Abstract
Antimicrobial resistance (AMR) poses a significant “One Health” challenge in the farming industry attributed to antimicrobial misuse and overuse, affecting the health of humans, animals, and the environment. Recognizing the crucial role of the environment in facilitating the transmission of AMR is imperative for addressing this global health issue. Despite its urgency, there remains a notable gap in understanding resistance levels in the environment. This scoping review aims to consolidate and summarize available evidence of AMR prevalence and resistance genes in dairy farm settings. This study was conducted following the PRISMA Extension checklist to retrieve relevant studies conducted in Asian countries between 2013 and 2023. An electronic literature search involving PubMed, ScienceDirect, Embase, and Scopus resulted in a total of 1126 unique articles that were identified. After a full-text eligibility assessment, 39 studies were included in this review. The findings indicate that AMR studies in dairy farm environments have primarily focused on selective bacteria, especially Escherichia coli and other bacteria such as Staphylococcus aureus, Klebsiella spp., and Salmonella spp. Antimicrobial resistance patterns were reported across 24 studies involving 78 antimicrobials, which predominantly consisted of gentamicin (70.8%), ampicillin (58.3%), and tetracycline (58.3%). This review emphasizes the current state of AMR in the environmental aspects of dairy farms across Asia, highlighting significant gaps in regional coverage and bacterial species studied. It highlights the need for broader surveillance, integration with antimicrobial stewardship, and cross-sector collaboration to address AMR through a One Health approach.
1. Introduction
Milk and dairy products are known for their health benefits and are considered vital components of a well-balanced diet [1,2]. The nutrients from milk and dairy products are essential for most people, especially vulnerable populations, including infants, school children, and the elderly [1,2,3]. Bovine milk is the most frequently consumed dairy product worldwide, contributing significantly to global milk production [4]. According to the Food and Agriculture Organization (FAO), the production of milk has surged by 60% from 1987 to 2017, increasing from 522 million tons to 828 million tons [5]. FAO projections indicate a continued rise in global demand for milk and dairy products, with a particular focus on Asian countries [5].
Notably, Asia experienced the highest milk output expansion in 2018, surpassing other regions. According to the 2020 FAO dairy report, Asia recorded the largest increase in milk production, followed by Europe and other regions. In 2020, milk output reached 379 million tons, primarily driven by growth in India, China, and Pakistan [6]. This growth was largely attributed to an increase in dairy herd numbers and improvements in milk collection processes, which particularly benefited smallholder dairy producers [7].
The use of antimicrobials is a major concern for food safety and quality in the dairy industry [8]. The practice of administering antimicrobials in food animals contributed a significant portion of the total antimicrobial consumption. Antimicrobials are often used in livestock as growth promoters, prophylactics, and therapeutics. Consequently, these activities lead to environmental pollution with antimicrobials, antimicrobial-resistant bacteria, and antibiotic resistance genes (ARGs) [9,10].
The presence of these antimicrobial resistance (AMR) drivers amplifies the likelihood of interaction between antimicrobials and bacteria, fostering natural selection or mutation that favors resistance [11]. Subsequently, horizontal gene transfer may occur among environmental bacteria, leading to the dissemination of resistance, including pathogenic bacteria [12]. Humans, whether directly or indirectly, are exposed to these antimicrobial-resistant bacteria through food or the environment, thereby increasing the potential for infection [13].
A major global health burden, particularly in low- and middle-income countries (LMICs) is caused by AMR [14]. Based on studies on important pathogens including Escherichia coli, Staphylococcus aureus, and Klebsiella pneumoniae, it was predicted the continuous rise in AMR could kill 10 million people worldwide annually by 2050, especially in Asia and African countries [15]. Furthermore, resistance to last-resort antimicrobials garnered the attention of many authorities worldwide in mitigating it [16].
Asia is highly susceptible to the threats of AMR, and among its regions, Southeast Asian nations are assumed to be at the greatest risk of the emergence and spread of AMR [17]. This is largely attributed to the escalating use of antimicrobials in livestock production, particularly for growth promotion and treatment, which is further complicated by the easy access to antimicrobials and challenges in accessing quality veterinary care and preventive services [15,17]. The impact of AMR in Asia extends beyond clinical settings by increasing the burden of chronic diseases and widening health inequities [17]. These events culminate into a disproportionate AMR burden driven by imprudent antimicrobial stewardship practices and underdeveloped surveillance systems [14].
Global estimates have also shown that India and China are among the top five antimicrobial-consuming countries in terms of food-producing animals [4,14]. Despite the lack of comprehensive estimates, the veterinary sector accounts for 70% of antimicrobial consumption based on the surveillance of 36 commonly prescribed antimicrobials [17]. More importantly, three countries in the Southeast Asia region (Indonesia, Vietnam, and Myanmar) were among the five countries projected to record the largest increase in antimicrobial usage by food animals [18]. These projections highlight the risk of the emergence of AMR in Asia, likely to be driven by worsening antimicrobial usage hotspots and intensified farming systems.
Despite the burden of AMR in Asian countries, a review of the antimicrobial-resistant bacteria and ARGs, particularly in dairy farms and the immediate environment, is currently lacking. A recent systematic review of antimicrobial usage in animal production revealed that only 17 of the 89 articles reviewed were carried out in LMICs, with only two articles from the Southeast Asian region [8].
To elucidate the burden of antimicrobial-resistant bacteria in the environment, especially due to antimicrobial usage in dairy cattle, this scoping review was conducted to explore the burden of antimicrobial-resistant bacteria in dairy farm environments. The review aims to summarize the distribution of antimicrobial-resistant bacteria and ARGs in various dairy farms’ environmental samples focusing on Asian countries. Specifically, we aimed to present the available evidence addressing the following research questions: (i) What is the pattern of AMR in environmental bacteria on dairy farms? (ii) Which ARGs are highly present in the environment and among environmental bacteria? (iii) What are the various methods used to detect the presence of ARGs and their patterns on dairy farms?
2. Results
2.1. Selection of Evidence Source
Our search strategy identified 1126 de-duplicated studies that underwent title and abstract screening. After initial screening, 114 studies were selected for full-text eligibility assessment, and of these, 78 studies were excluded due to the absence of environmental samples or because they entailed only milk residue, clinical, or experimental studies. Overall, a total of 39 studies were selected in this scoping review, with three additional studies identified through reference scanning. Figure 1 shows the Prisma flow diagram based on the PRISMA 2020 flow diagram with some modifications [19].
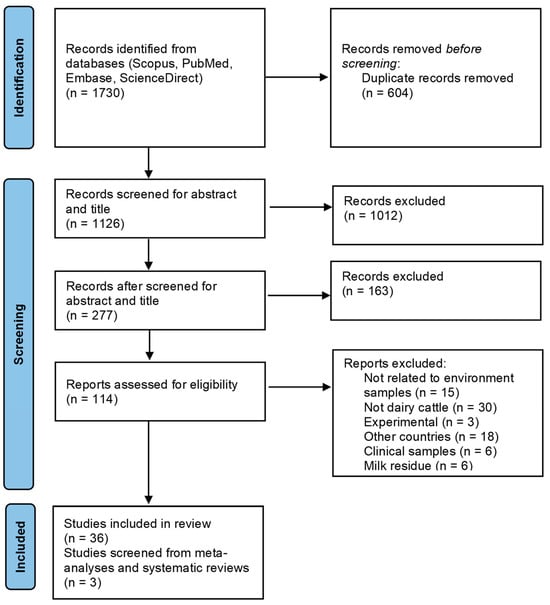
Figure 1.
PRISMA flow chart of literature selection based on inclusion/exclusion criteria to identify the occurrence of AMR in the environment of dairy farms.
2.2. Overall Characteristics and Results of Sources of Evidence
Of the thirty-nine included studies, twenty-four (61.5%) were conducted in China, followed by India (four studies, 10.3%), Bangladesh (two studies, 5.1%), Thailand (two studies, 5.1%), and Indonesia (two studies, 5.1%). Single studies were conducted in Malaysia, Iran, Japan, Pakistan, and South Korea (Figure 2). Despite the presence of forty-eight countries and three dependencies in Asia, only 19.6% of these nations published studies related to AMR in the dairy farm environment. Most studies were published between 2019 and 2023 (69.2%; n = 27) compared to those published between 2013 and 2018 (30.8%; n = 12). The most published year was 2021 with nine (38.5%) articles.
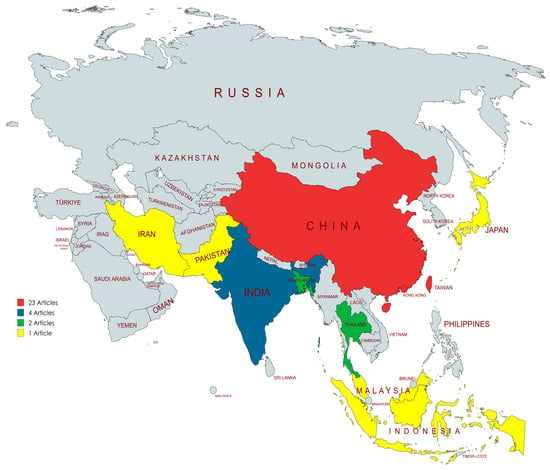
Figure 2.
Asian countries with the number of articles. Map lines delineate study areas and do not necessarily depict accepted national boundaries.
The analyzed environmental samples comprised soil, water, manure, and effluent. Specifically, 26 studies (66.7%) included manure or feces as part of the environmental samples, 14 (35.9%) included soil, 14 (35.9%) focused on drinking or pipe water, and 10 (25.6%) considered effluent or wastewater. The summary of study characteristics is shown in Table 1.

Table 1.
Summary of study characteristics (n = 39).
2.3. Bacteria Analysis
Among the 39 studies included, 25 (64.1%) focused on the analysis of one or more bacteria. The most commonly studied bacteria were E. coli (n = 17, 68.0%), followed by S. aureus (n = 3, 12.0%), Klebsiella species (n = 2, 8.0%), and Salmonella species (n = 2, 8.0%). Table 2 summarizes the number of studies related to environmental bacteria, AMR testing methods, and ARGs. Notably, only one study analyzed bacterial presence but did not detect AMR, focusing instead on ARGs [20]. Several studies (n = 14, 35.9%) did not study the environmental bacteria as shown in Table 3.

Table 2.
Types of environmental bacteria analyzed, AMR testing methods, and ARGs detection based on the number of studies involved.

Table 3.
The environmental matrix and characteristics of analysis conducted by the included studies.
2.4. Methods of Antimicrobial Susceptibility Testing
Various methods were used for antimicrobial susceptibility testing among the recovered isolates across studies, with the majority aligning with established standards such as CLSI, EUCAST, and others. Across the 24 studies, the predominant choice was the diffusion method (n = 15, 62.5%), followed by the dilution method to determine the minimum inhibitory concentration (MIC), either through broth medium (n = 8, 33.3%) or agar dilution (n = 1, 4.2%) (Table 2).
Overall, eight studies (33.3%) implemented agar supplemented with antimicrobials for screening purposes. One study reported the use of an E-test, a method that incorporates the principles of both dilution and diffusion approaches [59]. The usage of VITEK-2, an automated system, in two studies (8%) demonstrated the adoption of advanced technologies. Nevertheless, there is an opportunity for improvement, particularly in the accurate and rapid identification of bacterial resistance to antimicrobials [60]. Several studies did not state the methods of detection utilized for bacterial isolation and identification [23,32,40,41,42,43,47,50,52,53,54,58].
2.5. Antimicrobial Resistance Patterns
A total of 78 antimicrobials were used across 24 studies to assess resistance patterns among the isolates recovered from dairy farm environments in Asia. However, the number of antimicrobials tested by each study varied from one to twenty-six. Gentamicin appeared as the most frequently tested antimicrobial (n = 17, 70.8%), followed by ampicillin (n = 14, 58.3%), tetracycline (n = 14, 58.3%), ciprofloxacin (n = 13, 54.2%), chloramphenicol (n = 12, 50%), cefotaxime (n = 11, 45.8%), and trimethoprim-sulfamethoxazole (n = 11, 45.8%). Most studies reported high resistance to tetracycline, ampicillin, and trimethoprim-sulfamethoxazole compared to the rest [21,22,24,25,28,29,33,35,36,37,44,45,46,48,51,57] (Table 4).

Table 4.
Major findings of the 39 included studies on AMR and ARGS in the environment of dairy farms.
Carbapenems, known for their broad-spectrum antibacterial activity, were tested in eleven studies on imipenem, nine studies on meropenem, and three studies on ertapenem, covering various bacteria. Most studies revealed high susceptibility (96.6–100%) to meropenem and imipenem, except for specific cases reporting resistance [22,28,29,33,36,39,44,45,46,48,56,57]. Aligning with the World Health Organization’s (WHO) priority list of antimicrobial-resistant bacteria [61], out of seventeen studies investigating E. coli, five reported extended-spectrum beta-lactamases (ESBL) isolates, exhibiting resistance to third-generation cephalosporins. Among the fecal samples, ESBL was reported at 66% (China), 47% (India), 25% (Indonesia), and 5.5% (Malaysia) [22,29,31,36].
Three studies focused on S. aureus, with one addressing methicillin-resistant S. aureus (MRSA) in 1.2% of environmental samples [34]. Two studies reported antimicrobial susceptibility testing for Salmonella species. One study reported the AMR rate in an aggregated manner, encompassing both environmental and milk samples [27]. Meanwhile, a 100% resistance to azithromycin was recorded among environmental samples, with higher resistance to erythromycin and tetracycline ranging from 83% to 93% across different samples within the environmental domain [46].
2.6. Antibiotic Resistance Genes (ARGs) Detection
Of the 32 studies (82.1%) involving ARG testing, 25 utilized polymerase chain reaction (PCR) testing, three concentrated on whole genome sequencing, and five employed metagenomic analysis. In line with antimicrobial detection preferences and findings, tet and sul genes were most commonly analyzed and found in abundance in the environmental samples [21,26,30,32,32,33,35,38,40,41,42,43,44,46,48,49] (Table 4).
Concerning E. coli strains, ESBL-associated genes were commonly tested, and blaTEM, blaSHV, and blaCTX were found in abundance [21,22,28,29,31,35,46,57]. Metagenomic sequencing, mainly used for the detection of ARGs in manure, soil, and wastewater, found that the predominant genes were tetracycline (tet), beta-lactams (blaTEM), sulfonamide (sul), and aminoglycoside genes [32,40,43,54,58]. ARGs were not explored in seven studies [24,25,27,36,37,45,58] as shown in Table 4. Most of the studies focused on gene detection [20,23,32,38,40,41,42,43,47,49,50,52,53,54,55,58], followed by those focusing on antimicrobial resistance testing [24,25,27,36,37,45,58] and those involving both aspects [21,22,28,29,30,31,33,34,35,39,44,46,48,51,57].
2.7. Comparative Analysis of AMR Patterns Across Environmental Samples Based on Country
Comparative analysis of AMR patterns was conducted across environmental samples in countries with at least two or more studies involving the same environmental sample.
China
Four studies in China assessed AMR patterns in fecal samples from dairy farm settings [24,32,38,58]. The diversity and abundance of ARGs in dairy feces were significantly higher compared to those detected in soil samples, with a high detection rate of tet(X) in feces at 71.4% [32,58].
Three studies focused on AMR patterns in manure samples. The positive detection rate for ARGs was higher than 80% in the studies (Wang et al., 2020; Wang et al., 2016; Yang et al., 2022) [49,50,53]. However, Yang et al. (2022) found that the abundance of the bla gene in dairy cattle feces was lower relative to the level in chicken and beef cattle farms [53].
As for wastewater, two studies recorded a significantly higher detection rate of ARGs in wastewater samples (100%) relative to soil samples, with the most abundant being sul1, sul2, tetM [22], and tet(X) [32]. Comparative analysis was not performed for feed, raw milk, and bedding samples, given that only one study was reported for each sample.
India
Among the four studies conducted in India, two involved cow dung/slurry [22,30], one involved soil samples [37] and one entailed floor and milking machine swabs [26]. However, all studies investigated the prevalence of E. coli and its resistant genes, except Gandhale et al. (2017) [26] who focused on S. aureus.
More than 50% of the isolates from cow dung/slurry were found to have resistant genes, especially tetA and sulII [30], while ESBLs were identified as the main cause of resistance to beta-lactam antimicrobials in E. coli [22].
Thailand
Two studies in Thailand also focused on AMR patterns in E. coli, particularly in water samples [28,39] and milk samples [39]. Both samples demonstrated resistant E. coli strains to ampicillin and carbenicillin, while E. coli from water samples possessed an ESBL phenotype and antimicrobial resistance bla genes [28,39].
Bangladesh
Two studies in this review explored AMR patterns from diverse environmental samples, including cow dung, water, feed, and soil, but the bacteria of interest differed between the studies, with Listeria spp. in one [45] and E. coli and Salmonella spp. in the other [46]. Comparative analysis was not feasible; nevertheless, the most predominant resistance gene identified in cow dung, soil, and water samples was tetA (80.5–84.7%) [46].
Indonesia
Two studies in Indonesia reported the AMR pattern of E. coli isolates from several environmental samples, including wastewater, feces, drinking water, milk, feed, and rinses of workers’ hands [25,36]. The occurrence of E. coli AMR ranged from 54% at the farm level to 99.1% at the subject level. The incidence of ESBL-producing E. coli was 22.8% in wastewater samples [25], whereas fecal samples recorded the highest positive results at 25% compared to the other samples [36].
2.8. Comparative Analysis of AMR Patterns Across Environmental Samples Between Countries
Comparative analysis between studies conducted in different countries was performed for only two environmental samples, fecal and wastewater, as they constituted the predominantly investigated samples in the reviewed studies.
Fecal samples
Fecal samples, including manure and cow dung, depicted consistent AMR patterns across the studies conducted in several countries, such as India, China, Bangladesh, Indonesia, and Japan [32,36,46,48,58]. The detection rate of ARGs, especially tetA in fecal samples, ranged from 71.4% to 84.7% [32,58], which was significantly higher compared to the detection rates in soil and water samples [46]. Resistant genes to E. coli were the predominantly investigated AMR pattern in the studies, with fecal samples recording significantly higher results for ESBLs, at 25% relative to soil and water samples [46].
In Japan, the highest frequency of antimicrobial-resistant E. coli strains isolated from environmental samples was from cow feces at 1.7%, with all strains resistant to tetracycline and possessing tetA [48]. Meanwhile, in Malaysia, fecal samples accounted for 5.5% of total samples positive for ESBL-producing E. coli [31].
Wastewater and drinking water samples
Consistent findings were observed in studies on AMR patterns in water samples, with resistant E. coli strains possessing an ESBL phenotype and antimicrobial resistance bla genes [28],38]. The incidence of ESBL-producing E. coli was 22.8% in wastewater samples [25], whereas wastewater and drinking water samples recorded the second and third-highest positive results at 16% and 10%, respectively [.
Likewise, wastewater samples depicted a significantly higher detection rate of ARGs compared to soil samples, with tet(A), tet(X), and tetM as the predominant resistance genes [23,29,32,46]. Suzuki et al. [48] also found an antimicrobial resistance rate of 8.3% (10/120) in the drainage from the studied dairy cattle barns, with the presence of tet(A) in all the tetracycline-resistant strains.
3. Discussion
Recognizing the environment as a crucial component of the “One Health” approach, continuous surveillance of this component is necessary, extending beyond the focus on human and animal health alone. This scoping review highlighted the research gap in the context of AMR within the environment domain in dairy farms around Asian countries. Despite dairy industries expanding in countries like India, China, Indonesia, the Philippines, Myanmar, Laos, and Vietnam, the majority of environmental AMR research is concentrated in China [62]. The lack of research highlights a crucial gap in understanding the extent and dynamics of AMR in dairy farm environments across broader Asian regions. Therefore, there is a need for comprehensive, regional studies to capture the full scope of AMR within these settings.
Most of the included studies focused on investigating the presence of AMR, specifically in E. coli, a bacterium that is commonly found in the intestinal tracts of mammals, including cows. The prominence of targeting E. coli in dairy farm environments can be attributed to its role as an indicator of fecal bacterium [63,64]. E. coli serves as a reliable indicator for addressing the AMR spread in various settings and is a common carrier of various ARGs [65]. However, this singular focus represents a major gap, as other significant pathogens, particularly the ESKAPE pathogens (Enterococcus spp., Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.), remain largely studied.
These ESKAPE pathogens are identified by the WHO as critical MDR pathogens due to their ability to evade common antimicrobials, posing significant challenges in infection treatment and demanding the urgent development of new antimicrobials [66]. The scarcity of research on these pathogens in dairy farm environments limits the ability to assess their prevalence, transmission, and potential health risk to humans and animals [67,68,69]. Due to their potential for developing and extending resistance, future studies should focus on expanding surveillance including these high-risk pathogens.
The findings of this review reveal significant variations in resistance rates and the presence of ARGs, influenced by differences in the methods used for AMR testing and the types of samples analyzed. Despite these variations, a consistent trend emerges throughout the studies, which is the association between AMR and the use of antimicrobial agents for treatment on dairy farms [29,35,44,70]. The use of antimicrobials in dairy and food animals plays a significant role in shaping the AMR patterns in the farm environment. The frequent use of antimicrobials exerts selective pressure, favoring resistant strains that persist in environmental reservoirs such as soil and effluent, thereby contributing to the spread of resistant bacteria on farms [29,36,70]. Additionally, the genetic diversity of ARGs also plays a key role in influencing AMR through horizontal gene transfer [71,72].
For instance, the widespread emergence of colistin resistance is of particular concern, leading many countries in Asia to ban the use of colistin, especially in animal feeds, due to its importance as the last line of defense in human medicine. Interestingly, studies that have explored colistin resistance reported either total susceptibility or a very low level of resistance to this antimicrobial [33,44,45,51,57]. Although some studies reported low or no resistance to colistin, the detection of carbapenem-resistant bacteria from environmental sources raises concern regarding the use of antimicrobials in these areas. Carbapenem resistance poses a particularly severe threat to public health, due to their ability to facilitate horizontal gene transfer, further exacerbating AMR spread [73]. These findings underscore the urgent need for stringent antimicrobial stewardship practices and surveillance efforts to mitigate the spread of multidrug-resistant pathogens in dairy farm environments [74].
However, there were several limitations to this review, and these need to be acknowledged. The heterogeneity in study methodologies across the included papers, including differences in AMR susceptibility screening techniques, makes it difficult to compare across studies. Varying susceptibility testing methods, including disk diffusion, broth microdilution, and molecular approaches, can lead to different resistance profiles and hence, different study outcomes. Excluding gray literature and non-English articles, those published in Chinese may have introduced publication bias, thus limiting the scope of this review. Given that China has contributed significantly to AMR research, the omission of Chinese-language studies may lead to the underrepresentation of some important findings. Thus, future reviews should attempt to include a wider variety of studies in order to provide a more holistic assessment of AMR patterns in dairy farm environments. In addition, since this scoping review did not include a quality assessment and risk of bias analysis, it is recommended that further studies carry out a systematic review or meta-analysis to provide a more comprehensive and detailed investigation of AMR.
4. Materials and Methods
This review was performed using the methodological framework elaborated by Arksey and O’Malley and following the Prisma Extension for Scoping Review (PRISMA-ScR) checklist by Tricco et al. [75,76,77]. Therefore, only qualitative and comparative analyses were carried out in line with the provisions of a scoping review. Prior research has also shown that a scoping review encompasses studies with different research designs, which reflect a high degree of heterogeneity between the studies [77]. Thus, scoping reviews are exempted from robust quality assessment and statistical analyses.
4.1. Search Terms and Strategy
This review focused on AMR in environmental bacteria found in dairy farms. The study period covered research published between 2013 and 2023 to capture the latest pattern of AMR and ARGs in dairy farm environments. The review protocol was developed following the PRISMA-ScR guidelines, and no prior registration was conducted. Three reviewers (Y.V., S.R., and S.A.T.) independently performed a systematic bibliographic search across four databases—PubMed, ScienceDirect, Embase, and Scopus. The search strategy was initially drafted by Y.V. and subsequently reviewed and validated by S.R. and S.A.T. to ensure accuracy and consistency.
A comprehensive search of the relevant studies was conducted across 48 countries and 3 dependencies in Asia according to the United Nations [78] (Supplementary Table S1). The keywords used for the literature search were as follows: “cow”, “dairy”, “cattle”, “farm”, “farming”, “industry”, “environment”, “soil”, effluent”, “water”, “manure”, “wastewater”, “antibiotic”, “antimicrobial”, “resistant”, and “resistance”, and the general Boolean operators used for all four databases were as follows: (cow OR cows OR dairy OR cattle) AND (farm OR farms OR industry OR farming) AND (environment OR environments OR soil OR effluent OR water OR wastewater OR manure OR land) AND (antibiotics OR antibiotic OR antimicrobial OR antimicrobials OR multidrug OR resistant OR resistance). Country terms were not included in search terms, as studies conducted in Asia were filtered manually during the study selection phase. The search was conducted across the four mentioned databases. Subsequently, the records from each database were imported into Zotero reference management software version 6.0.37. Consequently, the software was used to remove duplicate entries of articles. All the records of the prospective articles were exported to a Microsoft Excel spreadsheet for study selection.
4.2. Eligibility Criteria
Independently, three reviewers (Y.V., S.R., and S.A.T.) completed the study selection based on the following inclusion criteria: (i) original, peer-reviewed, and primary research articles; (ii) studies addressing AMR in environmental bacteria found in dairy farm settings; (iii) published in English; (iv) published between 2013 and 2023; and (v) conducted in Asian countries (48 countries and 3 dependencies). Meanwhile, the exclusion criteria included (i) meta-analyses and all types of review articles; (ii) clinical, diagnostic, or health facility-based studies; and (iii) studies not involving environmental samples from dairy farms (e.g., studies focusing only on milk, dairy products, or human infections without environmental involvement). However, reference lists from these meta-analyses and systematic reviews were screened for additional relevant studies, leading to the inclusion of three additional articles.
4.3. Study Selection Process
A multi-step process was used to identify the studies. First, the title and abstract were screened based on the relevance to AMR in dairy farm environments in Asian countries. Next, full-text screening was conducted to ensure that the studies investigated AMR in environmental bacteria in dairy farms, were conducted in an Asian country, and reported the presence of AMR or ARGs. The study selection and screening process was performed independently by three reviewers (Y.V., S.R., and S.A.T.). Any disagreements were settled by discussion.
4.4. Data Management and Charting
All the data were extracted and charted using Microsoft Excel, including the study inclusion criteria and key variables. The data that were systematically extracted from each study included the author’s name, year of publication, study location, title of the research, research objectives, bacteria analyzed, types of environmental samples analyzed, methods used for AMR or ARG detection, antimicrobial agents tested, presence and percentage of resistance, ARG detected, and summary of key findings (Supplementary Table S2).
4.5. Data Extraction and Synthesis
Using a descriptive and narrative analysis style, specific data were collected from each study based on the subjects’ characteristics, types of environmental samples, country in which the study was conducted, methods used, and reported resistance patterns. Table 2 depicts the tested antimicrobials and ARGs based on their reported presence and percentage of resistance. Key findings were synthesized to identify trends, research gaps, and methodological limitations in the current literature on AMR in dairy farm environments.
5. Conclusions
This scoping review aimed to address the current status of AMR in the environmental domain of dairy farms across Asian regions. While AMR surveillance is an expanding area of research, significant knowledge gaps remain, particularly in regional coverage and limited focus on bacteria species. Thus, future studies should broaden geographical coverage and investigate emerging pathogens. Additionally, integrating environmental surveillance with antimicrobial stewardship is critical for mitigating the spread of resistance and protecting both human and animal health. Collaborative efforts among veterinary, agricultural, and public health sectors are essential in tackling AMR from a One Health perspective, ensuring sustainable dairy farming practices while safeguarding public health.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics14050436/s1, Table S1: List of countries in the Asian Region; Table S2: The details on the included articles.
Author Contributions
Conceptualization, Y.V. and Z.Z.; Methodology, Y.V.; Data curation, Y.V., S.S.A.T. and S.R.; Validation, Y.V., S.S.A.T. and S.R.; Formal analysis, Y.V., S.S.A.T., S.A.R., R.M. and S.R.; Writing—original draft, Y.V., S.S.A.T. and S.R.; Visualization, Y.V. and S.R.; Supervision, Z.Z.; Project administration, Z.Z.; Writing—review and editing, Z.Z., S.A.R. and R.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Health, Malaysia.
Data Availability Statement
The data included in this review are referenced in this article.
Acknowledgments
The authors would like to thank the Director-General of Health Malaysia for his permission to publish this article.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AMR | Antimicrobial resistance |
| FAO | Food and Agriculture Organization |
| ARGs | Antibiotic resistance genes |
| LMICs | Low- and middle-income countries |
| E. coli | Escherichia coli |
| S. aureus | Staphylococcus aureus |
| MIC | Minimum inhibitory concentration |
| WHO | World Health Organization |
| ESBL | Extended-spectrum beta-lactamases |
| PRISMA-ScR | Prisma Extension for Scoping Review |
References
- Gasmalla, M.A.A.; Tessema, H.A.; Salaheldin, A.; Alahmad, K.; Hassanin, H.A.; Aboshora, W. Health Benefits of Milk and Functional Dairy Products. MOJ Food Process. Technol. 2017, 4, 108–111. [Google Scholar] [CrossRef][Green Version]
- Özer, B.H.; Kirmaci, H.A. Functional Milks and Dairy Beverages. Int. J. Dairy Technol. 2010, 63, 1–15. [Google Scholar] [CrossRef]
- Givens, D.I. MILK Symposium Review: The Importance of Milk and Dairy Foods in the Diets of Infants, Adolescents, Pregnant Women, Adults, and the Elderly*. J. Dairy Sci. 2020, 103, 9681–9699. [Google Scholar] [CrossRef] [PubMed]
- Claeys, W.L.; Verraes, C.; Cardoen, S.; De Block, J.; Huyghebaert, A.; Raes, K.; Dewettinck, K.; Herman, L. Consumption of Raw or Heated Milk from Different Species: An Evaluation of the Nutritional and Potential Health Benefits. Food Control 2014, 42, 188–201. [Google Scholar] [CrossRef]
- Cuong, N.V.; Padungtod, P.; Thwaites, G.; Carrique-Mas, J.J. Antimicrobial Usage in Animal Production: A Review of the Literature with a Focus on Low- and Middle-Income Countries. Antibiotics 2018, 7, 75. [Google Scholar] [CrossRef]
- Morgan, N. Introduction: Dairy Development in Asia. Available online: https://www.fao.org/4/i0588e/I0588E02.htm (accessed on 31 January 2025).
- FAO. Dairy Market Review 2019. Available online: https://openknowledge.fao.org/server/api/core/bitstreams/ca99822d-2fac-4096-b9e8-f9ff0602faaa/content (accessed on 14 November 2022).
- Sadiq, M.B.; Syed-Hussain, S.S.; Ramanoon, S.Z.; Saharee, A.A.; Ahmad, N.I.; Mohd Zin, N.; Khalid, S.F.; Naseeha, D.S.; Syahirah, A.A.; Mansor, R. Knowledge, Attitude and Perception Regarding Antimicrobial Resistance and Usage among Ruminant Farmers in Selangor, Malaysia. Prev. Vet. Med. 2018, 156, 76–83. [Google Scholar] [CrossRef]
- Medina-Pizzali, M.L.; Hartinger, S.M.; Salmon-Mulanovich, G.; Larson, A.; Riveros, M.; Mäusezahl, D. Antimicrobial Resistance in Rural Settings in Latin America: A Scoping Review with a One Health Lens. Int. J. Environ. Res. Public Health 2021, 18, 9837. [Google Scholar] [CrossRef]
- Singer, A.C.; Shaw, H.; Rhodes, V.; Hart, A. Review of Antimicrobial Resistance in the Environment and Its Relevance to Environmental Regulators. Front. Microbiol. 2016, 7, 1728. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 481–511. [Google Scholar] [CrossRef]
- Ashbolt, N.J.; Amézquita, A.; Backhaus, T.; Borriello, P.; Brandt, K.K.; Collignon, P.; Coors, A.; Finley, R.; Gaze, W.H.; Heberer, T.; et al. Human Health Risk Assessment (HHRA) for Environmental Development and Transfer of Antibiotic Resistance. Environ. Health Perspect. 2013, 121, 993–1001. [Google Scholar] [CrossRef]
- Lundborg, C.S.; Tamhankar, A.J. Antibiotic Residues in the Environment of South East Asia. BMJ 2017, 358, j2440. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Malijan, G.M.; Howteerakul, N.; Ali, N.; Siri, S.; Kengganpanich, M.; Nascimento, R.; Booton, R.D.; Turner, K.M.E.; Cooper, B.S.; Meeyai, A. A Scoping Review of Antibiotic Use Practices and Drivers of Inappropriate Antibiotic Use in Animal Farms in WHO Southeast Asia Region. One Health 2022, 15, 100412. [Google Scholar] [CrossRef] [PubMed]
- Samreen; Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental Antimicrobial Resistance and Its Drivers: A Potential Threat to Public Health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar] [CrossRef]
- Kakkar, M.; Chatterjee, P.; Chauhan, A.S.; Grace, D.; Lindahl, J.; Beeche, A.; Jing, F.; Chotinan, S. Antimicrobial Resistance in South East Asia: Time to Ask the Right Questions. Glob. Health Action 2018, 11, 1483637. [Google Scholar] [CrossRef] [PubMed]
- Hosain, M.Z.; Kabir, S.M.L.; Kamal, M.M. Antimicrobial Uses for Livestock Production in Developing Countries. Vet World 2021, 14, 210–221. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Xi, X.; Zhang, J.; Kwok, L.; Huo, D.; Feng, S.; Zhang, H.; Sun, T. Microbial Pollution Tracking of Dairy Farm with a Combined PCR-DGGE and qPCR Approach. Curr. Microbiol. 2015, 71, 678–686. [Google Scholar] [CrossRef]
- Ali, A.; Fontana, H.; Sano, E.; Li, R.; Humayon, M.; Rahman, S.; Lincopan, N.; Mohsin, M. Genomic Features of a High-Risk Mcr-1.1-Positive Escherichia Coli ST10 Isolated from Cattle Farm Environment. Environ. Sci. Pollut. Res. Int. 2021, 28, 54147–54152. [Google Scholar] [CrossRef]
- Borah, V.; Roy, M.; Saikia, K. High Prevalence of Antibiotic Resistance in Escherichia Coli Isolated from Fecal Sample of Cows and Assessment of Antibacterial Efficacy of Indigenous Medicinal Plants from Assam, India. Austin. J. Biotechnol. Bioeng. 2014, 1, 6. [Google Scholar]
- Chen, B.; Hao, L.; Guo, X.; Wang, N.; Ye, B. Prevalence of Antibiotic Resistance Genes of Wastewater and Surface Water in Livestock Farms of Jiangsu Province, China. Environ. Sci. Pollut. Res. 2015, 22, 13950–13959. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Liu, X.; Dietrich, R.; Märtlbauer, E.; Cao, J.; Ding, S.; Zhu, K. Characterization of Bacillus Cereus Isolates from Local Dairy Farms in China. FEMS Microbiol. Lett. 2016, 363, fnw096. [Google Scholar] [CrossRef] [PubMed]
- Dameanti, F.N.A.E.P.; Yanestria, S.M.; Widodo, A.; Effendi, M.H.; Plumeriastuti, H.; Tyasningsih, W.; Sutrisno, R.; Akramsyah, M.A. Incidence of Escherichia Coli Producing Extended-Spectrum Beta-Lactamase in Wastewater of Dairy Farms in East Java, Indonesia. Biodiversitas 2023, 24, 1143–1150. [Google Scholar] [CrossRef]
- Gandhale, D.; Kolhe, R.; Nalband, S.; Deshpande, P.; Jagtap, U.; Dhandore, C.; Bhave, S.; Jadhav, S.; Muglikar, D.; Kolhe, S. Molecular Types and Antimicrobial Resistance Profile of Staphylococcus Aureus Isolated from Dairy Cows and Farm Environments. Turk. J. Vet. Anim. Sci. 2017, 41, 713–724. [Google Scholar] [CrossRef]
- Halimi, H.A.; Seifi, H.A.; Rad, M. Bovine Salmonellosis in Northeast of Iran: Frequency, Genetic Fingerprinting and Antimicrobial Resistance Patterns of Salmonella Spp. Asian Pac. J. Trop. Biomed. 2014, 4, 1–7. [Google Scholar] [CrossRef]
- Hinthong, W.; Pumipuntu, N.; Santajit, S.; Kulpeanprasit, S.; Buranasinsup, S.; Sookrung, N.; Chaicumpa, W.; Aiumurai, P.; Indrawattana, N. Detection and Drug Resistance Profile of Escherichia Coli from Subclinical Mastitis Cows and Water Supply in Dairy Farms in Saraburi Province, Thailand. PeerJ 2017, 2017, e3431. [Google Scholar] [CrossRef]
- Huang, S.; Tian, P.; Kou, X.; An, N.; Wu, Y.; Dong, J.; Cai, H.; Li, B.; Xue, Y.; Liu, Y.; et al. The Prevalence and Characteristics of Extended-Spectrum β-Lactamase Escherichia Coli in Raw Milk and Dairy Farms in Northern Xinjiang, China. Int. J. Food Microbiol. 2022, 381, 109908. [Google Scholar] [CrossRef]
- Jindal, P.; Bedi, J.; Singh, R.; Aulakh, R.; Gill, J. Phenotypic and Genotypic Antimicrobial Resistance Patterns of Escherichia Coli and Klebsiella Isolated from Dairy Farm Milk, Farm Slurry and Water in Punjab, India. Environ. Sci. Pollut. Res. 2021, 28, 28556–28570. [Google Scholar] [CrossRef]
- Kamaruzzaman, E.A.; Aziz, S.A.; Bitrus, A.A.; Zakaria, Z.; Hassan, L. Occurrence and Characteristics of Extended-Spectrum β-Lactamase-Producing Escherichia Coli from Dairy Cattle, Milk, and Farm Environments in Peninsular Malaysia. Pathogens 2020, 9, 1007. [Google Scholar] [CrossRef]
- Kang, J.; Liu, Y.; Chen, X.; Xu, F.; Wang, H.; Xiong, W.; Li, X. Metagenomic Insights into the Antibiotic Resistomes of Typical Chinese Dairy Farm Environments. Front. Microbiol. 2022, 13, 990272. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.; Lu, X.; Peng, K.; Liu, Y.; Xiao, X.; Song, H.; Wang, Z. Occurrence and Characterization of NDM-1-Producing Shewanella Spp. and Acinetobacter Portensis Co-Harboring Tet(X3) in a Chinese Dairy Farm. Antibiotics 2022, 11, 1422. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.-K.; Nam, H.-M.; Jang, G.-C.; Lee, H.-S.; Jung, S.-C.; Kim, T.-S. Transmission and Persistence of Methicillin-Resistant Staphylococcus Aureus in Milk, Environment, and Workers in Dairy Cattle Farms. Foodborne Pathog. Dis. 2013, 10, 731–736. [Google Scholar] [CrossRef]
- Liu, C.; Liu, Y.; Feng, C.; Wang, P.; Yu, L.; Liu, D.; Sun, S.; Wang, F. Distribution Characteristics and Potential Risks of Heavy Metals and Antimicrobial Resistant Escherichia Coli in Dairy Farm Wastewater in Tai’an, China. Chemosphere 2021, 262, 127768. [Google Scholar] [CrossRef] [PubMed]
- Maulana, K.Y.; Pichpol, D.; Farhani, N.R.; Widiasih, D.A.; Unger, F.; Punyapornwithaya, V.; Meeyam, T. Antimicrobial Resistance Characteristics of Extended Spectrum Beta Lactamase (Esbl)-Producing Escherichia Coli from Dairy Farms in the Sleman District of Yogyakarta Province, Indonesia. Vet. Integr. Sci. 2021, 19, 525–535. [Google Scholar] [CrossRef]
- Parul; Bist, B.; Sharma, B.; Jain, U. Virulence Associated Factors and Antibiotic Sensitivity Pattern of Escherichia Coli Isolated from Cattle and Soil. Vet. World 2014, 7, 369–372. [Google Scholar] [CrossRef]
- Peng, S.; Zhang, H.; Song, D.; Chen, H.; Lin, X.; Wang, Y.; Ji, L. Distribution of Antibiotic, Heavy Metals and Antibiotic Resistance Genes in Livestock and Poultry Feces from Different Scale of Farms in Ningxia, China. J. Hazard. Mater. 2022, 440, 129719. [Google Scholar] [CrossRef]
- Pumipuntu, N.; Pumipuntu, S. Detection of Antimicrobial Resistance Genes of Carbapenem-Resistant Enterobacteriaceae in Escherichia Coli Isolated from the Water Supply of Smallholder Dairy Farms in Saraburi and Maha Sarakham, Thailand. Int. J. One Health 2020, 6, 1–5. [Google Scholar] [CrossRef]
- Qi, Z.; Jin, S.; Guo, X.; Tong, H.; Ren, N.; You, S. Distribution and Transmission of β-Lactamase Resistance Genes in Meal-to-Milk Chain on Dairy Farm. Environ. Pollut. 2023, 331, 121831. [Google Scholar] [CrossRef]
- Qi, Z.; Le, Z.; Han, F.; Qi, Y.; Liu, R. β-Lactamase Genes Transmission Influenced by Tetracycline, Sulfonamide and β-Lactams Antibiotics Contamination in the on-Site Farm Soil. Ecotoxicol. Environ. Saf. 2022, 241, 113753. [Google Scholar] [CrossRef]
- Qi, Z.; Qi, Y.; Le, Z.; Han, F.; Li, F.; Yang, H.; Zhang, T.; Feng, Y.; Liu, R.; Sun, Y. The Interactions Between Antibiotic Resistance Genes and Heavy Metal Pollution Under Co-Selective Pressure Influenced the Bio-Enzyme Activity. Front. Chem. 2021, 9, 691565. [Google Scholar] [CrossRef]
- Qiu, T.; Huo, L.; Guo, Y.; Gao, M.; Wang, G.; Hu, D.; Li, C.; Wang, Z.; Liu, G.; Wang, X. Metagenomic Assembly Reveals Hosts and Mobility of Common Antibiotic Resistome in Animal Manure and Commercial Compost. Environ. Microbiomes 2022, 17, 42. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, M.; He, Z.; Geng, X.; Tang, M.; Hao, R.; Wang, S.; Shang, R.; Wang, X.; Zhang, H.; Pu, W. The Emergence of Multi-Drug Resistant and Virulence Gene Carrying Escherichia Coli Strains in the Dairy Environment: A Rising Threat to the Environment, Animal, and Public Health. Front. Microbiol. 2023, 14, 1197579. [Google Scholar] [CrossRef] [PubMed]
- Shourav, A.H.; Hasan, M.; Ahmed, S. Antibiotic Susceptibility Pattern of Listeria Spp. Isolated from Cattle Farm Environment in Bangladesh. J. Agric. Food Res. 2020, 2, 100082. [Google Scholar] [CrossRef]
- Sobur, M.A.; Sabuj, A.A.M.; Sarker, R.; Rahman, A.M.M.T.; Kabir, S.M.L.; Rahman, M.T. Antibiotic-Resistant Escherichia Coli and Salmonella Spp. Associated with Dairy Cattle and Farm Environment Having Public Health Significance. Vet. World 2019, 12, 984–993. [Google Scholar] [CrossRef]
- Sun, M.; Ye, M.; Wu, J.; Feng, Y.; Wan, J.; Tian, D.; Shen, F.; Liu, K.; Hu, F.; Li, H.; et al. Positive Relationship Detected between Soil Bioaccessible Organic Pollutants and Antibiotic Resistance Genes at Dairy Farms in Nanjing, Eastern China. Environ. Pollut. 2015, 206, 421–428. [Google Scholar] [CrossRef]
- Suzuki, Y.; Hiroki, H.; Xie, H.; Nishiyama, M.; Sakamoto, S.H.; Uemura, R.; Nukazawa, K.; Ogura, Y.; Watanabe, T.; Kobayashi, I. Antibiotic-Resistant Escherichia Coli Isolated from Dairy Cows and Their Surrounding Environment on a Livestock Farm Practicing Prudent Antimicrobial Use. Int. J. Hyg. Environ. Health 2022, 240, 113930. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Guo, X.; Yan, Z.; Wang, W.; Chen, B.; Ge, F.; Ye, B. A Comprehensive Analysis on Spread and Distribution Characteristic of Antibiotic Resistance Genes in Livestock Farms of Southeastern China. PLoS ONE 2016, 11, e0156889. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Fu, Y.; Chen, Y.; Wang, Y.; Ye, D.; Wang, C.; Hu, X.; Zhou, L.; Du, J.; et al. Association of Florfenicol Residues with the Abundance of Oxazolidinone Resistance Genes in Livestock Manures. J. Hazard. Mater. 2020, 399, 123059. [Google Scholar] [CrossRef]
- Wu, X.; Liu, J.; Feng, J.; Shabbir, M.A.B.; Feng, Y.; Guo, R.; Zhou, M.; Hou, S.; Wang, G.; Hao, H.; et al. Epidemiology, Environmental Risks, Virulence, and Resistance Determinants of Klebsiella Pneumoniae from Dairy Cows in Hubei, China. Front. Microbiol. 2022, 13, 858799. [Google Scholar] [CrossRef]
- Yang, F.; Tian, X.; Han, B.; Zhao, R.; Li, J.; Zhang, K. Tracking High-Risk β-Lactamase Gene (Bla Gene) Transfers in Two Chinese Intensive Dairy Farms. Environ. Pollut. 2021, 274, 116953. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, Z.; Li, Z.; Han, B.; Zhang, K.; Yang, P.; Ding, Y. Prevalence of High-Risk β-Lactam Resistance Genes in Family Livestock Farms in Danjiangkou Reservoir Basin, Central China. Int. J. Environ. Res. Public Health 2022, 19, 6036. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ma, C.; Zhang, W.; Li, W.; Yu, J.; Xue, D.; Wu, X.; Deng, G. Shifts in Microbial Community, Pathogenicity-Related Genes and Antibiotic Resistance Genes during Dairy Manure Piled Up. Microb. Biotechnol. 2020, 13, 1039–1053. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, B.; Guo, Y.; Wang, J.; Zhao, P.; Liu, J.; He, K. Colistin Resistance Prevalence in Escherichia Coli from Domestic Animals in Intensive Breeding Farms of Jiangsu Province. Int. J. Food Microbiol. 2019, 291, 87–90. [Google Scholar] [CrossRef]
- Zhao, Q.; Hu, P.; Li, Q.; Zhang, S.; Li, H.; Chang, J.; Jiang, Q.; Zheng, Y.; Li, Y.; Liu, Z.; et al. Prevalence and Transmission Characteristics of Listeria Species from Ruminants in Farm and Slaughtering Environments in China. Emerg. Microbes Infect. 2021, 10, 356–364. [Google Scholar] [CrossRef]
- Zheng, B.; Feng, C.; Xu, H.; Yu, X.; Guo, L.; Jiang, X.; Song, X. Detection and Characterization of ESBL-Producing Escherichia Coli Expressing Mcr-1 from Dairy Cows in China. J. Antimicrob. Chemother. 2019, 74, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Wang, C.; Zhao, Q.; Wang, Y.; Huo, M.; Wang, J.; Wang, S. Prevalence and Dissemination of Antibiotic Resistance Genes and Coselection of Heavy Metals in Chinese Dairy Farms. J. Hazard. Mater. 2016, 320, 10–17. [Google Scholar] [CrossRef]
- Benkova, M.; Soukup, O.; Marek, J. Antimicrobial Susceptibility Testing: Currently Used Methods and Devices and the near Future in Clinical Practice. J. Appl. Microbiol. 2020, 129, 806–822. [Google Scholar] [CrossRef]
- Reller, L.B.; Weinstein, M.; Jorgensen, J.H.; Ferraro, M.J. Antimicrobial Susceptibility Testing: A Review of General Principles and Contemporary Practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Oliveros, M.C.R. The Dairy Industry in Southeast Asia: Perspective, Challenges and Opportunities. IOP Conf. Ser. Earth Environ. Sci. 2019, 372, 012068. [Google Scholar] [CrossRef]
- Savin, M.; Bierbaum, G.; Hammerl, J.A.; Heinemann, C.; Parcina, M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. ESKAPE Bacteria and Extended-Spectrum-β-Lactamase-Producing Escherichia Coli Isolated from Wastewater and Process Water from German Poultry Slaughterhouses. Appl. Environ. Microbiol. 2020, 86, e02748-19. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Silva, V.; Dapkevicius, M.d.L.E.; Caniça, M.; Tejedor-Junco, M.T.; Igrejas, G.; Poeta, P. Escherichia coli as Commensal and Pathogenic Bacteria among Food-Producing Animals: Health Implications of Extended Spectrum β-Lactamase (ESBL) Production. Animals 2020, 10, 2239. [Google Scholar] [CrossRef]
- Messele, Y.E.; Alkhallawi, M.; Veltman, T.; Trott, D.J.; McMeniman, J.P.; Kidd, S.P.; Low, W.Y.; Petrovski, K.R. Phenotypic and Genotypic Analysis of Antimicrobial Resistance in Escherichia Coli Recovered from Feedlot Beef Cattle in Australia. Animals 2022, 12, 2256. [Google Scholar] [CrossRef]
- WHO. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed; Geneva; 2017. Available online: https://www.who.int/zh/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 18 November 2022).
- Denissen, J.; Reyneke, B.; Waso-Reyneke, M.; Havenga, B.; Barnard, T.; Khan, S.; Khan, W. Prevalence of ESKAPE Pathogens in the Environment: Antibiotic Resistance Status, Community-Acquired Infection and Risk to Human Health. Int. J. Hyg. Environ. Health 2022, 244, 114006. [Google Scholar] [CrossRef]
- Patil, A.; Banerji, R.; Kanojiya, P.; Saroj, S.D. Foodborne ESKAPE Biofilms and Antimicrobial Resistance: Lessons Learned from Clinical Isolates. Pathog. Glob. Health 2021, 115, 339–356. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [PubMed]
- Arbab, S.; Ullah, H.; Wang, W.; Li, K.; Akbar, A.; Zhang, J. Isolation and Identification of Infection-Causing Bacteria in Dairy Animals and Determination of Their Antibiogram. J. Food Qual. 2021, 2021, 2958304. [Google Scholar] [CrossRef]
- Liu, J.; Niu, J.; Han, Z.; Bi, C.; Mehmood, K.; Farraj, D.A.A.; Alzaidi, I.; Iqbal, R.; Qin, J. Complete Genome of Multi-Drug Resistant Staphylococcus Aureus in Bovine Mastitic Milk in Anhui, China. Pak. Vet. J. 2023, 43, 456–462. [Google Scholar] [CrossRef]
- Habib, S.; Gibbon, M.J.; Couto, N.; Kakar, K.; Habib, S.; Samad, A.; Munir, A.; Fatima, F.; Mohsin, M.; Feil, E.J. The Diversity, Resistance Profiles and Plasmid Content of Klebsiella Spp. Recovered from Dairy Farms Located around Three Cities in Pakistan. Antibiotics 2023, 12, 539. [Google Scholar] [CrossRef]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef]
- Cella, E.; Giovanetti, M.; Benedetti, F.; Scarpa, F.; Johnston, C.; Borsetti, A.; Ceccarelli, G.; Azarian, T.; Zella, D.; Ciccozzi, M. Joining Forces against Antibiotic Resistance: The One Health Solution. Pathogens 2023, 12, 1074. [Google Scholar] [CrossRef] [PubMed]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]
- Arksey, H.; O’Malley, L. Scoping Studies: Towards a Methodological Framework. Int. J. Soc. Res. Methodol. Theory Pract. 2005, 8, 19–32. [Google Scholar] [CrossRef]
- Murphy, C.; Fajt, V.; Scott, H.; Foster, M.; Wickwire, P.; Mcewen, S. Scoping Review to Identify Potential Non-Antimicrobial Interventions to Mitigate Antimicrobial Resistance in Commensal Enteric Bacteria in North American Cattle Production Systems. Epidemiol. Infect. 2015, 144, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Worldometer. How Many Countries in Asia? Available online: https://www.worldometers.info/geography/how-many-countries-in-asia/ (accessed on 1 February 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).