Abstract
Colistin, a polymyxin antibiotic used as a last-resort treatment for serious infections, exhibits efficacy against multidrug-resistant organisms. Colistin-resistant bacteria limit the treatment options and increase the risk of untreatable infections. In this study, we investigated various antimicrobial-resistant Escherichia coli strains isolated from broiler cecal feces. In the primary screening using CHROMagar C3GR and ESBL, 147 E. coli isolates were obtained from 231 broiler cecal samples at five domestic poultry farms in Japan in 2024. Of the 147 isolates tested for antimicrobial susceptibility, 20 (13.6%) showed resistance to colistin. Moreover, whole-genome sequencing detected the colistin resistance gene, mcr-1.1 (phosphoethanolamine transferase), in the colistin-resistant E. coli strains isolated from the tested five poultry farms. Multilocus sequence typing revealed that all strains belonged to ST1485, indicating that the cloned strains had spread to multiple poultry farms. Subsequent core-genome comparison analysis with global ST1485 strains indicated that the ST1485 isolates in this study were highly identical, whereas the global strains were distinct. The complete genome sequence of BroCaecum-55 contained mcr-1.1 in a 62,716 bp IncI2 replicon plasmid (pBroCa-55-p2). In conclusion, mcr-1.1-positive colistin-resistant E. coli strains, which are rarely reported in Japan, were isolated from Japanese broilers, indicating that colistin resistance persisted even after the ban on colistin use as a feed additive in Japan in 2018. Our findings highlight the need for continuous monitoring of colistin-resistant bacteria in livestock to reduce the transmission risk to humans.
1. Introduction
Increased spread of antimicrobial-resistant bacteria and associated infections is a serious issue worldwide. The United Nations World Health Organization (WHO) estimated 700,000 deaths worldwide due to antimicrobial resistance (AMR) in 2019. In the absence of proper control, this number is estimated to increase to 10 million by 2050, exceeding the number of cancer-related deaths.
AMR is a major issue impacting humans, livestock, the environment, and the food industry. Although initially developed against human infectious diseases, antimicrobials are also widely used for animals, including livestock (cattle, pigs, and chickens), aquatic animals (farmed fish), and pet animals (dogs and cats). In 2021, the total amount of antimicrobials used in Japan [1] was 1795 tons, with 800 tons used as animal antimicrobials and 226 tons used as antimicrobial feed additives. Although it enhances the livestock production safety, antimicrobial use also increases the prevalence of antimicrobial-resistant bacteria in livestock.
WHO proposed a Global Action Plan on AMR at its General Assembly in May 2015, requesting all member countries to develop their own action plans within two years [2]. Therefore, the Ministerial Conference on Countermeasures to Combat Internationally Threatening Infectious Diseases held by the Cabinet Office in Japan revealed the Action Plan for AMR Control 2016–2020 in April 2016. Subsequently, the Action Plan for AMR Control 2023–2027 [3] was published based on the five pillars of the WHO Global Action Plan on AMR: (1) public awareness and education, (2) trend surveillance and monitoring, (3) infection prevention and control, (4) proper use of antimicrobial agents, and (5) research and development and drug discovery. Recently, international collaboration was added as the sixth pillar to the plan. The action plan outlines strategies and specific initiatives for all six areas.
The Action Plan for AMR Control emphasizes the importance of AMR control using a one-health approach. The AMR Action Plan 2023–2027 clearly highlights the importance of promoting AMR countermeasures and implementing trend surveys on antimicrobial-resistant bacteria isolated from humans, animals, food, and the environment. The recent spread of antimicrobial-resistant bacteria has posed major public health concerns on food hygiene, particularly livestock hygiene, in humans. In Japan, the Japanese Veterinary Antimicrobial Resistance Monitoring system monitors the antimicrobial use in livestock and AMR of field strains, foodborne pathogenic bacteria, and indicator bacteria. Additionally, the Japan Nosocomial Infections Surveillance system monitors the trends in and use of antimicrobial-resistant bacteria.
Previously, third-generation cephalosporin antibiotic ceftiofur was used to prevent bacterial infections during the vaccination of fertilized broiler eggs [4]. However, it increased the prevalence of third-generation cephem-resistant bacteria. Subsequently, the poultry industry in Quebec, Canada voluntarily restricted the use of ceftiofur, leading to the decrease in the prevalence of resistant bacteria [4]. However, the prevalence of resistant bacteria increased again upon the reintroduction of ceftiofur, indicating a causal relationship between increased cephem use and prevalence of cephem-resistant bacteria. Owing to concerns regarding the increased prevalence of cephalosporin-resistant bacteria, the poultry industry voluntarily restricted the use of ceftiofur in Japan in 2010.
Colistin (CL) exhibits bactericidal activity against gram-negative rods, such as Escherichia coli and Pseudomonas aeruginosa, and is among the few treatment options available for multidrug-resistant bacteria [5]. It is a critically important antimicrobial in human medicine. It is also widely used in veterinary medicine to prevent and treat infectious diseases and promote growth. However, its use in livestock has been re-evaluated in many countries owing to the emergence of plasmid-mediated CL resistance gene (mcr-1)-carrying gram-negative bacteria [6]. mcr-1 encodes a CL resistance protein by adding phosphoethanolamine to lipid A of lipopolysaccharide, thereby altering the polarity of the outer membrane and reducing its affinity for CL [6]. This gene was first reported as a novel factor for CL resistance in China in 2016 [6]. It is present on the plasmid and poses a transmission risk to various bacteria, including Enterobacteriaceae members [7]. The spread of this CL resistance gene to multidrug-resistant bacteria, such as multidrug-resistant Pseudomonas aeruginosa and Acinetobacter that do not respond to carbapenems, will make effective treatment challenging [8].
mcr-1-positive Salmonella and E. coli strains have been retrospectively identified in bovine mastitis and swine bacteremia cases in 2012 and 2013, respectively [9], by searching the whole-genome sequence data archives of Japan. mcr-1-positive E. coli have also been isolated from healthy food-producing animals [10], with approximately 10% positive detection of mcr-1 in broilers in 2014 and increasing detection thereafter. Pathogenic swine isolates exhibited high mcr-1 positivity (50%) in 2014 [11], and multiple mcr variants (mcr-1, -3, and -5) were detected in the diseased swine in Japan [12]. To date, CL resistance gene variants (mcr-1–10) have been mainly detected in Enterobacteriaceae members, such as E. coli, Salmonella, and Klebsiella pneumoniae [7]. In Japan, the mcr-1-positive E. coli clinical isolate, E. coli ST5702, was first detected in 2017 [13]. K. pneumoniae and E. coli clinical isolates have also been detected in other regions of Japan [14].
In Japan, the Food Safety Commission conducted a risk assessment in 2017 focusing on the prevalence of antimicrobial-resistant E. coli due to CL use and indicated the risk of CL resistance as “moderate”. Subsequently, the designation of CL as a feed additive was revoked and its use was prohibited by the Ministry of Agriculture, Forestry, and Fisheries based on the Risk Management Measures Guidelines for Antimicrobial Feed Additives on 1 July 2018 [15]. Additionally, “limited use as a second-line antimicrobial” risk management measure was proposed for CL as a veterinary antimicrobial based on the Risk Management Measures Guidelines for Veterinary Antimicrobials.
In this study, we examined the antimicrobial-resistant E. coli strains in the cecal stools of livestock, particularly broilers, and determined their transmission risk to humans.
2. Materials and Methods
2.1. Samples
According to the one-health approach, broiler cecal stools were selected for this study owing to the high antimicrobial use in livestock and ease of AMR analysis of the intestinal microbiota as cecal stools are small and less contaminated.
In total, 231 cecal stool samples of poultry from five poultry farms (A–E) collected by domestic meat hygiene laboratories in 2024 were included in this study. Specifically, 18, 67, 41, 61, and 44 fecal samples were obtained from poultry farms A, B, C, D, and E, respectively. The sample size depended on the shipping amount for each farm size.
2.2. Evaluation of Bacteria
2.2.1. Isolation and Identification of E. coli Strains
Specific E. coli strains were isolated using CHROMagar C3GR (bioMérieux, Marcy-l’Étoile, France) and tested using CHROMagar ESBL (bioMérieux) to detect the extended-spectrum β-lactamase (ESBL)- and AmpC β-lactamase-producing bacteria. The cecum of the broilers was aseptically incised (approximately 0.5 cm), and cecal feces were collected using a sterile swab. The feces were inoculated into a screening medium and incubated at 35 ± 1 °C for 16–18 h. E. coli strains were isolated for further screening. Basically, one strain was isolated from one broiler fecal sample, and multiple strains were isolated from the same fecal sample in 18 cases. The strains were further identified using API20E (bioMérieux) for Enterobacteriaceae and other gram-negative rods, according to the manufacturer’s instructions.
2.2.2. Antimicrobial Susceptibility
Next, minimum inhibitory concentrations (MICs) were determined using the broth microdilution method with the Clinical and Laboratory Standards Institute (CLSI)-compliant Dry Plate DP45 (Eiken Chemical Co., Ltd., Tokyo, Japan), according to the manufacturer’s instructions. Susceptible (S), intermediate (I), and resistant (R) strains were determined according to CLSI document M100-ED34 [16]. The following 2-fold serial broth dilutions of antimicrobials were tested in DP45: piperacillin (PIPC, 2–64 µg/mL), tazobactam (TAZ, 4 µg/mL constant)/PIPC (2–64 µg/mL), cefepime (0.5–16 µg/mL), ceftazidime (1–32 µg/mL), cefozopran (0.5–16 µg/mL), CL (1–4 µg/mL), fosfomycin (32–128 µg/mL), imipenem (0.5–16 µg/mL), meropenem (MEPM, 0.5–16 µg/mL), gentamicin (GM, 1–8 µg/mL), amikacin (AMK, 4–32 µg/mL), tobramycin (TOB, 1–8 µg/mL), minocycline (MINO, 1–8 µg/mL), levofloxacin (LVFX, 0.5–4 µg/mL), ciprofloxacin (CPFX, 0.25–2 µg/mL), doripenem (1–8 µg/mL), aztreonam (AZT, 2–16 µg/mL), and trimethoprim/sulfamethoxazole (TMP/SMX, 0.5/9.5–2/38 µg/mL).
2.2.3. PCR Testing of ESBL Genes
All obtained E. coli strains were tested for ESBL genes by the multiplex PCR method using the Cica Geneus ESBL Genotype Detection Kit 2 (Kanto Kagaku, Tokyo, Japan) covering the detection as follows: CTX-M-1 group, CTX-M-2 group, CTX-M-8 group, CTX-M-9 group, CTX-M-25 group, CTX-M chimera, ESBL-type GES, TEM, SHV.
2.3. Molecular Biology Techniques
2.3.1. DNA Extraction
DNA was extracted from E. coli strains for next-generation sequencing (NGS) analysis. E. coli suspensions were prepared in 1% TE sodium dodecyl sulfate and inactivated at 65 °C for 2 h. Then, the inactivated bacterial solution was used to purify the genomic DNA using the MinElute PCR Purification Kit (QIAGEN, Venlo, The Netherlands), according to the manufacturer’s instructions.
2.3.2. NGS Analysis
To determine the whole-genome sequences of the isolates, NGS libraries were constructed using the QIASeq FX DNA library kit (QIAGEN). To remove the adapter dimers interfering with NGS decoding, a library with an insert length of 350 bp was recovered via 1% TAE agarose electrophoresis and gel recovery. The library was sequenced via 250-mer paired-end MiSeq using the MiSeq Reagent Kit v2 (Illumina, San Diego, CA, USA) for 500 cycles.
Oxford Nanopore Technologies that provide long reads for complete genome determination were used. A library was constructed using the Ligation Sequencing gDNA-Native Barcoding Kit 24 V14 (Oxford Nanopore Technologies, Oxford, UK), according to the manufacturer’s instructions. Then, the library was loaded onto flow cell R10.4.1 in MinION Mk1b and analyzed using MinKNOW v24.11.8 (Oxford Nanopore Technologies).
2.3.3. Genome Informatics Analysis
CLC genome workbench v.24 was used for adaptor trimming of Illumina data and subsequent de novo assembly. Draft genome assembly was annotated using the DNA Fast Annotation and Structure Tool (https://dfast.ddbj.nig.ac.jp/, accessed on 15 November 2024) [17]. Then, hybrid assembly of the Illumina short reads and Nanopore long reads was performed using Unicycler v.0.4.8 [18]. AMR genes were detected using ResFinder v.4.6.0 (http://genepi.food.dtu.dk/resfinder, accessed on 15 November 2024) [19], and multilocus sequence typing (MLST; Achtman; https://pubmlst.org/multilocus-sequence-typing, accessed on 15 November 2024) was performed using the genome sequence. Sequence types (STs) were determined by comparing with the sequences of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) [20]. Pan-genome analysis was performed using Roary, which identifies the core and accessory genomes from the annotated data [21]. Core-genome analysis was performed using Parsnp v.1.7.4 (fast reference-based single nucleotide polymorphism [SNP] calling and core-genome alignment) to align the SNPs and core genomes from genome sequences [22]. A circular plasmid map was generated using Proksee (https://proksee.ca/, accessed on 8 January 2025) [23]. Additionally, comprehensive plasmid analysis was performed using the Plasmid Sequence Database (PLSDB) v2024_05_31_v2 (https://ccb-microbe.cs.uni-saarland.de/plsdb2025/, accessed on 20 January 2025) [24].
3. Results
3.1. Antimicrobial Susceptibility of the E. coli Isolates Obtained from Broiler Feces
To investigate third-generation cephalosporin-resistant E. coli strains in broilers, 231 broiler cecal samples were obtained from five domestic poultry farms (A, B, C, D, and E) in Japan in 2024. Using CHROMagar C3GR and ESBL, 147 isolates were obtained during primary screening. Notably, the 147 strains used in the analysis were isolated from individual broilers, but in 18 cases, multiple strains were isolated from a single individual. Specifically, 11 (61.1%) of 18 specimens, 30 (44.8%) of 67 specimens, 22 (53.7%) of 41 specimens, 28 (45.9%) of 61 specimens, and 32 (72.7%) of 44 specimens were obtained from the A, B, C, D, and E poultry farms, respectively.
Although the E. coli strains were obtained using CHROMagar C3GR and ESBL, MICs of the tested antimicrobials (Table S1) revealed that all 147 isolates were susceptible to TAZ/PIP, cefozopran, AZT, imipenem, meropenem, and doripenem. AMR rates were as follows: PIPC (64 isolates; 43.5%), cefepime (one isolate; 0.7%), ceftazidime (nine isolates; 6.1%), CL (20 isolates; 13.6%), fosfomycin (one isolate; 0.7%), GM (47 isolates; 31.9%), AMK (one isolate; 0.7%), TOB (12 isolates; 8.2%), MINO (10 isolates; 6.8%), LVFX (19 isolates; 12.9%), CPFX (20 isolates; 13.6%), and TMP/SMX (67 isolates; 45.6%). Many strains were resistant to aminoglycoside antimicrobial agents, such as GM, and some multidrug-resistant strains were resistant to fluoroquinolone antimicrobial agents. Thirteen isolates (8.8%) were resistant to aminoglycosides and fluoroquinolones, and 19 isolates (12.9%) were resistant to CL. Unexpectedly, PCR detection of ESBL using a multiplex PCR kit suggested that all strains were negative for CTX-M groups and ESBL-type GES.
Strains were classified as S, I, and R strains based on the MIC breakpoint suggested by CLSI M100 ED34 [16]. Subsequently, susceptibility profiles of the 147 isolates were clustered via UPGMA (Figure 1). Based on their susceptibility profiles, the isolates were classified into the following five major clusters (clusters 1–5; Figure 1) with specific resistance patterns: cluster 1 (resistant to GM and TMP/SMX), cluster 2 (almost susceptible), cluster 3 (resistant to GM and TOB), cluster 4 (resistant to PIPC, CL, and ST), and cluster 5 (resistant to PIPC, GM, TOB, MINO, LVFX, CPFX, and TMP/SMX). CL R or I phenotype was observed only in cluster 4, indicating strains in this cluster showed notable AMR in the livestock.
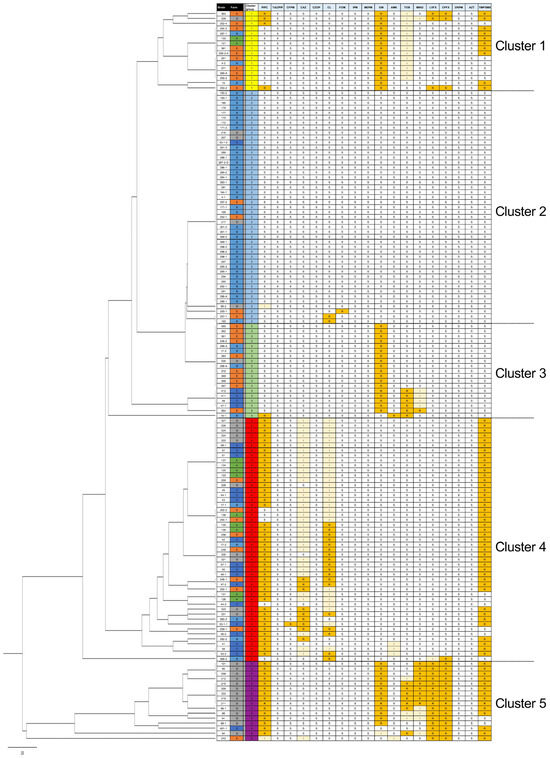
Figure 1.
Unweighted pair group method with arithmetic mean (UPGMA) clustering of the antimicrobial susceptibility profiles of 147 broiler-derived Escherichia coli isolates. Antimicrobial susceptibility was determined using the broth microdilution method, and the obtained antimicrobial profiles (susceptible [S], intermediate [I], and resistant [R]) were clustered via UPGMA. R and I strains are highlighted by orange and light orange background, respectively. Strains in cluster 4 showed a notable colistin (CL)-resistant phenotype (Table S1).
To characterize the AMR genes in notable CL-resistant strains, we performed whole-genome sequencing of the strains in cluster 4, with at least one strain from each poultry farm A, B, C, D, and E, respectively. Additionally, two CAZ-resistant strains (BroCaecum-53-1-1 and BroCaecum-323) were included, and in total, seven strains were selected for whole-genome analysis (Table 1).

Table 1.
Antimicrobial resistance genes in the genome sequences of Escherichia coli ST1485 strains identified in this study.
3.2. Whole-Genome Sequencing of Antimicrobial-Resistant E. coli Strains
Whole-genome sequencing revealed that seven strains belonged to ST1485 and were resistant to CL, harboring the CL resistance gene, mcr-1.1 (seven of seven isolates: 100%; Table 1). Notably, all strains harboring mcr-1.1 also carried blaCMY-2, aph(6)-Id, aph(3″)-Ib, qnrS1, sul2, and dfrA14 (Table 1).
3.3. Comparative Genome Analysis of E. coli ST1485 Strains
As the CL-resistant strains belonged to ST1485 (Table 1), we performed comparative genome analysis using ST1485 E. coli isolated worldwide using the PubMLST database (https://pubmlst.org/bigsdb?db=pubmlst_escherichia_isolates. acessed on 15 November 2024). Core-genome SNP analysis was performed using Parsnp v.1.7.4 [22]. Subsequently, SNP network analysis was performed using population analysis with reticulate trees v.1.7 software [25].
We observed over nine thousand nucleotide variations between the Japanese (BroCaecum-xxx) and China-3468 strains; however, both types belonged to ST1485 and were mcr-1.1-positive strains (Figure 2A). Other global strains also showed clear differences from the Japanese strain (BroCaecum-xxx; Figure 2A), indicating that the E. coli ST1485 are not the key genome feature to show the CL resistance with mcr-1.1.
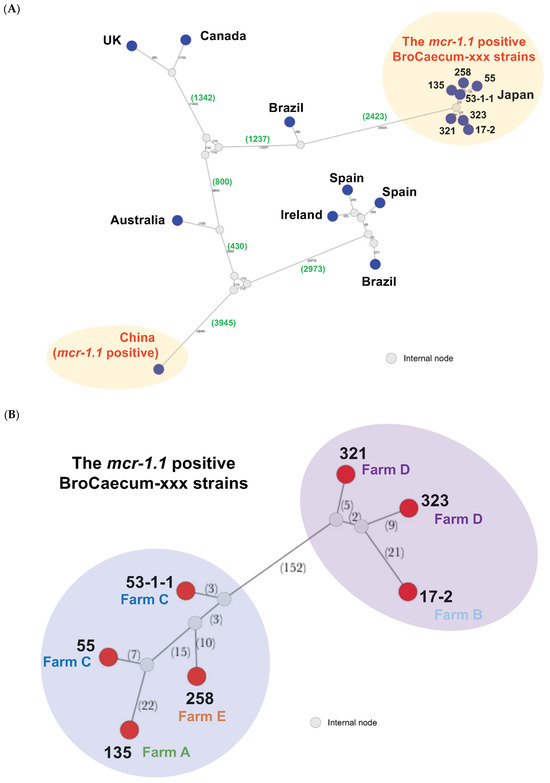
Figure 2.
Core-genome single nucleotide polymorphism (SNP) network analysis of E. coli ST1485 isolates, including the mcr-1.1-positive colistin-resistant isolates. (A) Global ST1485 strains available from pubMLST database were included for the core-genome analysis. Seven Japanese isolates (BroCaecum-xxx) and one strain from China were mcr-1.1-positive. Dark blue nodes indicate the locations where the isolates were detected, and hypothetical internal nodes are shown in gray. Numbers of genomic SNPs are indicated in parentheses. (B) Subsequent core-genome analysis for seven Japanese isolates (BroCaecum-xxx). Red circle nodes indicate the strains, and hypothetical internal nodes are shown in gray. Numbers of genomic SNPs are indicated in parentheses.
To evaluate the clonality of the Japanese strains (BroCaecum-xxx), SNP network analysis was performed using the seven mcr-1.1-positive CL-resistant E. coli strains isolated in this study. These seven isolates were clonal and classified into two clusters (cluster A: 17-2, 321, and 323; cluster B: 53-1-1, 55, 135, and 258), with 152 nucleotide variations (Figure 2B). Thus far, these clusters are unique to the E. coli ST1485 strain carrying mcr-1.1.
Similar to core-genome analysis, pan-genome analysis using Roary also indicated that the Japanese strain (BroCaecum-xxx) was different from the China-3468 strain due to the low similarity in their pan-genomes (Figure 3).
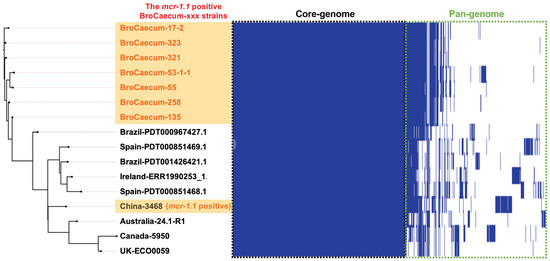
Figure 3.
Pan-genome analysis of E. coli ST1485 strains using Roary. The mcr-1.1-positive BroCaecum-xxx strains were closely clustered in both pan-genome and core-genome SNP analyses (Figure 2B).
3.4. Complete Genome Sequence of the mcr-1.1-Positive E. coli BroCaecum-55 Strain
To reveal the horizontal gene acquisition mechanism of mcr-1.1, the complete genome sequence of BroCaecum-55 (mcr-1.1-positive E. coli strain) was determined via hybrid assembly of the Illumina short reads (250-mer PE; 870,640 reads) and Nanopore long reads (mean length: 4897 bp; 559,728 reads; Table 2). Its circular complete genome sequence indicated that BroCaecum-55 contained four plasmids: 176,133 bp, 62,716 bp, 5875 bp, and 3373 bp (Table 2). Notably, mcr-1.1 was localized on the second largest plasmid with the IncI2 replicon (pBroCa-55-p2).

Table 2.
Complete genome sequence of the E. coli BroCaecum-55 strain.
The circular plasmid map of pBroCa-55-p2 in Figure 4 shows mcr-1.1 located at 18,135–19,760 bp. Zinc transporters involved in zinc tolerance were localized in the exogenous gene regions most likely acquired via horizontal gene transfer.
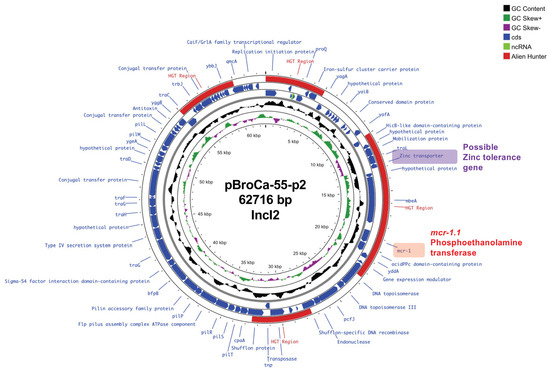
Figure 4.
Circular map of the pBroCa-55-p2 plasmid from the BroCaecum-55 strain was analyzed using Proksee. Different circles indicate the scale (kbp), GC skew, GC content (average 42.8%), coding sequence (CDS) in forward orientation, CDS in reverse orientation, potential horizontal gene transfer (HGT) region predicted by the Alien Hunter program, and CDS annotation.
The plasmid homology search using PLSDB revealed that pBroCa-55-p2 shared the highest identity (99.5%) with the E. coli plasmids (pEC507_2 and pEC521_1) isolated from wild boars in Japan (Kagoshima and Iwate Prefectures, respectively) in 2012.
4. Discussion
This study demonstrated the significantly high isolation efficiency (13.6%; 20 of 147 E. coli isolates) of CL-resistant E. coli strains from broiler feces; however, CL-resistant strains have not been detected in broilers since July 2018 due to the restrictions on CL use as a feed additive in Japan [15]. According to the annual report of the Japanese Veterinary Antimicrobial Resistance Monitoring system of Japan, CL resistance rate in broilers was 3.3% in 2017, 0% in 2018 and 2019, 0.8% (one of 121 isolates) in 2020, and 0% in 2021 [26] (Figure S1), indicating no increase in CL resistance rates. Moreover, CL resistance rate of swine E. coli in Japan was 2.4% in 2017, 6.0% in 2018, 2.5% in 2019, 2.2% in 2020, and 2.0% in 2021 [26] (Figure S1). In Japan, mcr-1 was detected in pigs before the 2019 study [27]. Notably, CL resistance and mcr-1 positivity rates in this study are higher than those reported in previous studies. In China, CL withdrawal policy in agriculture has significantly reduced CL resistance in animals and humans [28,29,30]. Therefore, the specific conditions in Japan are possibly due to increased CL resistance, as observed in this study.
In this study, mcr-1.1-positive CL-resistant E. coli strains were isolated from five poultry farms. MLST (Atchman) revealed that all strains belonged to ST1485. Comparative core-genome analysis indicated that the Japanese mcr-1.1-positive CL-resistant strains were different from other global ST1485 strains by several thousand SNPs (Figure 2A). Moreover, only a few SNPs were observed among the Japanese strains, indicating that they were clonal domestically disseminating strains in Japan (Figure 2B) that were not recently introduced from other countries. A plasmid sequence search against PLSDB revealed notable nucleotide identity (99.5%) between the pBroCa-55-p2 and Japanese wild boar E. coli plasmids (pEC507_2 and pEC521_1), further confirming the persistence of mcr-1-positive E. coli strains in Japanese livestock and wild animals.
CL use as a feed additive has been prohibited by law in Japan since 2018 [15]. Despite this, CL resistance is prevalent possibly due to the presence of a resistance gene close to mcr-1.1, which is essential for bacterial survival, on the plasmid, with mcr-1.1 consistently maintained via continued selection pressure. Here, we determined the complete genome sequence of BroCaecum-55 and found mcr-1.1 and the zinc transporter gene in the IncI2 replicon plasmid (pBroCa-55-p2; Figure 4). Zinc products, such as zinc oxide, are used as antimicrobial feed additives to reduce the impact of CL withdrawal [31]. As zinc transporters are involved in zinc tolerance, mcr-1.1 on the same plasmid is maintained via heavy metal selection, even when CL use is prohibited. In the veterinary field, Cu is used as a feed additive in swine at levels potentially leading to selection pressure on Enterobacterales. Arai et al. revealed that Salmonella 4,[5],12:i:-ST34 carrying copper tolerance genes shows high persistence and colonization of the intestine [32], leading to its recent dominance among Salmonella STs.
Except the CL resistance gene mcr-1.1, we could not isolate any ESBL-producing Enterobacterales in this study. In 2012, voluntary cessation of ceftiofur use by the Japanese poultry industry annually decreased the prevalence of cephalosporin-resistant Salmonella from 29.2% in 2012 to 10.5% in 2015 [33]. The cefotaxime resistance rate in broiler-derived E. coli further decreased annually from 4.7% in 2017 to 2.1% in 2021 [26] (Figure S1). Therefore, ESBL-producing bacteria are rarely detected in livestock. In contrast, the CPFX resistance rate has slightly increased annually from 12.0% in 2017 to 14.5% in 2021 [26] (Figure S1), warranting careful monitoring and control.
5. Conclusions
In this study, mcr-1.1-positive CL-resistant E. coli strains, which are rarely reported in Japan, were isolated from Japanese broilers, indicating their clonal distribution and prevalence in farms. Development of new antimicrobial agents has reduced the use of CL in clinical practice, thereby reducing the risk of CL resistance in humans. However, improper CL use may lead to a similar situation as that observed in 2015, with mcr-1.1 complicating the control of multidrug-resistant organisms. The sample size from five poultry farms (A–E) was very small in this study, warranting further investigations with larger sample sizes from various farms. In addition, as limitations of this study, longitudinal data collection and investigation of environmental and feed-related factors are needed to accurately assess CL resistance in Japan. Overall, our findings highlight the importance of the continuous monitoring of CL-resistant bacteria in livestock to reduce the transmission risk to humans.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antibiotics14040360/s1, Figure S1: Antimicrobial resistance (AMR) one-health approach for animals with antimicrobial-resistant Escherichia coli; Table S1: Antimicrobial susceptibility profiles of 147 Escherichia coli strains isolated in this study; Table S2: Whole-genome sequence information of E. coli ST1485 strains.
Author Contributions
Conceptualization, T.K. and M.K.; methodology, M.K.; validation, K.K.; formal analysis, K.K. and M.K.; investigation, K.K. and M.K.; resources, T.M.; data curation, K.K. and M.K.; writing—original draft preparation, T.K. and M.K.; writing—review and editing, T.K. and M.K.; visualization, M.K.; supervision, T.K. and M.K.; project administration, T.M. and M.K.; funding acquisition, T.K. and M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Research Program on Emerging and Reemerging Infectious Diseases of the Japan Agency for Medical Research and Development (grant number JP24fk0108666s0502), JSPS KAKENHI (grant number JP24K10568), and Kumamoto Health Science University Special Fellowship P&P (grant number 2022-C-02).
Institutional Review Board Statement
Not applicable. Broiler cecal feces were collected from the animals after slaughter.
Informed Consent Statement
Not applicable.
Data Availability Statement
All nucleotide sequence data analyzed in this study have been deposited into the DNA Data Bank of Japan (DDBJ) Sequence Read Archive under accession numbers PRJDB19974 and SAMD00874694–SAMD00874701. The complete and draft genome sequences have also been deposited into DDBJ and are listed in Table 2 and Table S2.
Acknowledgments
We would like to express our deepest gratitude to the Kumamoto Prefectural Meat Inspection Center for providing the materials used in this study and Johoku Livestock Health Center (Kumamoto, Japan) for providing useful information for this study. We would also like to thank the undergraduate students (Eika Nakagawa, Renma Nakajima, Hina Honda, Yui Matsuda, and Yukina Yokota) at our laboratory for their assistance with this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| MIC | Minimum inhibitory concentration |
| CLSI | Clinical and Laboratory Standards Institute |
| S | Susceptible |
| I | Intermediate |
| R | Resistant |
| ESBL | Extended-spectrum β-lactamase |
| PIPC | Piperacillin |
| TAZ/PIPC | Tazobactam/piperacillin |
| CFPM | Cefepime |
| CAZ | Ceftazidime |
| CZOP | Cefozopran |
| CL | Colistin |
| FOM | Fosfomycin |
| IPM | Imipenem |
| MEPM | Meropenem |
| GM | Gentamicin |
| AMK | Amikacin |
| TOB | Tobramycin |
| MINO | Minocycline |
| LVFX | Levofloxacin |
| CPFX | Ciprofloxacin |
| DRPM | Doripenem |
| AZT | Aztreonam |
| TMP/SMX | Trimethoprim/sulfamethoxazole |
| MLST | Multilocus sequence typing |
| ST | Sequence type |
| AMR | Antimicrobial resistance |
| ARGs | Antimicrobial resistant genes |
| JVARM | Japanese Veterinary Antimicrobial Resistance Monitoring System |
| JANIS | Japan Nosocomial Infections Surveillance |
| UPGMA | Unweighted pair group method with arithmetic mean |
| SNPs | Single nucleotide polymorphisms |
| PLSDB | Plasmid Sequence Database |
| NGS | Next-generation sequencing |
| DDBJ | DNA Data Bank of Japan |
References
- Antimicrobial Resistance (AMR) One Health Platform System for Animals Antimicrobial. Available online: https://amr-onehealth-platform.ncgm.go.jp/resistantBacteria/202 (accessed on 20 January 2025).
- World Health Organization. Global Action Plan on Antimicrobial Resistance. 2015. Available online: https://www.who.int/publications/i/item/9789241509763 (accessed on 20 January 2025).
- National Action Plan on Antimicrobial Resistance (AMR) (2023–2027) Japan. Available online: https://www.kantei.go.jp/jp/singi/kokusai_kansen/pdf/action_plan.pdf (accessed on 20 January 2025).
- Dutil, L.; Irwin, R.; Finley, R.; Ng, L.K.; Avery, B.; Boerlin, P.; Bourgault, A.M.; Cole, L.; Daignault, D.; Desruisseau, A.; et al. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg. Infect. Dis. 2010, 16, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Food Health Effects Assessment of Drug-Resistant Bacteria Related to Colistin Sulfate for Use in Livestock. Food Safety Commission Working Group on Drug-Resistant Bacteria. 2016. Available online: https://www.fsc.go.jp/senmon/sonota/amr_wg/amr_info.data/161122_colistin_draft_report.pdf (accessed on 20 January 2025).
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Martiny, H.M.; Munk, P.; Brinch, C.; Szarvas, J.; Aarestrup, F.M.; Petersen, T.N. Global Distribution of mcr Gene Variants in 214K Metagenomic Samples. mSystems 2022, 7, e0010522. [Google Scholar] [CrossRef] [PubMed]
- WHO. Critically Important Antimicrobials for Human Medicine: 6th Revision. Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 20 January 2025).
- Suzuki, S.; Ohnishi, M.; Kawanishi, M.; Akiba, M.; Kuroda, M. Investigation of a plasmid genome database for colistin-resistance gene mcr-1. Lancet Infect. Dis. 2016, 16, 284–285. [Google Scholar] [CrossRef]
- Kawanishi, M.; Abo, H.; Ozawa, M.; Uchiyama, M.; Shirakawa, T.; Suzuki, S.; Shima, A.; Yamashita, A.; Sekizuka, T.; Kato, K.; et al. Prevalence of Colistin Resistance Gene mcr-1 and Absence of mcr-2 in Escherichia coli Isolated from Healthy Food-Producing Animals in Japan. Antimicrob. Agents Chemother. 2017, 61, e02057-16. [Google Scholar] [CrossRef]
- Kusumoto, M.; Ogura, Y.; Gotoh, Y.; Iwata, T.; Hayashi, T.; Akiba, M. Colistin-Resistant mcr-1-Positive Pathogenic Escherichia coli in Swine, Japan, 2007–2014. Emerg. Infect. Dis. 2016, 22, 1315–1317. [Google Scholar] [CrossRef]
- Fukuda, A.; Sato, T.; Shinagawa, M.; Takahashi, S.; Asai, T.; Yokota, S.I.; Usui, M.; Tamura, Y. High prevalence of mcr-1, mcr-3 and mcr-5 in Escherichia coli derived from diseased pigs in Japan. Int. J. Antimicrob. Agents 2018, 51, 163–164. [Google Scholar] [CrossRef]
- Tada, T.; Uechi, K.; Nakasone, I.; Shimada, K.; Nakamatsu, M.; Kirikae, T.; Fujita, J. Emergence of a colistin-resistant Escherichia coli clinical isolate harboring mcr-1 in Japan. Int. J. Infect. Dis. 2017, 63, 21–22. [Google Scholar] [CrossRef]
- Tada, T.; Uechi, K.; Nakasone, I.; Nakamatsu, M.; Satou, K.; Hirano, T.; Kirikae, T.; Fujita, J. Emergence of IncX4 plasmids encoding mcr-1 in a clinical isolate of Klebsiella pneumoniae in Japan. Int. J. Infect. Dis. 2018, 75, 98–100. [Google Scholar] [CrossRef]
- Revocation of Designation of Feed Additive “Colistin Sulfate” in Ministry of Agriculture, Forestry and Fisheries Japan. Available online: https://www.maff.go.jp/j/syouan/tikusui/siryo/attach/pdf/additive-13.pdf (accessed on 20 January 2025).
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 34th Edition. Available online: https://clsi.org/standards/products/microbiology/documents/m100/ (accessed on 20 January 2025).
- Tanizawa, Y.; Fujisawa, T.; Kaminuma, E.; Nakamura, Y.; Arita, M. DFAST and DAGA: Web-based integrated genome annotation tools and resources. Biosci. Microbiota Food Health 2016, 35, 173–184. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Molano, L.G.; Hirsch, P.; Hannig, M.; Muller, R.; Keller, A. The PLSDB 2025 update: Enhanced annotations and improved functionality for comprehensive plasmid research. Nucleic Acids Res 2025, 53, D189–D196. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D.; Nakagawa, S. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar]
- Antimicrobial Resistance (AMR) One Health Platform System for Animals Antimicrobial-Resistance. Available online: https://amr-onehealth-platform.ncgm.go.jp/resistantBacteria/201 (accessed on 20 January 2025).
- Yoshizawa, N.; Hikoda-Kogiku, Y.; Tamamura-Andoh, Y.; Kusumoto, M. mcr-1 remains detectable in various Escherichia coli lineages isolated from healthy swine after withdrawal of colistin use on the farm. J. Vet. Med. Sci. 2023, 85, 536–540. [Google Scholar] [CrossRef]
- Shen, C.; Zhong, L.L.; Zhong, Z.; Doi, Y.; Shen, J.; Wang, Y.; Ma, F.; Ahmed, M.; Zhang, G.; Xia, Y.; et al. Prevalence of mcr-1 in Colonized Inpatients, China, 2011–2019. Emerg. Infect. Dis. 2021, 27, 2502–2504. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Zhang, R.; Chen, Y.; Shen, Y.; Hu, F.; Liu, D.; Lu, J.; Guo, Y.; Xia, X.; et al. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: An epidemiological comparative study. Lancet Infect. Dis. 2020, 20, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, T.; Wang, C.; Liang, G.; Lu, Q.; Wen, G.; Guo, Y.; Cheng, Y.; Wang, Z.; Shao, H.; et al. Prevalence of colistin resistance gene mcr-1 in Escherichia coli isolated from chickens in central China, 2014 to 2019. J. Glob. Antimicrob. Resist. 2022, 29, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Japanese Association of Swine Practicing Veterinarians (JASV). Collection of Case Studies on Efforts to De-antimicrobial Feed Additives. 2017. Available online: https://www.e-jasv.com/alic.pdf (accessed on 8 January 2025).
- Arai, N.; Shibahara, T.; Nishiura, R.; Tamamura-Andoh, Y.; Nishiura, H.; Muneta, Y.; Sawada, H.; Watanabe-Yanai, A.; Iwata, T.; Akiba, M.; et al. ICEmST contributes to colonization of Salmonella in the intestine of piglets. Sci. Rep. 2024, 14, 31407. [Google Scholar] [CrossRef]
- Shigemura, H.; Matsui, M.; Sekizuka, T.; Onozuka, D.; Noda, T.; Yamashita, A.; Kuroda, M.; Suzuki, S.; Kimura, H.; Fujimoto, S.; et al. Decrease in the prevalence of extended-spectrum cephalosporin-resistant Salmonella following cessation of ceftiofur use by the Japanese poultry industry. Int. J. Food Microbiol. 2018, 274, 45–51. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).