Antibacterial and Antibiofilm Activities of Hydralazine, an Antihypertensive Drug: In Vitro and In Silico Approaches
Abstract
1. Introduction
2. Results
2.1. Microdilution Test
2.2. HDZ Effect on Growth Curve
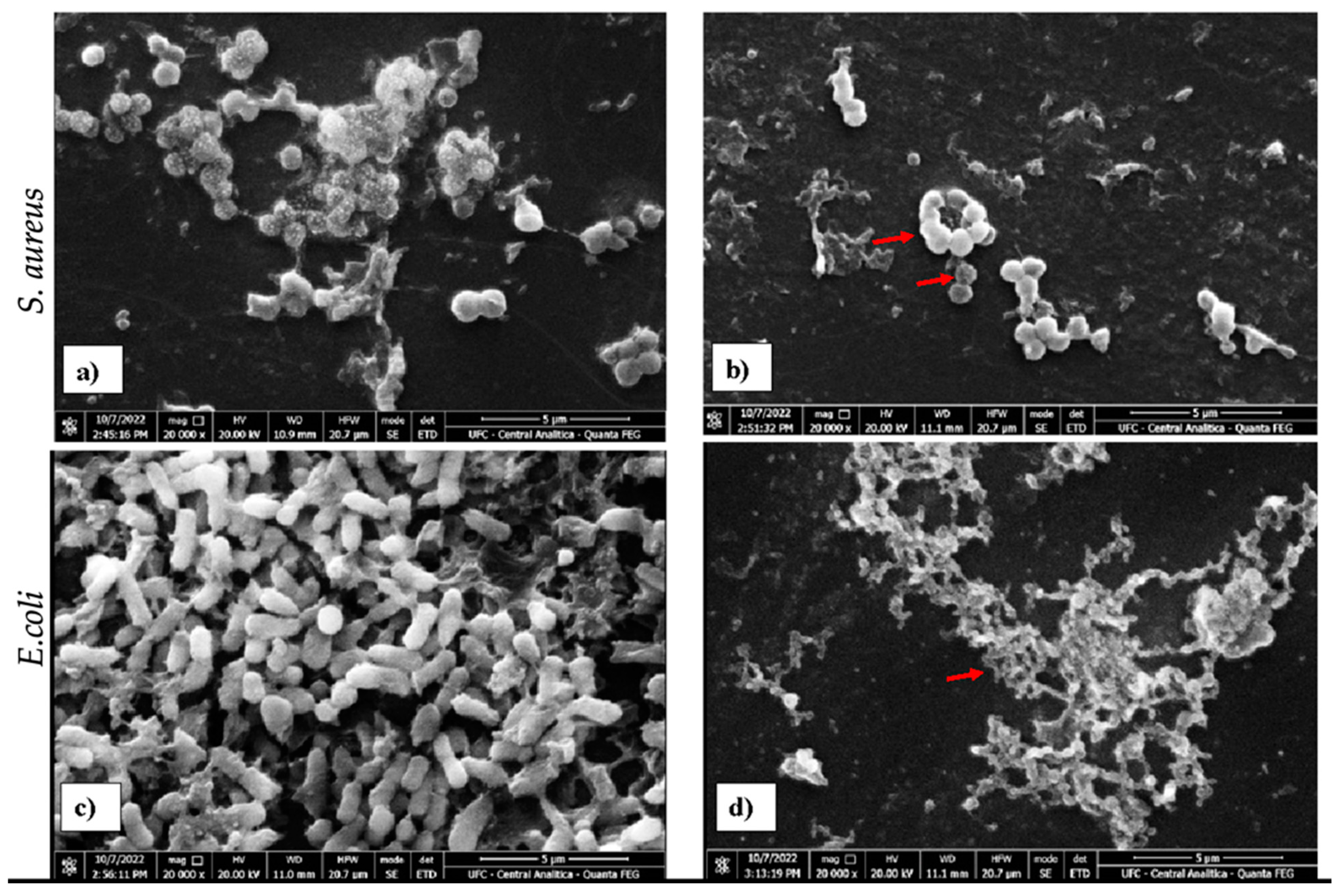
2.3. Scanning Electron Microscopy
2.4. Modulatory Effect of Hydralazine and Gentamicin Combined
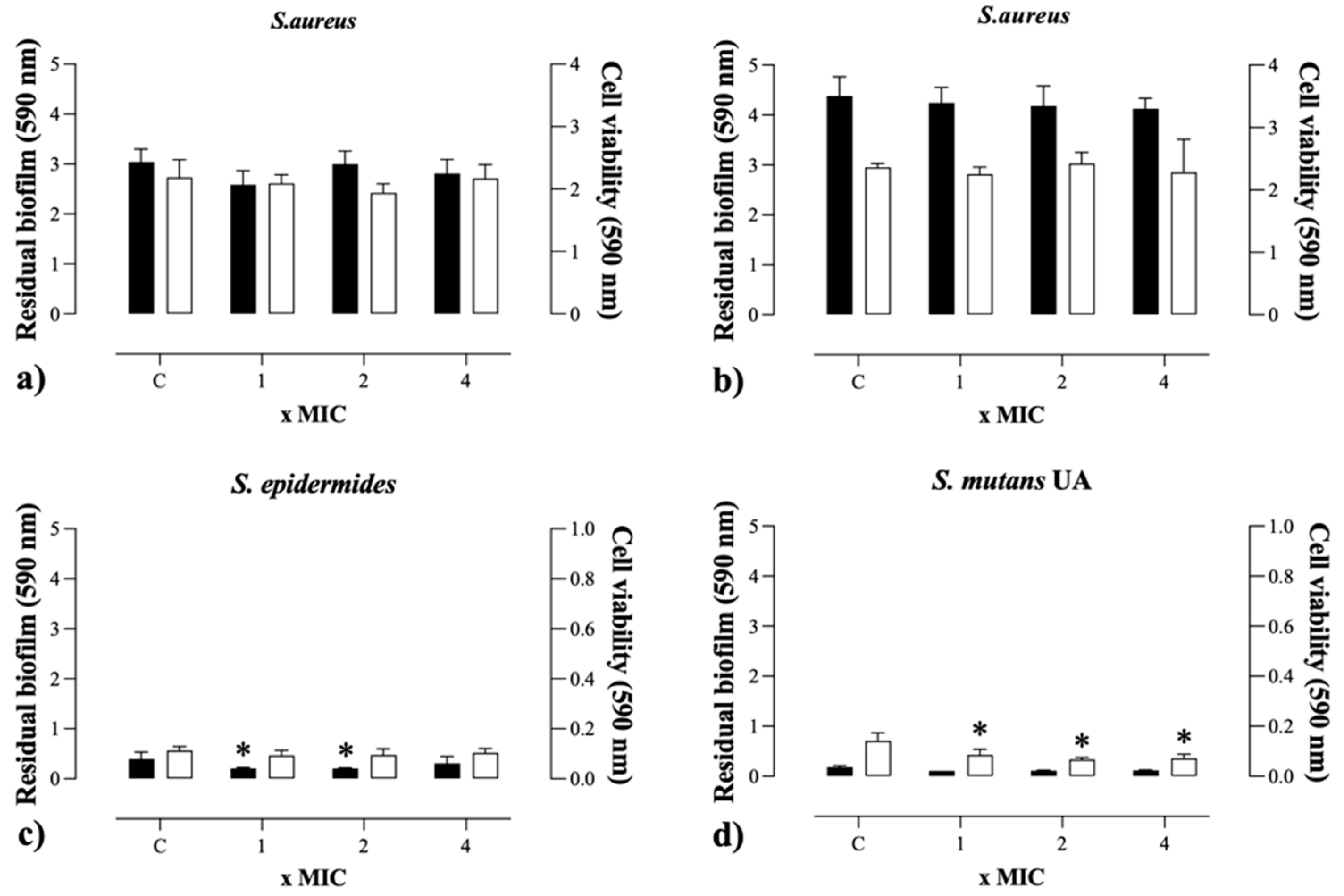
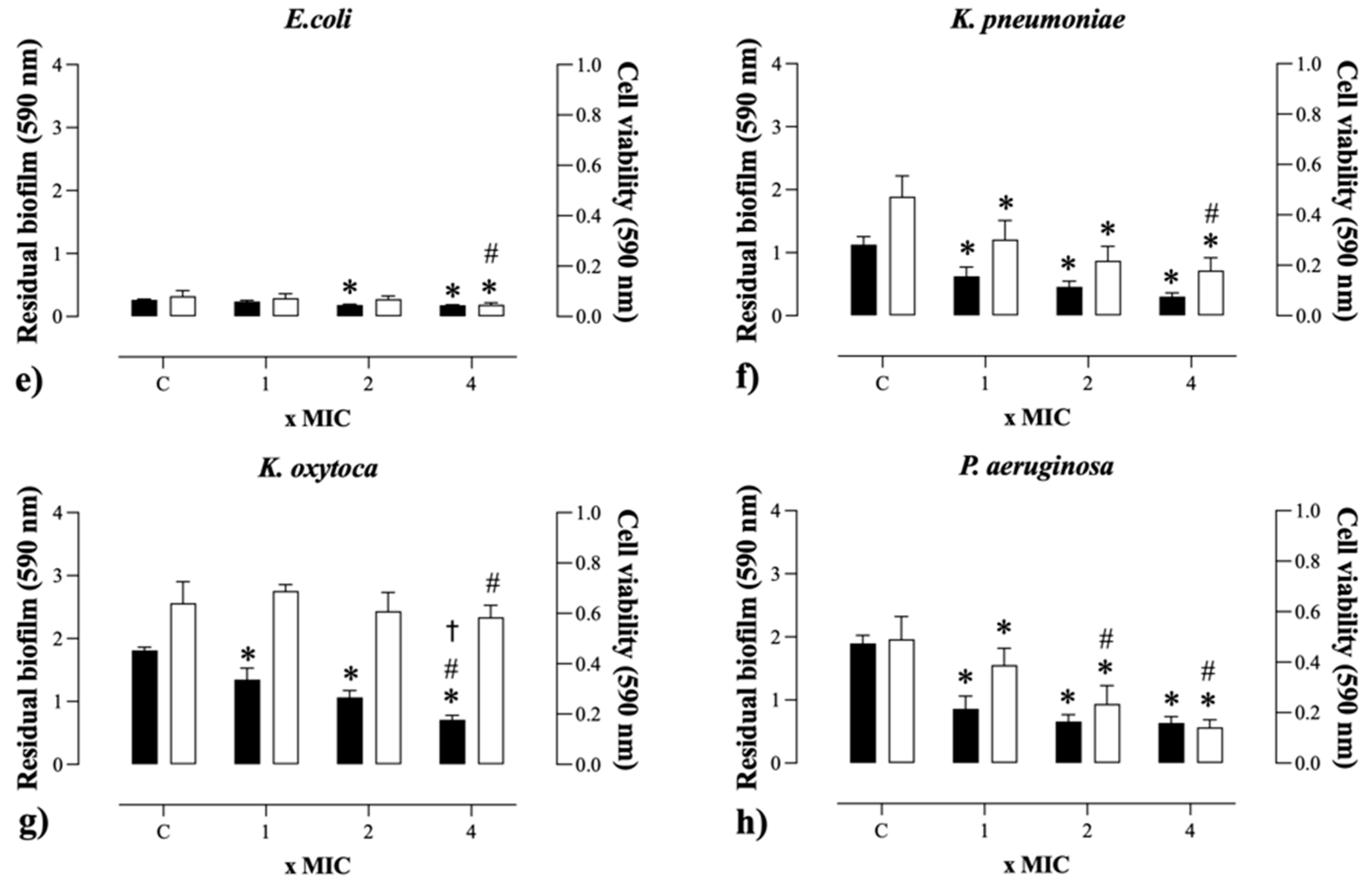
2.5. Antibiofilm Activity
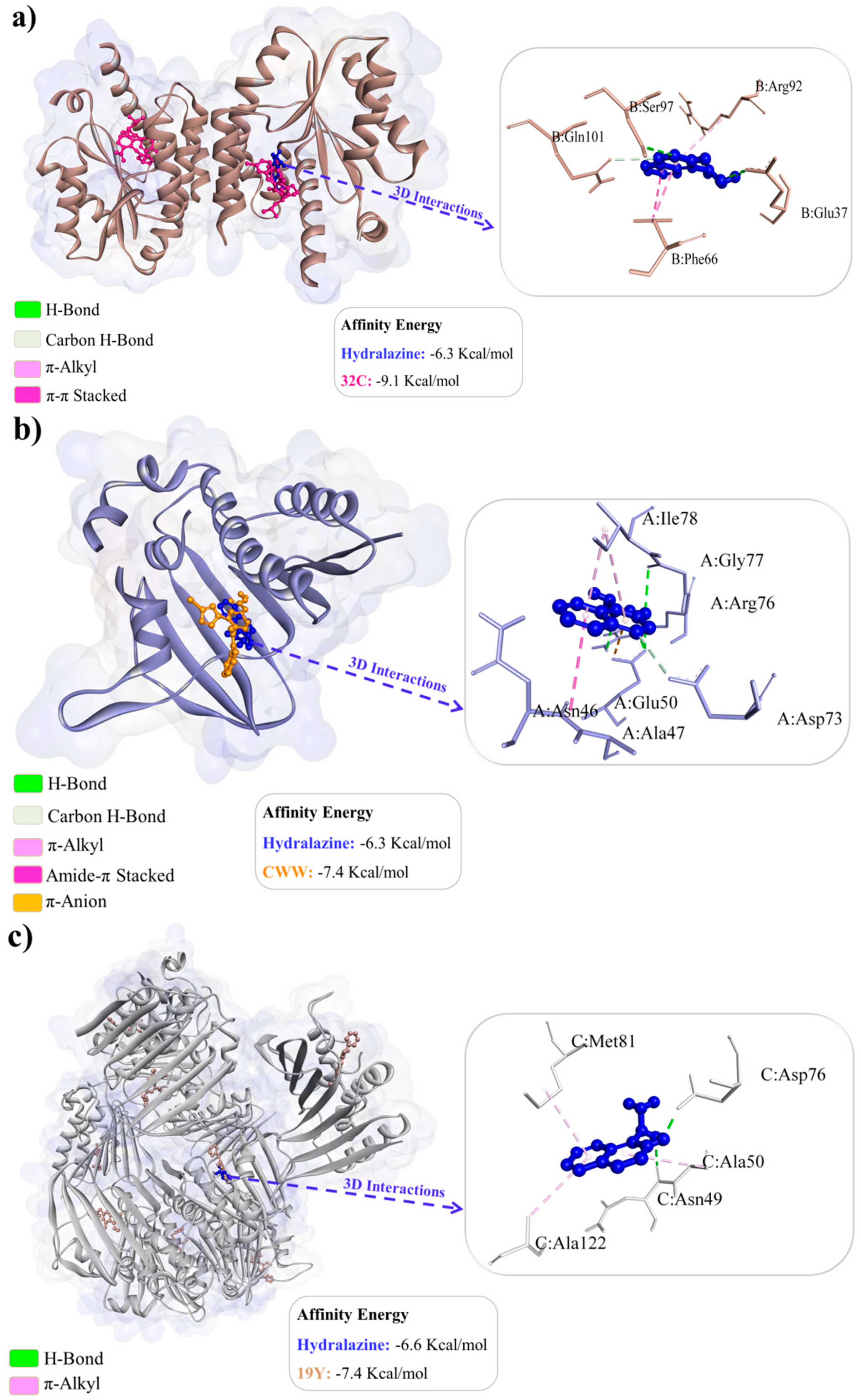
2.6. Molecular Docking
3. Discussion
4. Materials and Methods
4.1. Antibacterial Solution
4.2. Microorganisms and Culture Conditions
4.3. Antibacterial Activity
4.4. Kinetic Growth Assay
4.5. Scanning Electron Microscopy
4.6. Checkerboard Assay
4.7. Antibiofilm Activity
4.8. Molecular Docking
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- David, V.D.; David, P. Multidrug Resistant Bacteria in the Community: Trends and Lessons Learned. Infect. Dis. Clin. N. Am. 2016, 30, 377–390. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Spellberg, B.; Blaser, M.; Guidos, R.J.; Helen, W.B.; John, S.B.; Barry, I.E.; Dale, G.; Lynfield, R.; Reller, L.B.; John, R.; et al. Combating antimicrobial resistance: Policy recommendations to save lives. Clin. Infect. Dis. 2011, 52, 397–428. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Ciofu, O.; Molin, S.; Givskov, M.; Hoiby, N. Applying insights from biofilm biology to drug development—Can a new approach be developed? Nat. Rev. Drug. Discov. 2013, 12, 791–808. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Sarkar, S.; Das, B.; Bhattacharjee, S.; Tribedi, P. Biofilm, pathogenesis and prevention--a journey to break the wall: A review. Arch. Microbiol. 2016, 198, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Sutherland, I. Biofilm exopolysaccharides: A strong and sticky framework. Microbiology 2001, 147, 3–9. [Google Scholar] [CrossRef]
- Bottalico, L.; Charitos, I.A.; Potenza, M.A.; Montagnani, M.; Santacroce, L. The war against bacteria, from the past to present and beyond. Expert. Rev. Anti-Infect. Ther. 2022, 20, 681–706. [Google Scholar] [CrossRef]
- Han, L.W.; Ryu, R.J.; Cusumano, M.; Easterling, T.R.; Phillips, B.R.; Risler, L.J.; Shen, D.D.; Hebert, M.F. Effect of N-Acetyltransferase 2 Genotype on the Pharmacokinetics of Hydralazine During Pregnancy. J. Clin. Pharmacol. 2019, 59, 1678–1689. [Google Scholar] [CrossRef]
- Langedijk, J.; Mantel-Teeuwisse, A.K.; Slijkerman, D.S.; Schutjens, M.H.D.B. Drug Repositioning and Repurposing: Terminol-ogy and Definitions in Literature. Drug Discov. Today 2015, 20, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Oprea, T.I.; Mestres, J. Drug Repurposing: Far Beyond New Targets for Old Drugs. AAPS J. 2012, 14, 759–763. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug Repurposing: Progress, Challenges and Recommendations. Nat. Rev. Drug Discov. 2018, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Qamar, A.S.; Zamir, A.; Khalid, S.; Ashraf, W.; Imran, I.; Hussain, I.; Rehman, A.U.; Saeed, H.; Majeed, A.; Alqahtani, F.; et al. A Review on the Clinical Pharmacokinetics of Hydralazine. Expert Opin. Drug Metab. Toxicol. 2022, 18, 707–714. [Google Scholar] [CrossRef]
- Xue, H.; Li, J.; Xie, H.; Wang, Y. Review of Drug Repositioning Approaches and Resources. Int. J. Biol. Sci. 2018, 14, 1232–1244. [Google Scholar] [CrossRef] [PubMed]
- Arce, C.; Segura-Pacheco, B.; Perez-Cardenas, E.; Taja-Chayeb, L.; Candelaria, M.; Dueñnas-Gonzalez, A. Hydralazine target: From blood vessels to the epigenome. J. Transl. Med. 2006, 4, 10. [Google Scholar] [CrossRef]
- Segura-pacheco, B.; Trejo-becerril, C.; Perez-cardenas, E.; Taja-chayeb, L.; Mariscal, I.; Chavez, A.; Acun, C.; Salazar, A.M.; Lizano, M.; Duen, A. Reactivation of Tumor Suppressor Genes by the Cardiovascular Drugs Hydralazine and Procainamide and Their Potential Use in Cancer Therapy. Clin. Cancer Res. 2003, 9, 1596–1603. [Google Scholar]
- Münzel, T.; Kurz, S.; Rajagopalan, S.; Thoenes, M.; Berrington, W.R.; Thompson, J.A.; Freeman, B.A.; Harrison, D.G. Hydralazine prevents nitroglycerin tolerance by inhibiting activation of a membrane-bound NADH oxidase. A new action for an old drug. J. Clin. Investig. 1996, 98, 1465–1470. [Google Scholar] [CrossRef]
- Chang, T.-T.; Chen, J.-W. Potential Impacts of Hydralazine as a Novel Antioxidant on Cardiovascular and Renal Disease—Beyond Vasodilation and Blood Pressure Lowering. Antioxidants 2022, 11, 2224. [Google Scholar] [CrossRef]
- Steven, S.; Münzel, T.; Daiber, A. Exploiting the Pleiotropic Antioxidant Effects of Established Drugs in Cardiovascular Disease. Int. J. Mol. Sci. 2015, 16, 18185–18223. [Google Scholar] [CrossRef]
- Chhunchha, B.; Kubo, E.; Krueger, R.R.; Singh, D.P. Hydralazine Revives Cellular and Ocular Lens Health-Span by Ameliorating the Aging and Oxidative-Dependent Loss of the Nrf2-Activated Cellular Stress Response. Antioxidants 2023, 12, 140. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sargisson, O.; Nguyen, D.T.; Parker, K.; Pyke, S.J.R.; Alramahi, A.; Thihlum, L.; Fang, Y.; Wallace, M.E.; Berzins, S.P.; et al. Effect of Hydralazine on Angiotensin II-Induced Abdominal Aortic Aneurysm in Apolipoprotein E-Deficient Mice. Int. J. Mol. Sci. 2023, 24, 15955. [Google Scholar] [CrossRef]
- Romão, I.C.; Siqueira, S.M.C.; Silva Abreu, F.O.M.d.; Santos, H.S.d. Hydralazine and Hydrazine Derivatives: Properties, Applications, and Repositioning Potential. Chem. Biodiversity 2024, e202401561. [Google Scholar] [CrossRef] [PubMed]
- Farha, M.A.; Brown, E.D. Drug Repurposing for Antimicrobial Discovery. Nat. Microbiol. 2019, 4, 565–577. [Google Scholar] [CrossRef]
- Zhang, W.; Ran, J.; Shang, L.; Zhang, L.; Wang, M.; Fei, C.; Chen, C.; Gu, F.; Liu, Y. Niclosamide as a repurposing drug against Gram-positive bacterial infections. J. Antimicrob. Chemother. 2022, 28, 3312–3320. [Google Scholar] [CrossRef]
- Liu, L.; Yu, J.; Shen, X.; Cao, X.; Zhan, Q.; Guo, Y.; Yu, F. Resveratrol enhances the antimicrobial effect of polymyxin B on Klebsiella pneumoniae and Escherichia coli isolates with polymyxin B resistance. BMC Microbiol. 2020, 20, 306. [Google Scholar] [CrossRef]
- Bartzatt, R.; Sule, P.; Galbadage, T.; Cirillo, J.D. Aromatic Hydrazide Compounds That Inhibit the Growth of Mycobacterium tuberculosis. J. Adv. Med. Pharm. Sci. 2020, 21, 1–11. [Google Scholar] [CrossRef]
- Agnihotri, S.; Mukherji, S.; Mukherji, S. Size-Controlled Silver Nanoparticles Synthesized over the Range 5-100 nm Using the Same Protocol and Their Antibacterial Efficacy. RSC Adv. 2014, 4, 3974–3983. [Google Scholar] [CrossRef]
- Vasconcelos, B.M.; Pereira, A.M.G.; Coelho, P.A.T.; Cavalcante, R.M.B.; Carneiro-Torres, D.S.; Bandeira, P.N.; Silva, F.F.; Rodrigues, T.H.S.; Gomes, G.A.; Carneiro, V.A. Enhancement of chlorhexidine activity against planktonic and biofilm forms of oral streptococci by two Croton spp. essential oils from the caatinga biome. Biofouling 2022, 38, 984–993. [Google Scholar] [CrossRef]
- Mingeot-Leclercq, M.-P.; Glupczynski, Y.; Tulkens, P.M. Aminoglycosides: Activity and resistance. Antimicrob. Agents Chemother. 1999, 43, 727–737. [Google Scholar] [CrossRef]
- White, R.L.; Burgess, D.S.; Manduru, M.; Bosso, J.A. Comparison of three different in vitro methods of detecting synergy: Time-kill, checkerboard, and E test. Antimicrob. Agents Chemother. 1996, 40, 1914–1918. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.; Mathews, K.H.; Pulanic, D.; Falconer, R.; Rudan, I.; Campbell, H.; Nair, H. Risk factors for severe acute lower respiratory infections in children: A systematic review and meta-analysis. Croat. Med. J. 2013, 54, 110–121. [Google Scholar] [CrossRef]
- Deja, E. Aminoglycoside antibiotics: Trying to find a place in this world. Clin. Microbiol. Newsl. 2024, 46, 4–10. [Google Scholar] [CrossRef]
- Kumar, K.A.; Mazumdar, K.; Dutta, N.K.; Karak, P.; Dastidar, S.G.; Ray, R. Evaluation of Synergism between the Aminoglycoside Antibiotic Streptomycin and the Cardiovascular Agent Amlodipine. Biol. Pharm. Bull. 2004, 27, 1116–1120. [Google Scholar] [CrossRef]
- Souza, C.P.d.; Mendes, N.M. Repovoamento de criadouros de Biomphalaria glabrata após tratamento com niclosamida. Rev. Inst. Med. Trop. 1991, 33, 297–302. [Google Scholar] [CrossRef]
- Vestergaard, M.; Ingmer, H. Propriedades antibacterianas e antifúngicas do resveratrol. Rev. Int. Agentes Antimicrob. 2019, 53, 716–723. [Google Scholar] [CrossRef]
- Kawatkar, S.P.; Keating, T.A.; Olivier, N.B.; Breen, J.N.; Green, O.M.; Guler, S.Y.; Hentemann, M.F.; Loch, J.T.; McKenzie, A.R.; Newman, J.V.; et al. Antibacterial inhibitors of Gram-positive thymidylate kinase: Structure-activity relationships and chiral preference of a new hydrophobic binding region. J. Med. Chem. 2014, 57, 4584–4597. [Google Scholar] [CrossRef]
- Narramore, S.; Stevenson, C.E.M.; Maxwell, A.; Lawson, D.M.; Fishwick, C.W.G. New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase. Bioorg. Med. Chem. 2019, 27, 3546–3550. [Google Scholar] [CrossRef]
- Leslie, W.T.; Michael, T.; Daniel, C.B.; Xiaoming, L.; Zhiyong, C.; Thanh, L.; Junhu, Z.; Christopher, J.C.; Mark, L.C.; Bryan, K.; et al. Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. Bioorg. Med. Chem. Lett. 2013, 23, 1529–1536. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; CLSI Document M07-A10; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Field, D.; Quigley, L.; O’Connor, P.M.; Rea, M.C.; Daly, K.; Cotter, P.D.; Hill, C.; Ross, R.P. Studies with bioengineered Nisin peptides highlight the broad-spectrum potency of Nisin V. Microb Biotechnol. 2010, 3, 473–486. [Google Scholar] [CrossRef]
- Black, C.; Al Mahmud, H.; Howle, V.; Wilson, S.; Smith, A.C.; Wakeman, C.A. Development of a Polymicrobial Checkerboard Assay as a Tool for Determining Combinatorial Antibiotic Effectiveness in Polymicrobial Communities. Antibiotics 2023, 12, 1207. [Google Scholar] [CrossRef] [PubMed]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). EUCAST Definitive Document E.DEF 2.1, August 2000: Determination of antimicrobial susceptibility test breakpoints. Clin. Microbiol. Infect. 2000, 6, 570–572. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shen, G.; Yang, L.; Lv, X.; Zhang, Y.; Hou, X.; Li, M.; Zhou, M.; Pan, L.; Chen, A.; Zhang, Z. Antibiofilm Activity and Mechanism of Linalool against Food Spoilage Bacillus amyloliquefaciens. Int. J. Mol. Sci. 2023, 24, 10980. [Google Scholar] [CrossRef]
- Slobodníková, L.; Fialová, S.; Rendeková, K.; Kováč, J.; Mučaji, P. Antibiofilm Activity of Plant Polyphenols. Molecules 2016, 21, 1717. [Google Scholar] [CrossRef] [PubMed]
- Csizmadia, P. MarvinSketch and MarvinView: Molecule Applets for the World Wide Web. In The 3rd International Electronic Conference on Synthetic Organic Chemistry, 1–30 September 1999; MDPI: Basel, Switzerland, 1999; p. 1775. [Google Scholar]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Halgren, T.A. Merck Molecular Force Field. I. Basis, Form, Scope, Parameterization, and Performance of MMFF94. J. Comput. Chem. 1994, 17, 490–519. [Google Scholar] [CrossRef]
- João, B.A.N.; Vitória, P.F.C.; Lavouisier, F.B.N.; Cecília, R.S.; Lívia, G.A.V.S.; Anderson, R.S.; Wildson, M.B.S.; Jacilene, S.; Emmanuel, S.M.; Bruno, C.C.; et al. Anti-MRSA activity of curcumin in planktonic cells and biofilms and determination of possible action mechanisms. Microb. Pathog. 2021, 155, 104892. [Google Scholar] [CrossRef]
- Abo-Salem, H.M.; Abd El Salam, H.A.; Abdel-Aziem, A.; Abdel-Aziz, M.S.; El-Sawy, E.R. Synthesis, Molecular Docking, and Biofilm Formation Inhibitory Activity of Bis(Indolyl)Pyridines Analogues of the Marine Alkaloid Nortopsentin. Molecules 2021, 26, 4112. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, G.; Pan, J.; Wang, Y. α-Glucosidase inhibition by luteolin: Kinetics, interaction and molecular docking. Int. J. Biol. Macromol. 2014, 64, 213–223. [Google Scholar] [CrossRef]
- Huey, R.; Morris, G.M.; Forli, S. Using AutoDock 4 and AutoDock vina with AutoDockTools: A Tutorial; The Scripps Research Institute Molecular Graphics Laboratory: La Jolla, CA, USA, 2012; p. 92037. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Marinho, E.M.; Batista, A.N.J.; Silva, J.; Rocha, S.C.; Cavalcanti, B.C.; Marinho, E.S.; Nobre, J.H.V. Virtual screening based on molecular docking of possible inhibitors of COVID-19 main protease. Microb Pathog. 2020, 148, 104365. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, D.; Davis, A.M.; Kleywegt, G.J.; Schmitt, S. An Alternative Method for the Evaluation of Docking Performance: RSR vs RMSD. J. Chem. Inf. Model. 2008, 48, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Shityakov, S.; Förster, C. In silico predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter. Adv. Appl. Bioinform. Chem. 2014, 7, 23–36. [Google Scholar] [CrossRef]
- Silva, J.; Rocha, M.N.; Marinho, E.M.; Márcia, M.M.; Emmanuel, S.M.; Hélcio, S.S. Evaluation of the ADME, toxicological analysis and molecular docking studies of the anacardic acid derivatives with potential antibacterial effects against Staphylococcus aureus. J. Anal. Pharm. Res. 2021, 10, 177–194. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System, Version 2.3; Schrödinger LLC: New York, NY, USA, 2020.
- Imberty, A.; Hardman, K.D.; Carver, J.P.; Perez, S. Molecular Modelling of Protein-Carbohydrate Interactions. Docking of Monosaccharides in the Binding Site of Concanavalin A. Glycobiology 1991, 1, 631–642. [Google Scholar] [CrossRef]

| Strains Bacterial | Hydralazine | ||
|---|---|---|---|
| MIC (µg/mL) | MBC (µg/mL) | MBC/MIC | |
| Staphylococcus aureus ATCC 6538 | 78.1 | 156.2 | 2 |
| Staphylococcus aureus ATCC 700698 | 78.1 | 156.2 | 2 |
| Staphylococcus epidermidis ATCC 12238 | 39.5 | 78.1 | 2 |
| Streptococcus mutans UA 159 | 625 | 1250 | 2 |
| Escherichia coli ATCC 11303 | 625 | 1250 | 2 |
| Klebsiella pneumoniae ATCC 700603 | 625 | 1250 | 2 |
| Klebsiella oxytoca ATCC 13182 | 156.2 | 312.5 | 2 |
| Pseudomonas aeruginosa ATCC 15442 | 156.2 | 312.5 | 2 |
| Strains | Comp. | MIC (µg/mL) | FICi | X-Fold Reduction | |
|---|---|---|---|---|---|
| Individual | Combined | ||||
| S. aureus ATCC 6538 | HDZ | 78.1 | 4.87 | 0.58 (Add) | 16 |
| GEN | 625 | 312.5 | 2 | ||
| S.aureus ATCC 700698 | HDZ | 78.1 | 9.76 | 0.18 (Syn) | 8 |
| GEN | 10 | 625 | 16 | ||
| S. epidermidis ATCC 12238 | HDZ | 39.5 | 2.45 | 0.56 (Add) | 16 |
| GEN | 625 | 312.5 | 2 | ||
| S. mutans UA 159 | HDZ | 625 | 156 | 0.74 (Add) | 5 |
| GEN | 10 | 5 | 2 | ||
| E. coli ATCC11303 | HDZ | 625 | 625 | 1.13 (Ind) | 1 |
| GEN | 10 | 1.25 | 8 | ||
| K. pneumoniae ATCC 700603 | HDZ | 625 | 312.5 | 0.56 (Add) | 2 |
| GEN | 20 | 1.25 | 16 | ||
| K. oxytoca ATCC 13182 | HDZ | 156.2 | 78.1 | 0.56 (Add) | 2 |
| GEN | 5 | 312.5 | 16 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, A.M.G.; da Silva, B.F.; Araujo, I.M.F.; Aguiar, F.K.C.; Coelho, P.A.T.; Costa, R.A.; Marinho, M.M.; Marinho, E.S.; Nunes, J.V.S.; Carneiro, V.A.; et al. Antibacterial and Antibiofilm Activities of Hydralazine, an Antihypertensive Drug: In Vitro and In Silico Approaches. Antibiotics 2025, 14, 286. https://doi.org/10.3390/antibiotics14030286
Pereira AMG, da Silva BF, Araujo IMF, Aguiar FKC, Coelho PAT, Costa RA, Marinho MM, Marinho ES, Nunes JVS, Carneiro VA, et al. Antibacterial and Antibiofilm Activities of Hydralazine, an Antihypertensive Drug: In Vitro and In Silico Approaches. Antibiotics. 2025; 14(3):286. https://doi.org/10.3390/antibiotics14030286
Chicago/Turabian StylePereira, Antônio Mateus Gomes, Benise Ferreira da Silva, Ingrid Maria Frota Araujo, Francisco Kauê Carvalho Aguiar, Paulo Adenes Teixeira Coelho, Renata Albuquerque Costa, Marcia Machado Marinho, Emmanuel Silva Marinho, João Victor Serra Nunes, Victor Alves Carneiro, and et al. 2025. "Antibacterial and Antibiofilm Activities of Hydralazine, an Antihypertensive Drug: In Vitro and In Silico Approaches" Antibiotics 14, no. 3: 286. https://doi.org/10.3390/antibiotics14030286
APA StylePereira, A. M. G., da Silva, B. F., Araujo, I. M. F., Aguiar, F. K. C., Coelho, P. A. T., Costa, R. A., Marinho, M. M., Marinho, E. S., Nunes, J. V. S., Carneiro, V. A., & Santos, H. S. d. (2025). Antibacterial and Antibiofilm Activities of Hydralazine, an Antihypertensive Drug: In Vitro and In Silico Approaches. Antibiotics, 14(3), 286. https://doi.org/10.3390/antibiotics14030286








