Observed Prevalence and Characterization of Fluoroquinolone-Resistant and Multidrug-Resistant Bacteria in Loggerhead Sea Turtles (Caretta caretta) from the Adriatic Sea
Abstract
1. Introduction
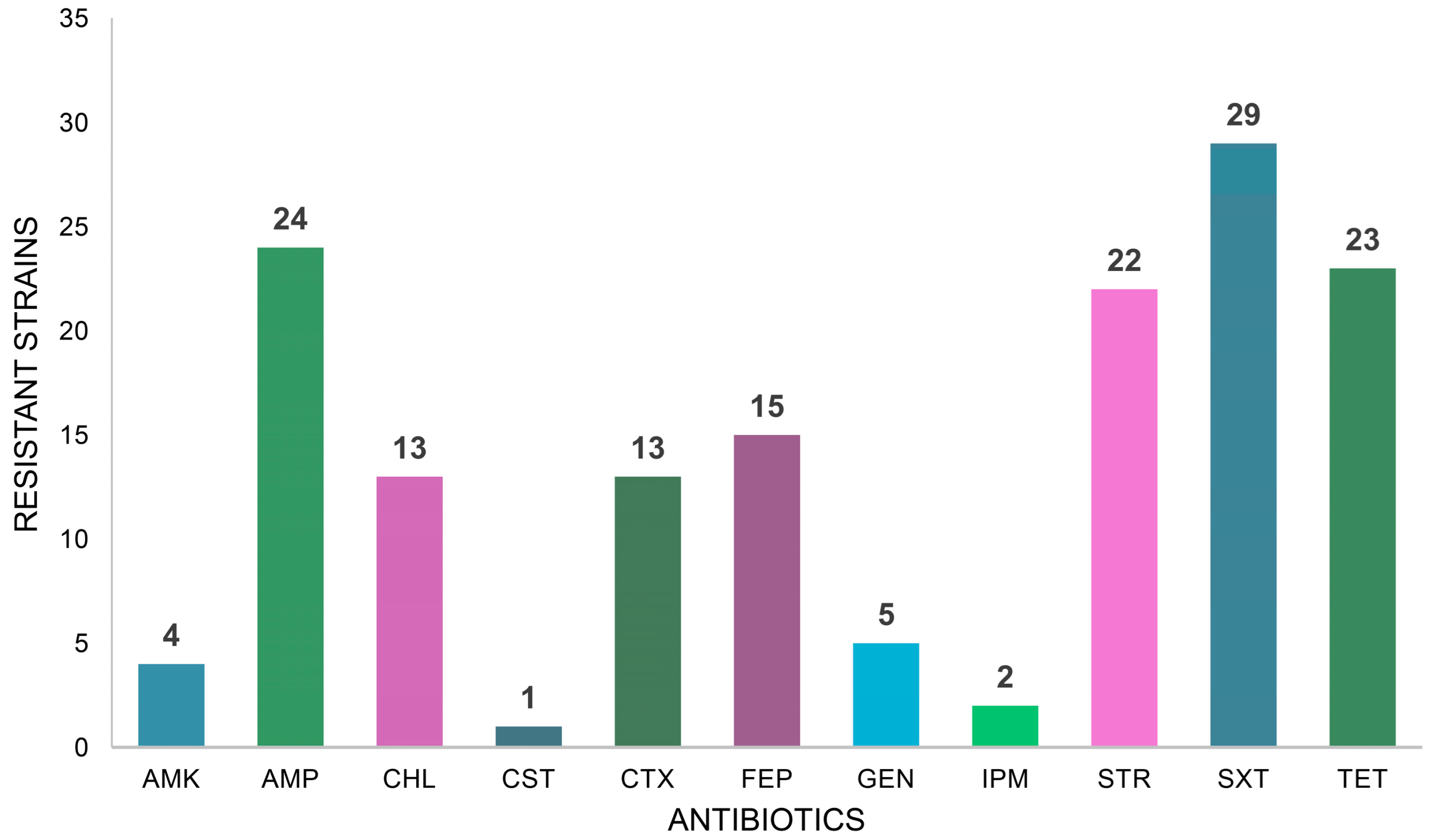
2. Results
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Bacterial Identification
4.3. Antimicrobial Susceptibility Test
4.4. Conjugation Experiments
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Action Plan on Antimicrobial Resistance; WHO Document Production Services: Geneva, Switzerland, 2015. [Google Scholar]
- World Health Organization. New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis. Available online: https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (accessed on 3 December 2024).
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States—2019; Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019. [Google Scholar]
- Endale, H.M.; Mathewos, M.; Abdeta, D. Potential causes of spread of antimicrobial resistance and preventive measures in One Health perspective—A review. Infect. Drug Resist. 2023, 16, 7515–7545. [Google Scholar] [CrossRef] [PubMed]
- Fatta-Kassinos, D.; Meric, S.; Nikolaou, A. Pharmaceutical residues in environmental waters and wastewater: Current state of knowledge and future research. Anal. Bioanal. Chem. 2011, 399, 251–275. [Google Scholar] [CrossRef]
- Guedes-Alonso, R.; Montesdeoca-Esponda, S.; Pacheco-Juárez, J.; Sosa-Ferrera, Z.; Santana-Rodríguez, J.J. A survey of the presence of pharmaceutical residues in wastewaters. Evaluation of their removal using conventional and natural treatment procedures. Molecules 2020, 25, 1639. [Google Scholar] [CrossRef] [PubMed]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low-and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef] [PubMed]
- Cabello, F.C.; Godfrey, H.P.; Tomova, A.; Ivanova, L.; Dölz, H.; Millanao, A.; Buschmann, A.H. Antimicrobial use in aquaculture re-examined: Its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 2013, 15, 1917–1942. [Google Scholar] [CrossRef]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Bürgmann, H.; Sørum, H.; Norström, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- Martinez, J.L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 2009, 157, 2893–2902. [Google Scholar] [CrossRef]
- Janecko, N.; Pokludova, L.; Blahova, J.; Svobodova, Z.; Literak, I. Implications of fluoroquinolone contamination for the aquatic environment—A review. Environ. Toxicol. Chem. 2016, 35, 2647–2656. [Google Scholar] [CrossRef]
- WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance. Critically Important Antimicrobials for Human Medicine: 6th Revision; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Committee for Medicinal Products for Veterinary Use, Committee for Medicinal Products for Human Use. Categorisation of Antibiotics in the European Union; European Medicines Agency: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Biswas, M.; Biswas, S.; Gupta, B.; Mascellino, M.T.; Rakshit, A.; Chakraborty, B. Changing paradigms in antibiotic resistance in Salmonella species with focus on fluoroquinolone resistance: A 5-year retrospective study of enteric fever in a tertiary care hospital in Kolkata, India. Antibiotics 2022, 11, 1308. [Google Scholar] [CrossRef] [PubMed]
- Khademi, F.; Sahebkar, A. Prevalence of fluoroquinolone-resistant Campylobacter species in Iran: A systematic review and meta-analysis. Int. J. Microbiol. 2020, 2020, 8868197. [Google Scholar] [CrossRef]
- Joel, E.O.; Akinlabi, O.C.; Olaposi, A.V.; Olowomofe, T.O.; Adekanmbi, A.O. High carriage of plasmid-mediated quinolone resistance (PMQR) genes by ESBL-producing and fluoroquinolone-resistant Escherichia coli recovered from animal waste dumps. Mol. Biol. Rep. 2024, 51, 424. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.C.; Jacoby, G.A. Topoisomerase inhibitors: Fluoroquinolone mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025320. [Google Scholar] [CrossRef]
- Ruiz, J. Transferable mechanisms of quinolone resistance from 1998 onward. Clin. Microbiol. Rev. 2019, 32, e00007-19. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, J.; Hu, Q.; Rao, X. Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 2016, 50, 10–17. [Google Scholar] [CrossRef]
- Van Bambeke, F.; Michot, J.M.; Van Eldere, J.; Tulkens, P.M. Quinolones in 2005: An update. Clin. Microbiol. Infect. 2005, 11, 256–280. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Yang, Y.; Li, F.; Li, X.; Liu, H.; Fazilani, S.A.; Guo, W.; Xu, G.; Zhang, X. The prevalence and mechanism of fluoroquinolone resistance in Escherichia coli isolated from swine farms in China. BMC Vet. Res. 2020, 16, 258. [Google Scholar] [CrossRef]
- Gestels, Z.; Baranchyk, Y.; Van den Bossche, D.; Laumen, J.; Abdellati, S.; Britto Xavier, B.; Manoharan-Basil, S.S.; Kenyon, C. Could traces of fluoroquinolones in food induce ciprofloxacin resistance in Escherichia coli and Klebsiella pneumoniae? An in vivo study in Galleria mellonella with important implications for maximum residue limits in food. Microbiol. Spectr. 2024, 12, e0359523. [Google Scholar] [CrossRef]
- Baquero, F.; Martínez, J.L.; Cantón, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef]
- Zhang, A.; Xiao, Y.; Han, Y.; Huang, Y.; Kan, B.; Liang, W. Characterization of quorum regulatory small RNAs in an emerging pathogen Vibrio fluvialis and their roles toward type VI secretion system VflT6SS2 modulation. Emerg. Microbes Infect. 2024, 13, 2396872. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, T.; Mori, N.; Kawai, F.; Sakurai, A.; Toyoda, M.; Mikami, Y.; Uehara, Y.; Furukawa, K. Vagococcus fluvialis as a causative pathogen of bloodstream and decubitus ulcer infection: Case report and systematic review of the literature. J. Infect. Chemother. 2021, 27, 359–363. [Google Scholar] [CrossRef] [PubMed]
- Wellington, E.M.; Boxall, A.B.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Zhao, Y.C.; Sun, Z.H.; Li, J.K.; Liu, J.K.; Cai, H.L.; Cao, W.; Yu, F.; Zhang, B.K.; Yan, M. Exploring the causes of the prevalence of vancomycin-resistant Enterococcus faecalis. Environ. Sci. Eur. 2024, 36, 92. [Google Scholar] [CrossRef]
- Gao, W.; Howden, B.P.; Stinear, T.P. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr. Opin. Microbiol. 2018, 41, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.R.; Pereira, A.P.; Novais, C.; Peixe, L. Multidrug-resistant high-risk Enterococcus faecium clones: Can we really define them? Int. J. Antimicrob. Agents 2021, 57, 106227. [Google Scholar] [CrossRef] [PubMed]
- Ono, S.; Muratani, T.; Matsumoto, T. Mechanisms of resistance to imipenem and ampicillin in Enterococcus faecalis. Antimicrob. Agents Chemother. 2005, 49, 2954–2958. [Google Scholar] [CrossRef]
- Teixeira, L.M.; Carvalho, M.G.; Merquior, V.L.; Steigerwalt, A.G.; Brenner, D.J.; Facklam, R.R. Phenotypic and genotypic characterization of Vagococcus fluvialis, including strains isolated from human sources. J. Clin. Microbiol. 1997, 35, 2778–2781. [Google Scholar] [CrossRef]
- Brunswick, J.; Spiro, J.; Wisniewski, P. Vagococcus: An under-recognized and emerging cause of antibiotic-resistant infection. IDCases 2024, 36, e01995. [Google Scholar] [CrossRef]
- Rodriguez Jimenez, A.; Guiglielmoni, N.; Goetghebuer, L.; Dechamps, E.; George, I.F.; Flot, J.F. Comparative genome analysis of Vagococcus fluvialis reveals abundance of mobile genetic elements in sponge-isolated strains. BMC Genom. 2022, 23, 618. [Google Scholar] [CrossRef] [PubMed]
- Rosenblueth, M.; Martínez, L.; Silva, J.; Martínez-Romero, E. Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst. Appl. Microbiol. 2004, 27, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.; Passet, V.; Rakotondrasoa, A.; Diallo, T.A.; Criscuolo, A.; Brisse, S. Description of Klebsiella africanensis sp. nov., Klebsiella variicola subsp. tropicalensis subsp. nov. and Klebsiella variicola subsp. variicola subsp. nov. Res. Microbiol. 2019, 170, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Medina, N.; Barrios-Camacho, H.; Duran-Bedolla, J.; Garza-Ramos, U. Klebsiella variicola: An emerging pathogen in humans. Emerg. Microbes Infect. 2019, 8, 973–988. [Google Scholar] [CrossRef]
- Chu, J.; Choi, J.; Ji, S.K.; Park, C.; Jung, S.H.; Park, S.H.; Lee, D.G. An outbreak of blaKPC-4- and blaVIM-1-producing Klebsiella pneumoniae and Klebsiella variicola at a single hospital in South Korea. Antimicrob. Resist. Infect. Control 2024, 13, 123. [Google Scholar] [CrossRef]
- Morris, S.; Cerceo, E. Trends, epidemiology, and management of multi-drug resistant Gram-negative bacterial infections in the hospitalized setting. Antibiotics 2020, 9, 196. [Google Scholar] [CrossRef]
- Abban, M.K.; Ayerakwa, E.A.; Mosi, L.; Isawumi, A. The burden of hospital acquired infections and antimicrobial resistance. Heliyon 2023, 9, e20561. [Google Scholar] [CrossRef]
- GBD 2021 Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance 1990-2021: A systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Singh, A.; Pratap, S.G.; Raj, A. Occurrence and dissemination of antibiotics and antibiotic resistance in aquatic environment and its ecological implications: A review. Environ. Sci. Pollut. Res. Int. 2024, 31, 47505–47529. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, J.; Zhang, W.; Feng, M.; Yu, X.; Ye, C. Neglected contributors to the transmission of bacterial antibiotic resistance in drinking water: Extracellular antibiotic resistance genes and the natural transformation. Sci. Total Environ. 2024, 953, 175970. [Google Scholar] [CrossRef] [PubMed]
- Circella, E.; Schiavone, A.; Barrasso, R.; Camarda, A.; Pugliese, N.; Bozzo, G. Pseudomonas azotoformans belonging to Pseudomonas fluorescens group as causative agent of blue coloration in carcasses of slaughterhouse rabbits. Animals 2020, 10, 256. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Madan, A. CAP3: A DNA sequence assembly program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Lee, J.H.; Jung, Y.; Kim, M.; Kim, S.; Kim, B.K.; Lim, Y.W. EzTaxon: A web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2259–2261. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standard Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 7th ed.; CLSI: Wayne, PA, USA, 2006; pp. 11–14. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Camarda, A.; Pugliese, N.; Pupillo, A.; Oliva, M.; Circella, E.; Dionisi, A.M.; Ricci, A.; Legretto, M.; Caroli, A.; Pazzani, C. Resistance genes, phage types and pulsed field gel electrophoresis pulsotypes in Salmonella enterica strains from laying hen farms in southern Italy. Int. J. Environ. Res. Public Health 2013, 10, 3347–3362. [Google Scholar] [CrossRef]
| Turtle | Isolate Code | Species | Resistance Profile 1 | ENR MIC (μg/mL) | CST MIC (μg/mL) 2 |
|---|---|---|---|---|---|
| A | FVBA1 | Escherichia coli | AMP-ENR-STR-SXT-TET | 32 | 2 |
| FVBA2 | Staphylococcus haemolyticus | AMP-ENR-STR-SXT-TET | 32 | NA | |
| C | FVBA15 | Escherichia coli | ENR-STR-SXT | 128 | 1 |
| D | FVBA5 | Pseudomonas mendocina | AMP-CHL-ENR-STR-SXT | 128 | 2 |
| E | FVBA6 | Pseudomonas mendocina | AMP-CHL-ENR-STR-SXT-TET | 128 | 2 |
| F | FVBA8 | Citrobacter portucalensis | AMP-CHL-ENR | 16 | 2 |
| G | FVBA10 | Escherichia coli | AMP-CTX-FEP-ENR | 128 | 2 |
| FVBA9 | Escherichia coli | AMP-CTX-ENR-SXT-TET | 128 | 2 | |
| H | FVBA11 | Escherichia coli | AMP-CTX-ENR-STR | 128 | 1 |
| FVBA12 | Escherichia coli | AMP-CTX-ENR-STR-SXT-TET | 256 | 1 | |
| I | FVBA16 | Vibrio fluvialis | AMP-ENR-SXT-TET | 64 | 1 |
| J | FVBA13 | Pseudomonas mendocina | AMP-CHL-ENR-SXT | 128 | 2 |
| M | FVBA19 | Morganella morganii | AMP-CST-ENR-TET | 128 | 256 |
| N | FVBA42 | Enterococcus faecium | AMP-CTX-ENR-FEP-GEN-IPM-STR | 64 | NA |
| FVBA46 | Vagococcus fluvialis | AMP-CTX-ENR-FEP-STR-SXT-TET | 32 | NA | |
| O | FVBA40 | Vagococcus fluvialis | AMK-CTX-ENR-FEP-STR-SXT-TET | 32 | NA |
| P | FVBA26 | Citrobacter freundii | AMP-CHL-ENR-SXT-TET | 32 | 2 |
| FVBA36 | Vagococcus fluvialis | CTX-ENR-FEP-STR-SXT-TET | 32 | NA | |
| FVBA25 | Citrobacter freundii | AMP-CHL-ENR-SXT-TET | 32 | 2 | |
| Q | FVBA37 | Vagococcus fluvialis | AMK-CHL-CTX-ENR-FEP-GEN-STR-SXT-TET | 32 | NA |
| FVBA38 | Vagococcus fluvialis | CHL-ENR-STR-SXT | 32 | NA | |
| R | FVBA27 | Aeromonas caviae | AMP-ENR-TET | 32 | 2 |
| FVBA33 | Vagococcus fluvialis | CHL-CTX-ENR-FEP-STR-SXT-TET | 32 | NA | |
| FVBA39 | Vagococcus fluvialis | AMK-ENR-FEP-GEN-STR-SXT-TET | 32 | NA | |
| S | FVBA29 | Klebsiella variicola subsp. variicola | AMP-CTX-ENR-FEP-GEN-STR-SXT-TET | 32 | 2 |
| FVBA32 | Vagococcus fluvialis | AMP-CTX-ENR-FEP-GEN-STR-SXT-TET | 32 | NA | |
| T | FVBA23 | Citrobacter freundii | AMP-CHL-ENR-SXT-TET | 32 | 2 |
| FVBA43 | Vagococcus fluvialis | ENR-STR-SXT | 32 | NA | |
| U | FVBA22 | Citrobacter freundii | AMP-CHL-ENR-SXT-TET | 32 | 2 |
| FVBA41 | Vagococcus fluvialis | CTX-ENR-FEP-STR-SXT-TET | 32 | NA | |
| V | FVBA44 | Vagococcus fluvialis | ENR-FEP-STR-SXT | 32 | NA |
| FVBA45 | Aeromonas veronii | AMP-ENR | 32 | 2 | |
| W | FVBA30 | Vagococcus fluvialis | ENR-FEP-STR-SXT | 32 | NA |
| X | FVBA47 | Pseudomonas mendocina | AMP-CHL-ENR-FEP-SXT-TET | 64 | 2 |
| Z | FVBA24 | Citrobacter freundii | AMP-CHL-ENR-IPM-SXT-TET | 32 | 2 |
| FVBA31 | Vagococcus fluvialis | AMK-ENR-FEP-STR-SXT | 32 | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, O.; Tinelli, A.; Soloperto, S.; Crescenzo, G.; Galante, D.; Calarco, A.; Tribuzio, M.; Manzulli, V.; Caioni, G.; Zizzadoro, C.; et al. Observed Prevalence and Characterization of Fluoroquinolone-Resistant and Multidrug-Resistant Bacteria in Loggerhead Sea Turtles (Caretta caretta) from the Adriatic Sea. Antibiotics 2025, 14, 252. https://doi.org/10.3390/antibiotics14030252
Lai O, Tinelli A, Soloperto S, Crescenzo G, Galante D, Calarco A, Tribuzio M, Manzulli V, Caioni G, Zizzadoro C, et al. Observed Prevalence and Characterization of Fluoroquinolone-Resistant and Multidrug-Resistant Bacteria in Loggerhead Sea Turtles (Caretta caretta) from the Adriatic Sea. Antibiotics. 2025; 14(3):252. https://doi.org/10.3390/antibiotics14030252
Chicago/Turabian StyleLai, Olimpia, Antonella Tinelli, Simona Soloperto, Giuseppe Crescenzo, Domenico Galante, Angela Calarco, Magda Tribuzio, Viviana Manzulli, Giulia Caioni, Claudia Zizzadoro, and et al. 2025. "Observed Prevalence and Characterization of Fluoroquinolone-Resistant and Multidrug-Resistant Bacteria in Loggerhead Sea Turtles (Caretta caretta) from the Adriatic Sea" Antibiotics 14, no. 3: 252. https://doi.org/10.3390/antibiotics14030252
APA StyleLai, O., Tinelli, A., Soloperto, S., Crescenzo, G., Galante, D., Calarco, A., Tribuzio, M., Manzulli, V., Caioni, G., Zizzadoro, C., Damiano, A., Camarda, A., & Pugliese, N. (2025). Observed Prevalence and Characterization of Fluoroquinolone-Resistant and Multidrug-Resistant Bacteria in Loggerhead Sea Turtles (Caretta caretta) from the Adriatic Sea. Antibiotics, 14(3), 252. https://doi.org/10.3390/antibiotics14030252






