, 309–316. [Google Scholar] [CrossRef]
- Collado, L.; Figueras, M.J. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 2011, 24, 174–192. [Google Scholar] [CrossRef]
- Hultman, J.; Tamminen, M.; Pärnänen, K.; Cairns, J.; Karkman, A.; Virta, M. Host range of antibiotic resistance genes in wastewater treatment plant influent and effluent. FEMS Microbiol. Ecol. 2018, 94, fiy038. [Google Scholar] [CrossRef]
- Yesilmen, S.; Vural, A.; Erka, M.E.; Yildirim, I.H. Prevalence and antimicrobial susceptibility of Arcobacter species in cow milk, water buffalo milk and fresh village cheese. Int. J. Food Microbiol. 2014, 188, 11–14. [Google Scholar] [CrossRef]
- Guo, X.; Tang, N.; Lei, H.; Fang, Q.; Liu, L.; Zhou, Q.; Song, C. Metagenomic analysis of antibiotic resistance genes in untreated wastewater from three different hospitals. Front. Microbiol. 2021, 12, 709051. [Google Scholar] [CrossRef]
- Pruden, A.; Larsson, D.G.J.; Amézquita, A.; Collignon, P.; Brandt, K.K.; Graham, D.W.; Lazorchak, J.M.; Suzuki, S.; Silley, P.; Snape, J.R.; et al. Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ. Health Perspect. 2013, 121, 878–885. [Google Scholar] [CrossRef]
- Uluseker, C.; Kaster, K.M.; Thorsen, K.; Basiry, D.; Shobana, S.; Jain, M.; Kumar, G.; Kommedal, R.; Pala-Ozkok, I. A review on occurrence and spread of antibiotic resistance in wastewaters and in wastewater treatment plants: Mechanisms and perspectives. Front. Microbiol. 2021, 12, 717809. [Google Scholar] [CrossRef]
- Ruehle, M.D.; Orias, E.; Pearson, C.G. Tetrahymena as a unicellular model eukaryote: Genetic and genomic tools. Genetics 2016, 203, 649–665. [Google Scholar] [CrossRef]

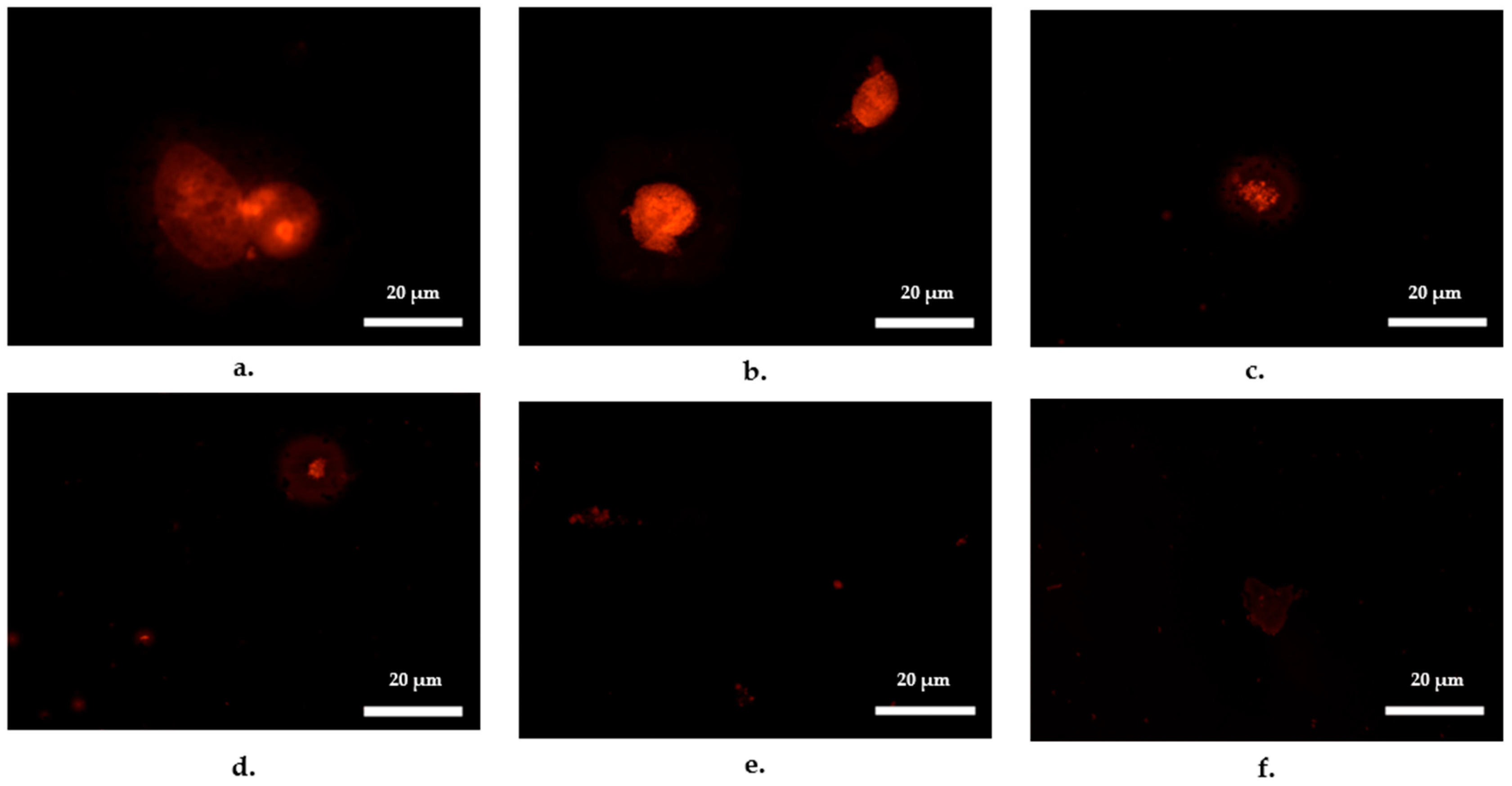
| Recipient Strain | Incubation Time, h | Treatment | Control | ||||
|---|---|---|---|---|---|---|---|
| Recipient Colony Count, RCC (CFU/mL) | Transconjugant Count, TC (CFU/mL) µ ± SEM, n = 16 | Gene Transfer Frequency (TC: RCC) | Recipient Colony Count, RCC (CFU/mL) | Transconjugant Count, TC (CFU/mL) µ ± SEM, n = 16 | Gene Transfer Frequency (TC: RCC) | ||
| ST02103Rif | 0 | 1.3 × 109 | 256 ± 14 | 2.0 × 10−7 | 1.3 × 109 | 228 ± 13 | 1.9 × 10−7 |
| 2 | 2.0 × 109 | 2002 ± 49 | 1.0 × 10−6 | 2.6 × 109 | 171 ± 12 | 6.7 × 10−8 | |
| 8 | 1.3 × 109 | 1198 ± 28 | 9.6 × 10−7 | 2.2 × 109 | 129 ± 12 | 6.3 × 10−8 | |
| 24 | 8.4 × 106 | 118 ± 11 | 2.0 × 10−5 | 2.0 × 106 | 37 ± 7 | 2.0 × 10−5 | |
| MW01105Rif | 0 | 1.0 × 109 | 39 ± 5 | 4.3 × 10−8 | 1.0 × 109 | 38 ± 5 | 4.1 × 10−8 |
| 2 | 2.7 × 109 | 66 ± 9 | 2.5 × 10−8 | 3.3 × 109 | 27 ± 5 | 8.4 × 10−9 | |
| 8 | 1.3 × 109 | 10 ± 2 | 6.9 × 10−9 | 3.0 × 109 | 19 ± 4 | 6.5 × 10−9 | |
| 24 | 4.1 × 106 | 0 | 0 | 2.6 × 106 | 6 ± 2 | 6.3 × 10−7 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olanrewaju, T.O.; Dooley, J.S.G.; Coleman, H.M.; McGonigle, C.; Arnscheidt, J. Bacterivorous Ciliate Tetrahymena pyriformis Facilitates vanA Antibiotic Resistance Gene Transfer in Enterococcus faecalis. Antibiotics 2025, 14, 448. https://doi.org/10.3390/antibiotics14050448
Olanrewaju TO, Dooley JSG, Coleman HM, McGonigle C, Arnscheidt J. Bacterivorous Ciliate Tetrahymena pyriformis Facilitates vanA Antibiotic Resistance Gene Transfer in Enterococcus faecalis. Antibiotics. 2025; 14(5):448. https://doi.org/10.3390/antibiotics14050448
Chicago/Turabian StyleOlanrewaju, Temilola O., James S. G. Dooley, Heather M. Coleman, Chris McGonigle, and Joerg Arnscheidt. 2025. "Bacterivorous Ciliate Tetrahymena pyriformis Facilitates vanA Antibiotic Resistance Gene Transfer in Enterococcus faecalis" Antibiotics 14, no. 5: 448. https://doi.org/10.3390/antibiotics14050448
APA StyleOlanrewaju, T. O., Dooley, J. S. G., Coleman, H. M., McGonigle, C., & Arnscheidt, J. (2025). Bacterivorous Ciliate Tetrahymena pyriformis Facilitates vanA Antibiotic Resistance Gene Transfer in Enterococcus faecalis. Antibiotics, 14(5), 448. https://doi.org/10.3390/antibiotics14050448








