Escherichia coli in Brazilian Poultry Fecal Samples: Co-Carriage of Fosfomycin and ESBL Resistance
Abstract
1. Introduction
2. Results
2.1. Higher Phenotypic Detection of Fosfomycin Resistance Compared to PCR Screening
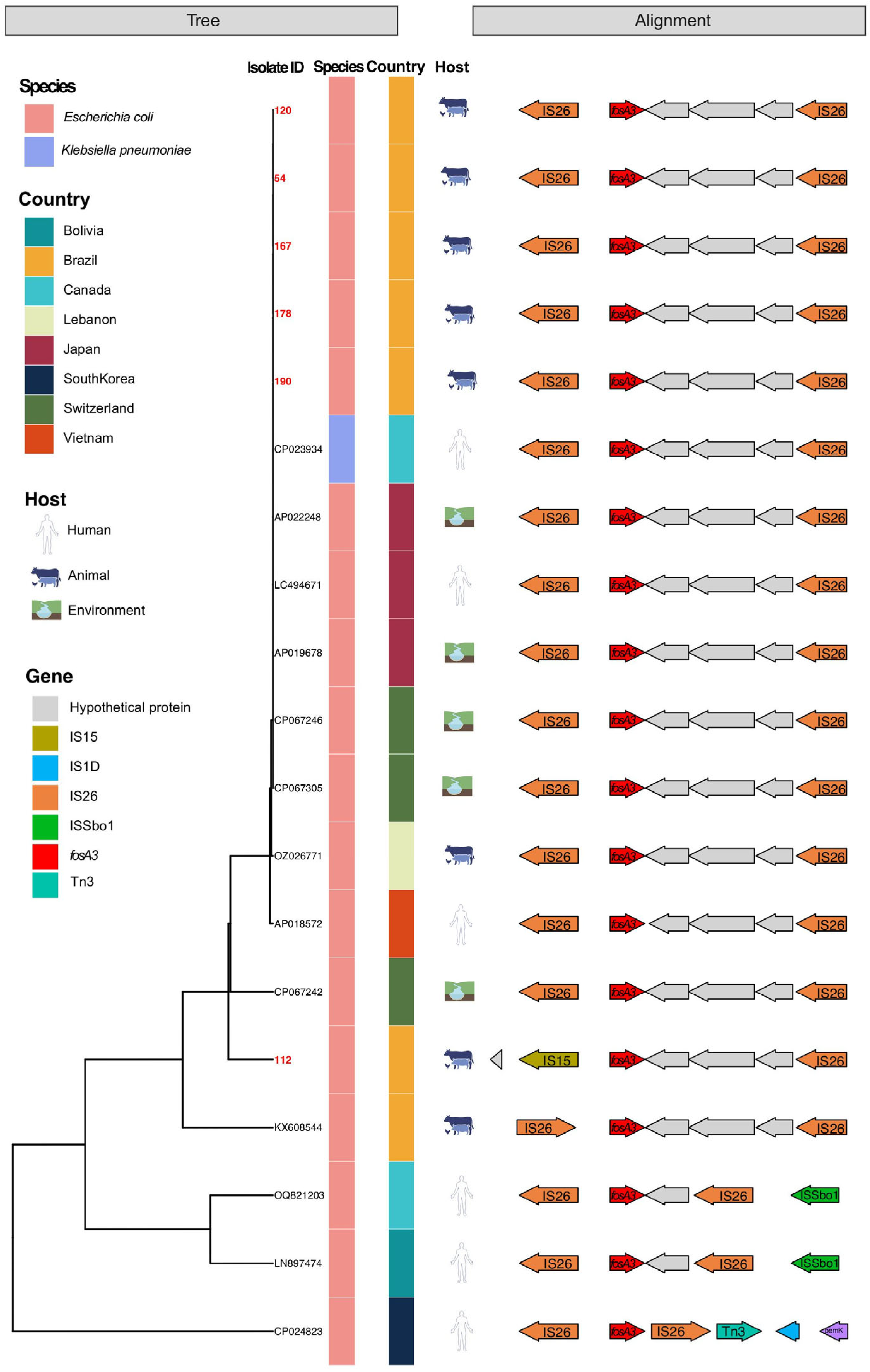
2.2. Genetic Diversity and Resistance Profiles in fosA3-Positive Isolates
2.3. Association of fosA3 and ESBL Genes with Host and Region
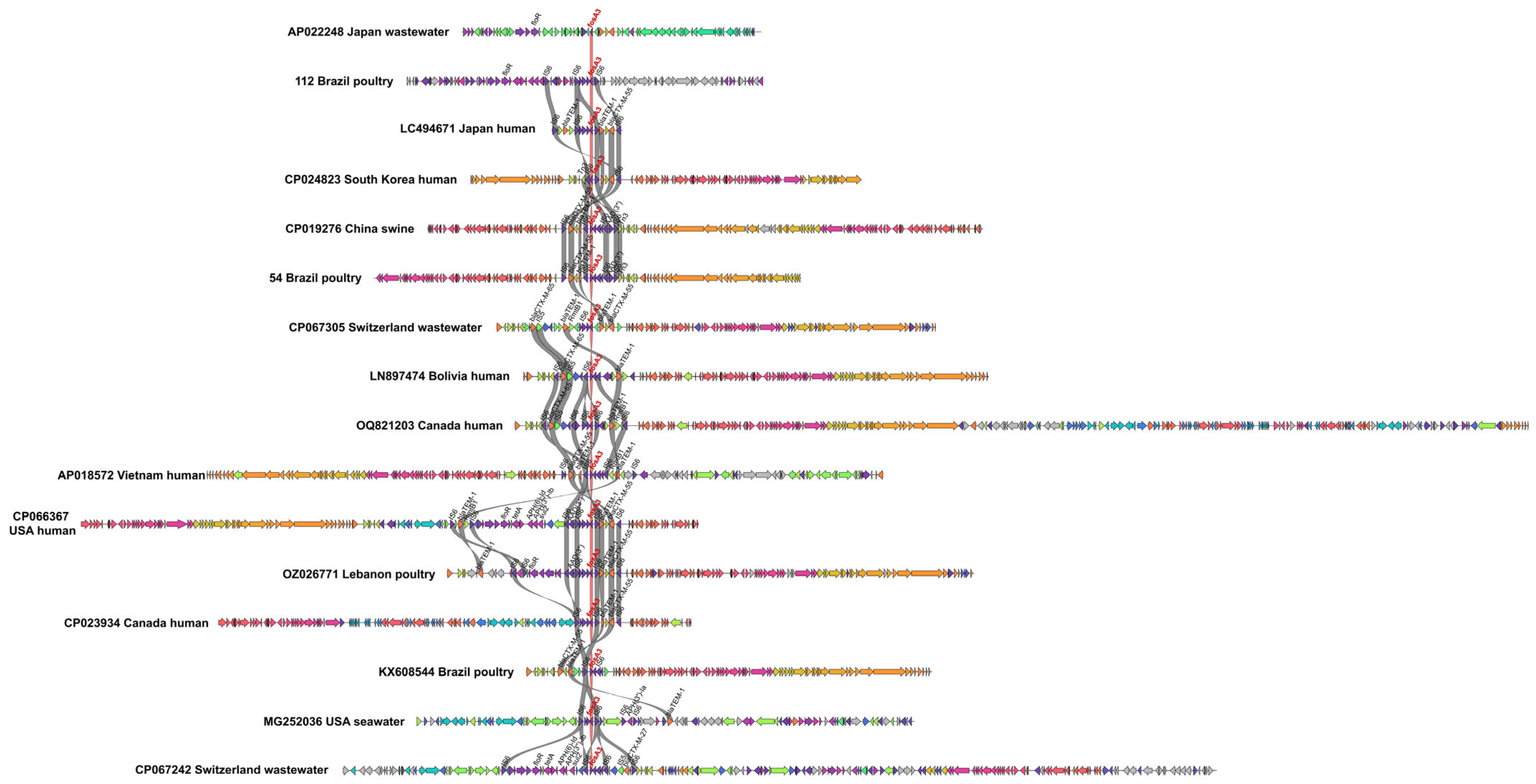
2.4. Shared Genetic Structures in fosA3-Positive Plasmids
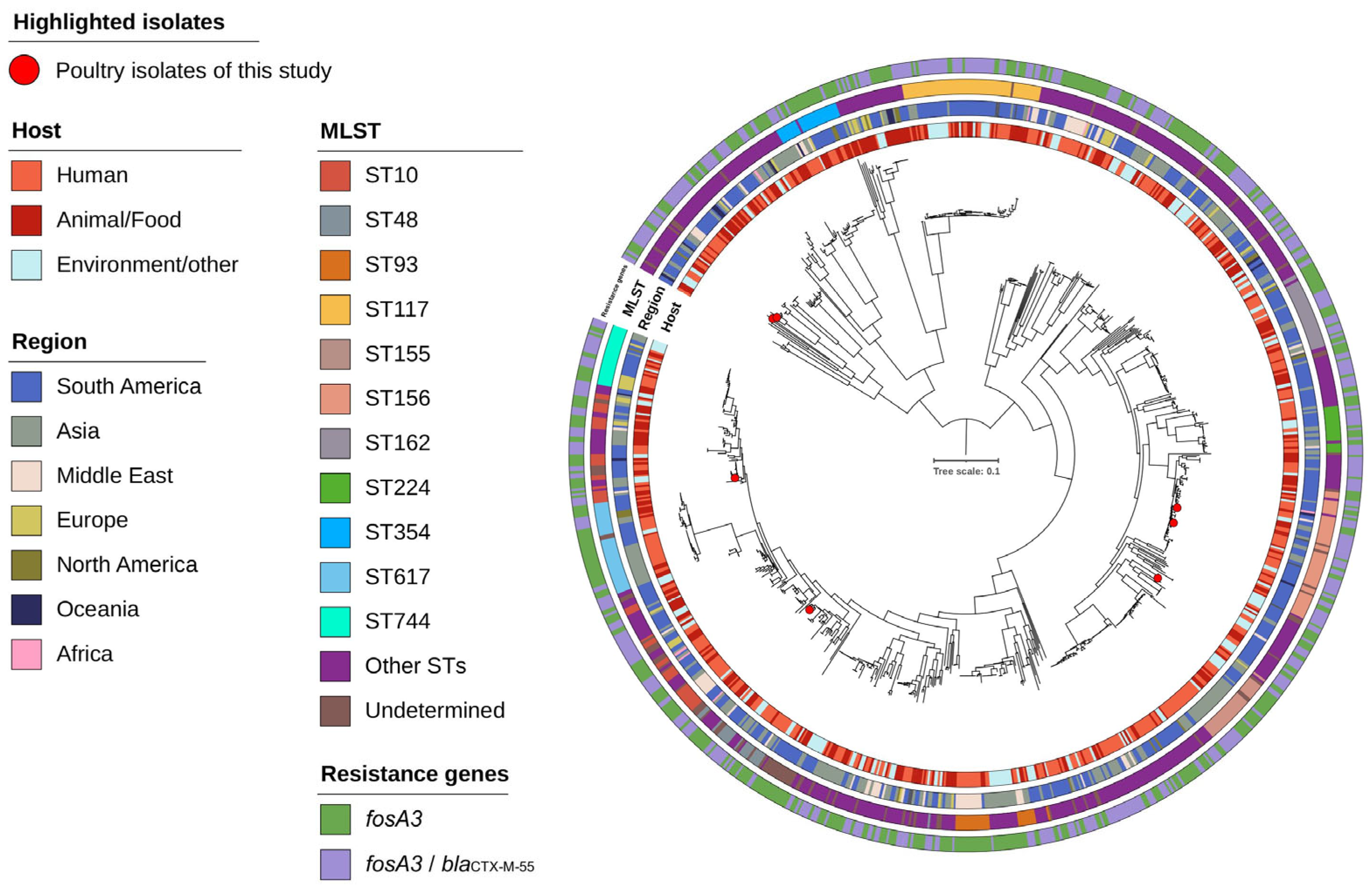
2.5. Phylogenetic Analysis of fosA3-Positive Isolates
3. Discussion
4. Materials and Methods
4.1. Selection of Production Systems
4.2. Bacterial Isolation and Identification
4.3. Evaluation of the Resistance Phenotype
4.4. Genotypic Characterization
4.5. Whole-Genome Sequencing and Plasmid Analysis
4.6. Global Comparative Genomic Analysis
4.7. Phylogenetic Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Falagas, M.E.; Vouloumanou, E.K.; Samonis, G.; Vardakas, K.Z. Fosfomycin. Clin. Microbiol. Rev. 2016, 29, 321–347. [Google Scholar] [CrossRef]
- Babiker, A.; Clarke, L.; Doi, Y.; Shields, R.K. Fosfomycin for treatment of multidrug-resistant pathogens causing urinary tract infection: A real-world perspective and review of the literature. Diagn. Microbiol. Infect. Dis. 2019, 95, 114856. [Google Scholar] [CrossRef] [PubMed]
- Zurfluh, K.; Treier, A.; Schmitt, K.; Stephan, R. Mobile fosfomycin resistance genes in Enterobacteriaceae—An increasing threat. Microbiol. Open 2020, 9, e1135. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Guerin, F. How is fosfomycin resistance developed in Escherichia coli? Future Microbiol. 2018, 13, 1693–1696. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.Y.; Lu, P.L.; Tseng, S.P. Update on fosfomycin-modified genes in Enterobacteriaceae. J. Microbiol. Immunol. Infect. 2019, 52, 9–21. [Google Scholar] [CrossRef]
- Cyoia, P.S.; Koga, V.L.; Nishio, E.K.; Houle, S.; Dozois, C.M.; de Brito, K.C.T.; de Brito, B.G.; Nakazato, G.; Kobayashi, R.K.T. Distribution of ExPEC Virulence factors, blaCTX-M, fosA3, and mcr-1 in Escherichia coli isolated from commercialized chicken carcasses. Front. Microbiol. 2019, 9, 3254. [Google Scholar] [CrossRef]
- Gazal, L.E.S.; Medeiros, L.P.; Dibo, M.; Nishio, E.K.; Koga, V.L.; Gonçalves, B.C.; Grassotti, T.T.; de Camargo, T.C.L.; Pinheiro, J.J.; Vespero, E.C.; et al. Detection of ESBL/AmpC-producing and fosfomycin-resistant Escherichia coli from different sources in poultry production in southern Brazil. Front. Microbiol. 2021, 11, 604544. [Google Scholar] [CrossRef]
- ABPA—Associação Brasileira de Proteína Animal. Annual Report of Brazilian Poultry Association; Brazilian Poultry Union: São Paulo, Brazil, 2023. [Google Scholar]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef]
- Dziva, F.; Stevens, M.P. Colibacillosis in poultry: Unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathol. 2008, 37, 355–366. [Google Scholar] [CrossRef]
- Ho, P.L.; Chan, J.; Lo, W.U.; Law, P.Y.; Li, Z.; Lai, E.L.; Chow, K.H. Dissemination of plasmid-mediated fosfomycin resistance fosA3 among multidrug-resistant Escherichia coli from livestock and other animals. J. Appl. Microbiol. 2013, 114, 695–702. [Google Scholar] [CrossRef]
- Xie, M.; Lin, D.; Chen, K.; Chan, E.W.; Yao, W.; Chen, S. Molecular characterization of Escherichia coli strains isolated from retail meat that harbor blaCTX-M and fosA3 Genes. Antimicrob. Agents Chemother. 2016, 60, 2450–2455. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jiang, W.; Men, S.; Kong, L.; Ma, S.; Yang, Y.; Wang, Y.; Yuan, Q.; Cheng, G.; Zou, W.; Wang, H. Prevalence of plasmid-mediated fosfomycin resistance gene fosA3 among CTX-M-producing Escherichia coli isolates from chickens in China. Foodborne Pathog. Dis. 2017, 14, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Mueller, L.; Cimen, C.; Poirel, L.; Descombes, M.C.; Nordmann, P. Prevalence of fosfomycin resistance among ESBL-producing Escherichia coli isolates in the community, Switzerland. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 945–949. [Google Scholar] [CrossRef]
- Peretz, A.; Naamneh, B.; Tkhawkho, L.; Nitzan, O. High rates of fosfomycin resistance in Gram-negative urinary isolates from Israel. Microb. Drug Resist. 2019, 25, 408–412. [Google Scholar] [CrossRef] [PubMed]
- Loras, C.; Mendes, A.C.; Peixe, L.; Novais, A.; Alos, J.I. Escherichia coli resistant to fosfomycin from urinary tract infections: Detection of the fosA3 gene in Spain. J. Glob. Antimicrob. Resist. 2020, 21, 414–416. [Google Scholar] [CrossRef]
- Coppola, N.; Cordeiro, N.F.; Trenchi, G.; Esposito, F.; Fuga, B.; Fuentes-Castillo, D.; Lincopan, N.; Iriarte, A.; Bado, I.; Vignoli, R. Imported One-Day-Old Chicks as Trojan Horses for Multidrug-Resistant Priority Pathogens Harboring mcr-9, rmtG and Extended-Spectrum β-Lactamase Genes. Appl. Environ. Microbiol. 2021, 88, e01675-21. [Google Scholar] [CrossRef]
- Hou, J.; Huang, X.; Deng, Y.; He, L.; Yang, T.; Zeng, Z.; Chen, Z.; Liu, J.H. Dissemination of the fosfomycin resistance gene fosA3 with CTX-M β-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob. Agents Chemother. 2012, 56, 2135–2138. [Google Scholar] [CrossRef]
- Sato, N.; Kawamura, K.; Nakane, K.; Wachino, J.; Arakawa, Y. First detection of fosfomycin resistance gene fosA3 in CTX-M-producing Escherichia coli isolates from healthy individuals in Japan. Microb. Drug Resist. 2013, 19, 477–482. [Google Scholar] [CrossRef]
- Galindo-Méndez, M.; Navarrete-Salazar, H.; Baltazar-Jiménez, F.; Muñoz-de la Paz, E.; Sánchez-Mawcinitt, M.F.; Gómez-Pardo, A.; Garza-González, E.; Ponce-de-León-Garduño, L.A.; Franco-Cendejas, R.; Morfín-Otero, R.; et al. Emergence of fosfomycin resistance by plasmid-mediated fos genes in uropathogenic ESBL-producing E. coli isolates in Mexico. Antibiotics 2022, 11, 1383. [Google Scholar] [CrossRef]
- Matamoros, S.; van Hattem, J.M.; Arcilla, M.S.; Willemse, N.; Melles, D.C.; Penders, J.; Vinh, T.N.; Thi Hoa, N.; Bootsma, M.C.J.; van Genderen, P.J.; et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci. Rep. 2017, 7, 15364. [Google Scholar] [CrossRef]
- Yao, H.; Wu, D.; Lei, L.; Shen, Z.; Wang, Y.; Liao, K. The detection of fosfomycin resistance genes in Enterobacteriaceae from pets and their owners. Vet. Microbiol. 2016, 193, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Lupo, A.; Saras, E.; Madec, J.Y.; Haenni, M. Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J. Antimicrob. Chemother. 2018, 73, 867–872. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Graziano, E.; Berruti, M.; Giacobbe, D.R. The role of fosfomycin for multidrug-resistant gram-negative infections. Curr. Opin. Infect. Dis. 2019, 32, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Oteo, J.; Orden, B.; Bautista, V.; Cuevas, O.; Arroyo, M.; Martinez-Ruiz, R.; Perez-Vazquez, M.; Alcaraz, M.; Garcia-Cobos, S.; Campos, J. CTX-M-15-producing urinary Escherichia coli O25b-ST131-phylogroup B2 has acquired resistance to fosfomycin. J. Antimicrob. Chemother. 2009, 64, 712–717. [Google Scholar] [CrossRef]
- Ho, P.L.; Chan, J.; Lo, W.U.; Lai, E.L.; Cheung, Y.Y.; Lau, T.C.K.; Chow, K.H. Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. J. Med. Microbiol. 2013, 62 Pt 11, 1707–1713. [Google Scholar] [CrossRef]
- Alrowais, H.; McElheny, C.L.; Spychala, C.N.; Sastry, S.; Guo, Q.; Butt, A.A.; Doi, Y. Fosfomycin resistance in Escherichia coli, Pennsylvania, USA. Emerg. Infect. Dis. 2015, 21, 2045–2047. [Google Scholar] [CrossRef]
- Falagas, M.E.; Athanasaki, F.; Voulgaris, G.L.; Triarides, N.A.; Vardakas, K.Z. Resistance to fosfomycin: Mechanisms, frequency and clinical consequences. Int. J. Antimicrob. Agents 2019, 53, 22–28. [Google Scholar] [CrossRef]
- Marchetti, M.V.; Hrabak, J.; Bitar, I. Fosfomycin resistance mechanisms in Enterobacterales: An increasing threat. Front. Cell Infect. Microbiol. 2023, 13, 1178547. [Google Scholar] [CrossRef]
- Liu, F.; Tian, A.; Wang, J.; Zhu, Y.; Xie, Z.; Zhang, R.; Jiang, S. Occurrence and molecular epidemiology of fosA3-bearing Escherichia coli from ducks in Shandong province of China. Poult. Sci. 2022, 101, 101620. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, M.; Cheng, Y.; Huang, Y.; Tan, Y.; Yang, Y.; Qian, Y.; Zhong, X.; Lu, Y.; Si, H. Spread and molecular characteristics of Enterobacteriaceae carrying fosA-like genes from farms in China. Microbiol. Spectr. 2022, 10, e0054522. [Google Scholar] [CrossRef]
- Saraiva, M.M.; Moreira Filho, A.L.; Freitas Neto, O.C.; Silva, N.M.; Givisiez, P.E.; Gebreyes, W.A.; Oliveira, C.J. Off-label use of ceftiofur in one-day chicks triggers a short-term increase of ESBL-producing E. coli in the gut. PLoS ONE 2018, 13, e0203158. [Google Scholar] [CrossRef] [PubMed]
- Rabello, R.F.; Bonelli, R.R.; Penna, B.A.; Albuquerque, J.P.; Souza, R.M.; Cerqueira, A.M.F. Antimicrobial resistance in farm animals in Brazil: An update overview. Animals 2020, 10, 552. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement. M100-S22; CLSI: Wayne, PA, USA, 2012. [Google Scholar]
- Chen, L.; Chen, Z.L.; Liu, J.H.; Zeng, Z.L.; Ma, J.Y.; Jiang, H.X. Emergence of RmtB methylase-producing Escherichia coli and Enterobacter cloacae isolates from pigs in China. J. Antimicrob. Chemother. 2007, 59, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar]
- Sherry, N.L.; Horan, K.A.; Ballard, S.A.; Gonçalves da Silva, A.; Gorrie, C.L.; Schultz, M.B.; Stevens, K.; Valcanis, M.; Sait, M.L.; Stinear, T.P.; et al. An ISO-certified genomics workflow for identification and surveillance of antimicrobial resistance. Nat. Commun. 2023, 14, 60. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Waters, N.R.; Abram, F.; Brennan, F.; Holmes, A.; Pritchard, L. Easy phylotyping of Escherichia coli via the EzClermont web app and command-line tool. Access Microbiol. 2020, 2, acmi000143. [Google Scholar] [CrossRef]
- Ingle, D.J.; Valcanis, M.; Kuzevski, A.; Tauschek, M.; Inouye, M.; Stinear, T.; Levine, M.M.; Robins-Browne, R.M.; Holt, K.E. In silico serotyping of E. coli from short read data identifies limited novel O-loci but extensive diversity of O:H serotype combinations within and between pathogenic lineages. Microb. Genom. 2016, 2, e000064, Erratum in Microb. Genom. 2017, 3, e000109. [Google Scholar] [CrossRef]
- Roer, L.; Tchesnokova, V.; Allesøe, R.; Muradova, M.; Chattopadhyay, S.; Ahrenfeldt, J.; Thomsen, M.C.F.; Lund, O.; Hansen, F.; Hammerum, A.M.; et al. Development of a web tool for Escherichia coli subtyping based on fimH alleles. J. Clin. Microbiol. 2017, 55, 2538–2543. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.-H. Clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [PubMed]
- Thorn, A.V.; Aarestrup, F.M.; Munk, P. Flankophile: A bioinformatic pipeline for prokaryotic genomic synteny analysis. Microbiol. Spectr. 2024, 12, e0241323. [Google Scholar] [CrossRef] [PubMed]
- Sahl, J.W.; Lemmer, D.; Travis, J.; Schupp, J.M.; Gillece, J.D.; Aziz, M.; Driebe, E.M.; Drees, K.P.; Hicks, N.D.; Williamson, C.H.D.; et al. NASP: An accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb. Genom. 2016, 2, e000074. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 1 November 2024).

| Integration Farm | Susceptible | Resistant |
|---|---|---|
| Integration 1 (N = 100) | 67% | 33% |
| Integration 2 (N = 100) | 85% | 15% |
| Integration 3 (N = 100) | 92% | 8% |
| Integration 4 (N = 100) | 80% | 20% |
| Integration Farms | Phenotypically Resistant Isolates | Positive isolates | ||
|---|---|---|---|---|
| fosA3 Positive Isolates | Of the Total Phenotypically Resistant Isolates | Of the Total Isolates (N = 400) | ||
| Integration 1 | 33 | 2 | 6.06% | 0.5% |
| Integration 2 | 15 | 9 | 60.00% | 2.25% |
| Integration 3 | 8 | 3 | 37.50% | 0.75% |
| Integration 4 | 19 | 2 | 10.53% | 0.5% |
| Total | 75 | 16 | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Juscele, F.; Saidenberg, A.B.S.; Christoffersen, L.E.B.; Edslev, S.M.; Hallstrøm, S.; Nacarato, J.R.; Barbosa, F.B.; Cunha, M.P.; Esposito, F.; Lincopan, N.H.; et al. Escherichia coli in Brazilian Poultry Fecal Samples: Co-Carriage of Fosfomycin and ESBL Resistance. Antibiotics 2025, 14, 269. https://doi.org/10.3390/antibiotics14030269
Juscele F, Saidenberg ABS, Christoffersen LEB, Edslev SM, Hallstrøm S, Nacarato JR, Barbosa FB, Cunha MP, Esposito F, Lincopan NH, et al. Escherichia coli in Brazilian Poultry Fecal Samples: Co-Carriage of Fosfomycin and ESBL Resistance. Antibiotics. 2025; 14(3):269. https://doi.org/10.3390/antibiotics14030269
Chicago/Turabian StyleJuscele, Felipe, Andre B. S. Saidenberg, Lars E. B. Christoffersen, Sofie M. Edslev, Søren Hallstrøm, Jessica R. Nacarato, Fernanda B. Barbosa, Marcos P. Cunha, Fernanda Esposito, Nilton H. Lincopan, and et al. 2025. "Escherichia coli in Brazilian Poultry Fecal Samples: Co-Carriage of Fosfomycin and ESBL Resistance" Antibiotics 14, no. 3: 269. https://doi.org/10.3390/antibiotics14030269
APA StyleJuscele, F., Saidenberg, A. B. S., Christoffersen, L. E. B., Edslev, S. M., Hallstrøm, S., Nacarato, J. R., Barbosa, F. B., Cunha, M. P., Esposito, F., Lincopan, N. H., Stegger, M., & Knöbl, T. (2025). Escherichia coli in Brazilian Poultry Fecal Samples: Co-Carriage of Fosfomycin and ESBL Resistance. Antibiotics, 14(3), 269. https://doi.org/10.3390/antibiotics14030269







