Co-Colonization of Non-difficile Clostridial Species in Antibiotic-Associated Diarrhea Caused by Clostridioides difficile
Abstract
1. Introduction
2. Results
2.1. Characteristics of CDI Patients Included
2.2. Characteristics of CDI + NDC Group
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. CDI Case Selection
4.3. CDI Detection
4.4. Ethics Statement
4.5. Stool Culture
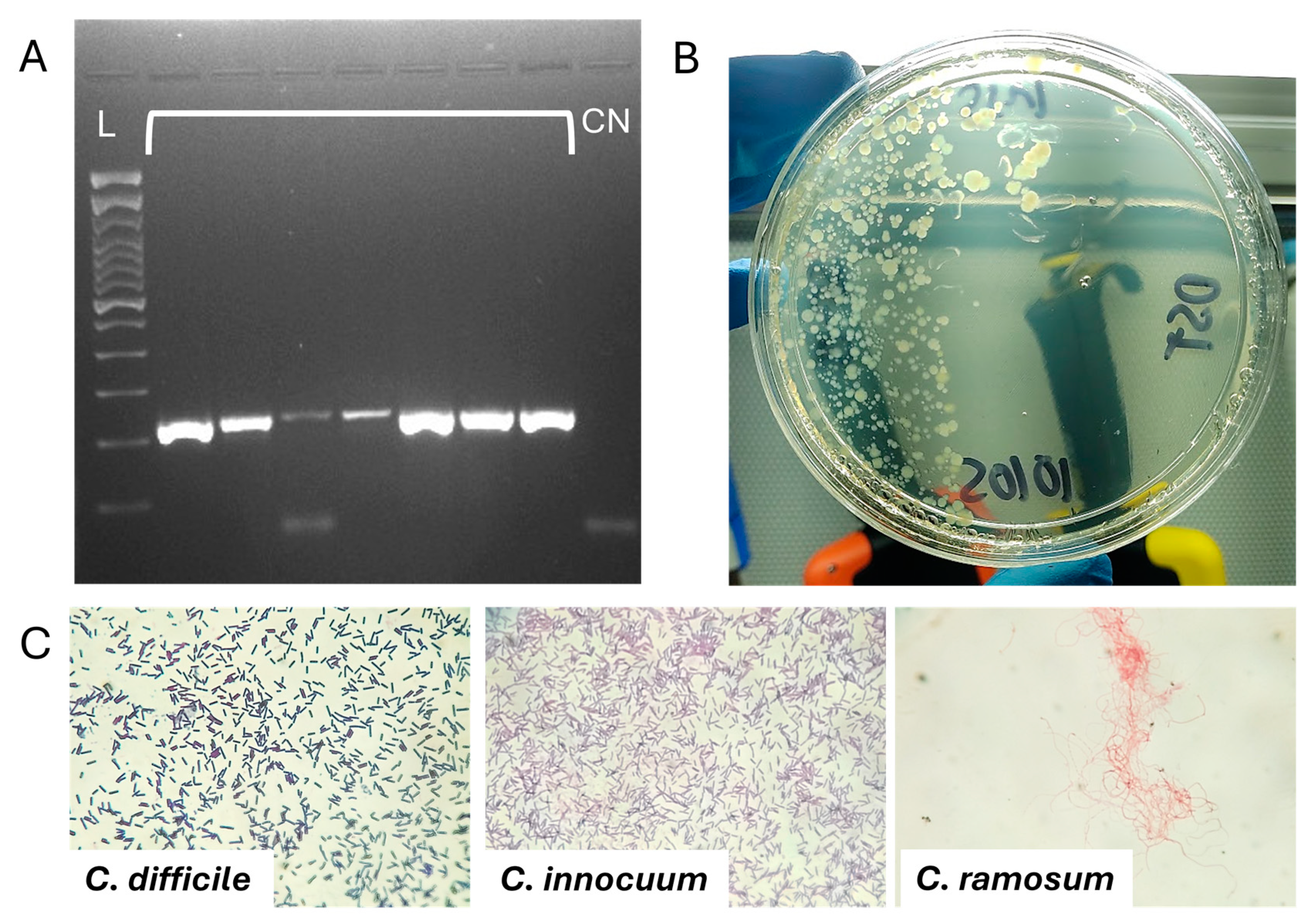
4.6. Clostridia Species Identification
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Motamedi, H.; Fathollahi, M.; Abiri, R.; Kadivarian, S.; Rostamian, M.; Alvandi, A. A worldwide systematic review and meta-analysis of bacteria related to antibiotic-associated diarrhea in hospitalized patients. PLoS ONE 2021, 16, e0260667. [Google Scholar] [CrossRef] [PubMed]
- Yutin, N.; Galperin, M.Y. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ. Microbiol. 2013, 15, 2631–2641. [Google Scholar] [CrossRef] [PubMed]
- Cherny, K.E.; Muscat, E.B.; Reyna, M.E.; Kociolek, L.K. Clostridium innocuum: Microbiological and clinical characteristics of a potential emerging pathogen. Anaerobe 2021, 71, 102418. [Google Scholar] [CrossRef] [PubMed]
- Lawson, P.A.; Perez, L.S.; Sankaranarayanan, K. Reclassification of Clostridium cocleatum, Clostridium ramosum, Clostridium spiroforme and Clostridium saccharogumia as Thomasclavelia cocleata gen. nov., comb. nov., Thomasclavelia ramosa comb. nov., gen. nov., Thomasclavelia spiroformis comb. nov. and Thomasclavelia saccharogumia comb. nov. Int. J. Syst. Evol. Microbiol. 2023, 73, 005694. [Google Scholar]
- Yamairi, K.; Niki, M.; Imoto, W.; Kuwabara, G.; Shibata, W.; Oshima, K.; Yamada, K.; Kaneko, Y.; Kakeya, H. Two cases of Clostridium ramosum bacteremia with intestinal perforation: The antimicrobial susceptibility of clinical strains. Anaerobe 2023, 80, 102695. [Google Scholar] [CrossRef]
- Shinzato, T.; Yonaha, T.; Oshiro, Y.; Ishiki, H. Clostridium ramosum bacteremia: A case series at a general acute care hospital. J. Infect. Chemother. 2023, 29, 78–81. [Google Scholar] [CrossRef]
- Milosavljevic, M.N.; Kostic, M.; Milovanovic, J.; Zaric, R.Z.; Stojadinovic, M.; Jankovic, S.M.; Stefanovic, S.M. Antimicrobial treatment of Erysipelatoclostridium ramosum invasive infections: A systematic review. Rev. Inst. Med. Trop. Sao Paulo 2021, 63, e30. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Le, P.-H.; Wang, Y.-H.; Chuang, T.-C.; Yeh, Y.-M.; Chiu, C.-H. Gut Colonization and Antibiotic-Associated Diarrhea by Clostridium innocuum in Children and Adults. Clin. Infect. Dis. 2022, 76, 369–371. [Google Scholar] [CrossRef]
- Luo, X.; Wang, X.; Wang, J.; Laranjo, M. Clostridium innocuum: More Important Than Ever. Int. J. Clin. Pract. 2024, 2024, 5797671. [Google Scholar] [CrossRef]
- Cobo, F.; Pérez-Carrasco, V.; Tarriño-León, M.; Aguilera-Franco, M.; García-Salcedo, J.A.; Navarro-Marí, J.M. Bacteremia due to Clostridium innocuum: Analysis of four cases and literature review. Anaerobe 2023, 83, 102771. [Google Scholar] [CrossRef]
- Haas, K.N.; Blanchard, J.L. Reclassification of the Clostridium clostridioforme and Clostridium sphenoides clades as Enterocloster gen. nov. and Lacrimispora gen. nov., including reclassification of 15 taxa. Int. J. Syst. Evol. Microbiol. 2020, 70, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Sermet, K.; Kipnis, E.; Duployez, C.; Wallet, F.; Dessein, R.; Le Guern, R. Answer to January 2022 Photo Quiz. J. Clin. Microbiol. 2022, 60, e00330-21. [Google Scholar] [CrossRef] [PubMed]
- Gajdács, M.; Spengler, G.; Urbán, E. Identification and Antimicrobial Susceptibility Testing of Anaerobic Bacteria: Rubik’s Cube of Clinical Microbiology? Antibiotics 2017, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Legaria, M.; García, S.; Tudanca, V.; Barberis, C.; Cipolla, L.; Cornet, L.; Famiglietti, A.; Stecher, D.; Vay, C. Clostridium ramosum rapidly identified by MALDI-TOF MS. A rare gram-variable agent of bacteraemia. Access Microbiol. 2020, 2, acmi000137. [Google Scholar] [CrossRef]
- Chiang-Ni, C.; Huang, J.-Y.; Hsu, C.-Y.; Lo, Y.-C.; Chen, Y.-Y.M.; Lai, C.-H.; Chiu, C.-H. Genetic diversity, biofilm formation, and Vancomycin resistance of clinical Clostridium innocuum isolates. BMC Microbiol. 2024, 24, 353. [Google Scholar] [CrossRef]
- Chia, J.-H.; Feng, Y.; Su, L.-H.; Wu, T.-L.; Chen, C.-L.; Liang, Y.-H.; Chiu, C.-H. Clostridium innocuum is a significant vancomycin-resistant pathogen for extraintestinal clostridial infection. Clin. Microbiol. Infect. 2017, 23, 560–566. [Google Scholar] [CrossRef]
- Bhattacharjee, D.; Flores, C.; Woelfel-Monsivais, C.; Seekatz, A.M. Diversity and Prevalence of Clostridium innocuum in the Human Gut Microbiota. mSphere 2022, 8, e00569-22. [Google Scholar] [CrossRef]
- Skinner, A.M.; Petrella, L.; Spandoni, S.; Serna-Perez, F.; Johnson, S. Can Clostridium innocuum Masquerade as Clostridioides difficile? Clin. Infect. Dis. 2022, 75, 1268–1269. [Google Scholar] [CrossRef]
- Cherny, K.; Balaji, A.; Mukherjee, J.; Goo, Y.; Hauser, A.; Ozer, E.; Satchell, K.; Bachta, K.; Kochan, T.; Mitra, S.; et al. Identification of Clostridium innocuum hypothetical protein that is cross-reactive with C. difficile anti-toxin antibodies. Anaerobe 2022, 75, 102555. [Google Scholar] [CrossRef]
- Cherny, K.E.; Muscat, E.B.; Balaji, A.; Mukherjee, J.; Ozer, E.A.; Angarone, M.P.; Hauser, A.R.; Sichel, J.S.; Amponsah, E.; Kociolek, L.K. Association Between Clostridium innocuum and Antibiotic-Associated Diarrhea in Adults and Children: A Cross-sectional Study and Comparative Genomics Analysis. Clin. Infect. Dis. 2022, 76, e1244–e1251. [Google Scholar] [CrossRef]
- Dehoux, P.; Marvaud, J.C.; Abouelleil, A.; Earl, A.M.; Lambert, T.; Dauga, C. Comparative genomics of Clostridium bolteae and Clostridium clostridioforme reveals species-specific genomic properties and numerous putative antibiotic resistance determinants. BMC Genom. 2016, 17, 819. [Google Scholar] [CrossRef] [PubMed]
- Stoeva, M.K.; Garcia-So, J.; Justice, N.; Myers, J.; Tyagi, S.; Nemchek, M.; McMurdie, P.J.; Kolterman, O.; Eid, J. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes 2021, 13, 1907272. [Google Scholar] [CrossRef] [PubMed]
- Cassir, N.; Benamar, S.; La Scola, B. Clostridium butyricum: From beneficial to a new emerging pathogen. Clin. Microbiol. Infect. 2016, 22, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Ariyoshi, T.; Hagihara, M.; Takahashi, M.; Mikamo, H. Effect of Clostridium butyricum on Gastrointestinal Infections. Biomedicines 2022, 10, 483. [Google Scholar] [CrossRef]
- Choi, M.H.; Kim, D.; Lee, K.H.; Kim, H.J.; Sul, W.J.; Jeong, S.H. Dysbiosis of the gut microbiota is associated with in-hospital mortality in patients with antibiotic-associated diarrhoea: A metagenomic analysis. Int. J. Antimicrob. Agents 2024, 64, 107330. [Google Scholar] [CrossRef]
- Lemee, L.; Dhalluin, A.; Testelin, S.; Mattrat, M.A.; Maillard, K.; Lemeland, J.F.; Pons, J.L. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (Toxin A), and tcdB (Toxin B) genes for toxigenic culture of Clostridium difficile. J. Clin. Microbiol. 2004, 42, 5710–5714. [Google Scholar] [CrossRef]
| Patient Characteristic | CDI + NDC (n = 7) | CDI (n = 21) | p-Value |
|---|---|---|---|
| Age (years) | 45.57 ± 23.02 | 45.33 ± 22.06 | 0.9807 a |
| Patient weight (kg) | 59.16 ± 9.55 | 72.38 ± 9.17 | 0.0030 a |
| BMI | 22.48 ± 4.21 | 26.41 ± 3.31 | 0.0179 a |
| Hospital LOS | 25.57 ± 17.90 | 32.00 ± 26.64 | 0.5592 a |
| Charlson Score | 2.85 ± 2.03 | 3.23 ± 3.46 | 0.7584 b |
| LOS in ICU | 0.83 ± 1.32 | 2.04 ± 6.02 | 0.7250 b |
| Bowel movements per day | 6.71 ± 2.81 | 4.90 ± 2.24 | 0.0967 a |
| Total leukocytes (Cel/µL) | 11.07 ± 4.05 | 12.11 ± 7.52 | 0.7324 a |
| Albumin (g/dL) | 2.11 ± 0.51 | 2.35 ± 0.77 | 0.6497 b |
| Creatinine (mg/dL) | 1.47 ± 1.51 | 3.47 ± 6.27 | 0.6027 b |
| ATLAS score | 3.28 ± 0.75 | 4.14 ± 1.59 | 0.1846 a |
| Days of antibiotic treatment | 11.14 ±1.95 | 8.65 ± 4.69 | 0.1888 a |
| Time to cessation of diarrhea | 5.57 ± 1.81 | 3.13 ± 0.65 | 0.0027 a |
| Patient Characteristic | CDI + NDC (n, %) | CDI (n, %) | OR (CI) | p-Value |
|---|---|---|---|---|
| Toxin A detectable | 6 (85.7) | 18 (100) | 0.11 (0.004–3.25) | 0.2800 |
| Toxin B detectable | 4 (57.1) | 16 (88.9) | 0.16 (0.02–1.35) | 0.1130 |
| PPIs | 6 (85.7) | 12 (57.1) | 4.50 (0.45–44.31) | 0.3642 |
| Leukocytes >16 K Cel/µL | 0 (0.0) | 8 (38.1) | 0.10 (0.005–2.10) | 0.0749 |
| Treatment with Metronidazole/Vancomycin | 3 (42.9) | 4 (19.0) | 3.18 (0.50–20.31) | 0.3183 |
| Treatment with Vancomycin | 5 (71.4) | 19 (90.5) | 0.26 (0.02–2.36) | 0.2530 |
| Antibiotic switch treatment | 2 (28.6) | 1 (5.6) | 6.80 (0.50–91.55) | 0.1796 |
| Previous Hospitalization | 7 (100) | 11 (52.4) | 13.70 (0.69–270.5) | 0.0302 |
| Hospitalization in ICU | 3 (42.9) | 4 (19.0) | 3.18 (0.50–20.31) | 0.3183 |
| Attributable mortality to CDI | 0 (0.0) | 3 (14.3) | 0.35 (0.01–7.69) | 0.5513 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salas-Treviño, D.; Flores-Treviño, S.; Cisneros-Rendón, C.; Domínguez-Rivera, C.V.; Camacho-Ortiz, A. Co-Colonization of Non-difficile Clostridial Species in Antibiotic-Associated Diarrhea Caused by Clostridioides difficile. Antibiotics 2025, 14, 397. https://doi.org/10.3390/antibiotics14040397
Salas-Treviño D, Flores-Treviño S, Cisneros-Rendón C, Domínguez-Rivera CV, Camacho-Ortiz A. Co-Colonization of Non-difficile Clostridial Species in Antibiotic-Associated Diarrhea Caused by Clostridioides difficile. Antibiotics. 2025; 14(4):397. https://doi.org/10.3390/antibiotics14040397
Chicago/Turabian StyleSalas-Treviño, Daniel, Samantha Flores-Treviño, Carlos Cisneros-Rendón, Cristian Valdemar Domínguez-Rivera, and Adrián Camacho-Ortiz. 2025. "Co-Colonization of Non-difficile Clostridial Species in Antibiotic-Associated Diarrhea Caused by Clostridioides difficile" Antibiotics 14, no. 4: 397. https://doi.org/10.3390/antibiotics14040397
APA StyleSalas-Treviño, D., Flores-Treviño, S., Cisneros-Rendón, C., Domínguez-Rivera, C. V., & Camacho-Ortiz, A. (2025). Co-Colonization of Non-difficile Clostridial Species in Antibiotic-Associated Diarrhea Caused by Clostridioides difficile. Antibiotics, 14(4), 397. https://doi.org/10.3390/antibiotics14040397







