Menadione as Antibiotic Adjuvant Against P. aeruginosa: Mechanism of Action, Efficacy and Safety
Abstract
1. Introduction
2. Results and Discussion
2.1. Antibacterial Activity of Menadione and Azithromycin Against Planktonic Bacteria
2.2. Activity of Menadione and Azithromycin Against P. aeruginosa Biofilms
2.2.1. Minimum Biofilm Eradication Concentration (MBEC)
2.2.2. Morphology of P. aeruginosa Biofilms
Untreated P. aeruginosa Biofilms
Azithromycin- and Menadione-Treated P. aeruginosa Biofilms
2.3. Antibacterial Mechanisms of Menadione
2.3.1. Induction of Reactive Oxygen Species
2.3.2. Disturbance of Bacterial Membrane Integrity
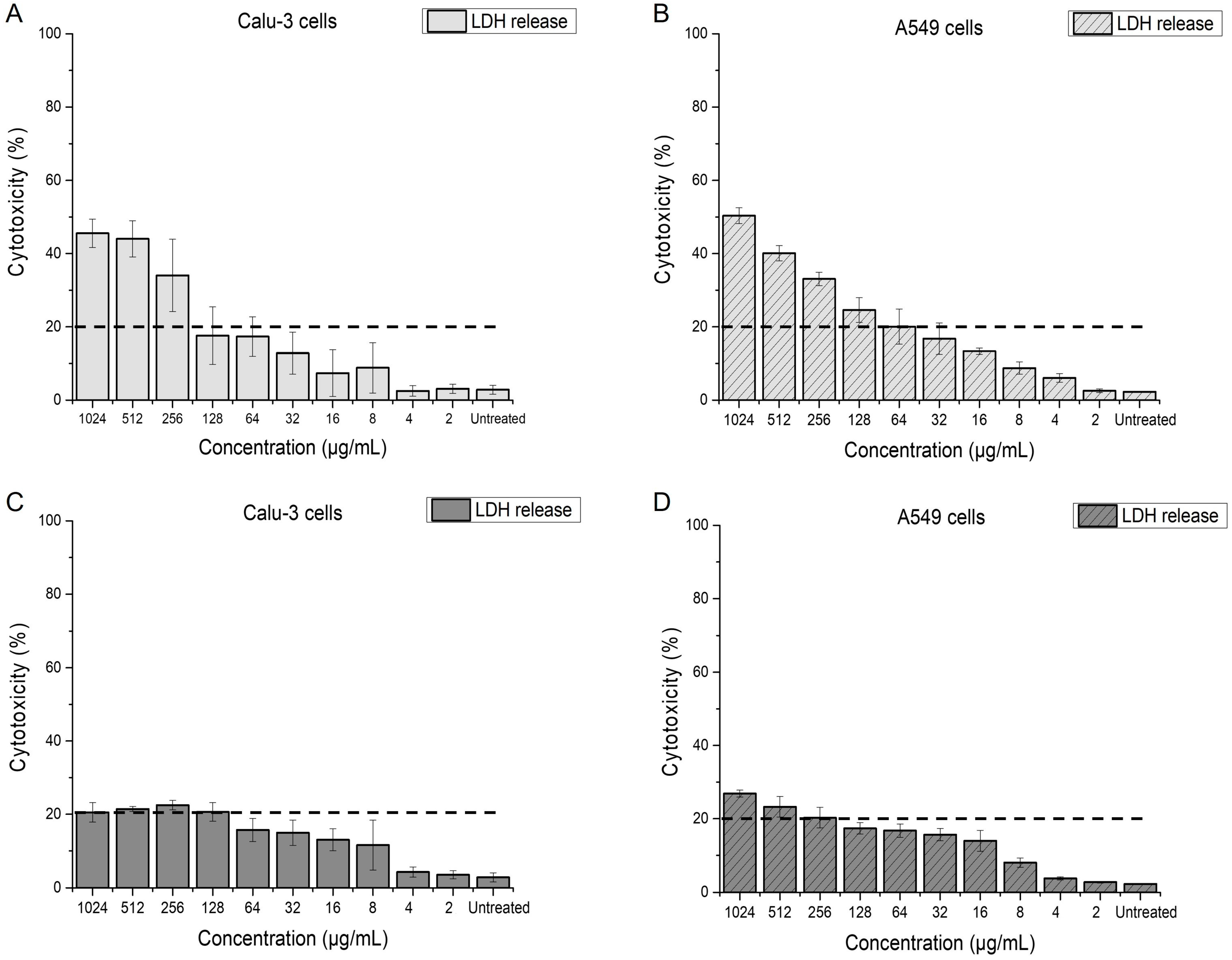
2.4. Effect of Menadione on Epithelial Lung Cells
2.4.1. Impact of Menadione on Membrane Integrity of Calu-3 and A549 Cells
2.4.2. ROS Induction in Calu-3 Cells
3. Materials and Methods
3.1. Bacterial Strains
3.2. Media and Growth Conditions
Maintenance of Bacterial Strains
3.3. Antimicrobial Activity of Azithromycin and Menadione
3.3.1. Checkerboard Assay-MIC
3.3.2. LC-MS Analysis
3.4. MBEC Assay
3.4.1. Checkerboard Assay-MBEC
3.4.2. Scanning Electron Microscopy (SEM)
3.5. Reactive Oxygen Species in Bacteria
3.6. mCherry Leakage from E. coli Cells
3.7. Cell Culture
3.8. Membrane Integrity Assay
3.9. Reactive Oxygen Species in Calu-3 Cells
4. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Poudel, A.N.; Zhu, S.; Cooper, N.; Little, P.; Tarrant, C.; Hickman, M.; Yao, G. The Economic Burden of Antibiotic Resistance: A Systematic Review and Meta-Analysis. PLoS ONE 2023, 18, e0285170. [Google Scholar] [CrossRef] [PubMed]
- Klinger-Strobel, M.; Lautenschläger, C.; Fischer, D.; Mainz, J.G.; Bruns, T.; Tuchscherr, L.; Pletz, M.W.; Makarewicz, O. Aspects of Pulmonary Drug Delivery Strategies for Infections in Cystic Fibrosis—Where Do We Stand? Expert. Opin. Drug Deliv. 2015, 12, 1351–1374. [Google Scholar] [CrossRef] [PubMed]
- Perikleous, E.P.; Gkentzi, D.; Bertzouanis, A.; Paraskakis, E.; Sovtic, A.; Fouzas, S. Antibiotic Resistance in Patients with Cystic Fibrosis: Past, Present, and Future. Antibiotics 2023, 12, 217. [Google Scholar] [CrossRef] [PubMed]
- Graeber, S.Y.; Mall, M.A. The Future of Cystic Fibrosis Treatment: From Disease Mechanisms to Novel Therapeutic Approaches. Lancet 2023, 402, 1185–1198. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas Aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef] [PubMed]
- CF Foundation Cystic Fibrosis Foundation. Available online: https://www.cff.org/managing-cf/antibiotics (accessed on 3 February 2025).
- Imperi, F.; Leoni, L.; Visca, P. Antivirulence Activity of Azithromycin in Pseudomonas Aeruginosa. Front. Microbiol. 2014, 5, 178. [Google Scholar] [CrossRef]
- Ong, T.; Ramsey, B.W. Cystic Fibrosis. JAMA 2023, 329, 1859. [Google Scholar] [CrossRef]
- Vitiello, A.; Blasi, F.; Sabbatucci, M.; Zovi, A.; Miele, F.; Ponzo, A.; Langella, R.; Boccellino, M. The Impact of Antimicrobial Resistance in Cystic Fibrosis. J. Clin. Med. 2024, 13, 1711. [Google Scholar] [CrossRef]
- De Boeck, K.; Amaral, M.D. Progress in Therapies for Cystic Fibrosis. Lancet Respir. Med. 2016, 4, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Ahmed, M.Z.; Rafique, S.; Almasoudi, S.E.; Shah, M.; Jalil, N.A.C.; Ojha, S.C. Recent Approaches for Downplaying Antibiotic Resistance: Molecular Mechanisms. Biomed. Res. Int. 2023, 2023, 1–27. [Google Scholar] [CrossRef]
- Aminin, D.; Polonik, S. 1,4-Naphthoquinones: Some Biological Properties and Application. Chem. Pharm. Bull. 2020, 68, 46–57. [Google Scholar] [CrossRef]
- Mone, N.; Bhagwat, S.; Sharma, D.; Chaskar, M.; Patil, R.; Zamboni, P.; Nawani, N.; Satpute, S. Naphthoquinones and Their Derivatives: Emerging Trends in Combating Microbial Pathogens. Coatings 2021, 11, 434. [Google Scholar] [CrossRef]
- Ohene-Agyei, T.; Mowla, R.; Rahman, T.; Venter, H. Phytochemicals Increase the Antibacterial Activity of Antibiotics by Acting on a Drug Efflux Pump. Microbiologyopen 2014, 3, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Sutormin, D.; Rubanova, N.; Logacheva, M.; Ghilarov, D.; Severinov, K. Single-Nucleotide-Resolution Mapping of DNA Gyrase Cleavage Sites across the Escherichia Coli Genome. Nucleic Acids Res. 2019, 47, 1373–1388. [Google Scholar] [CrossRef]
- Paul, P.; Chakraborty, P.; Chatterjee, A.; Sarker, R.K.; Dastidar, D.G.; Kundu, T.; Sarkar, N.; Das, A.; Tribedi, P. 1,4-Naphthoquinone Accumulates Reactive Oxygen Species in Staphylococcus Aureus: A Promising Approach towards Effective Management of Biofilm Threat. Arch. Microbiol. 2021, 203, 1183–1193. [Google Scholar] [CrossRef]
- Andrade, J.C.; Morais Braga, M.F.B.; Guedes, G.M.M.; Tintino, S.R.; Freitas, M.A.; Quintans, L.J.; Menezes, I.R.A.; Coutinho, H.D.M. Menadione (Vitamin K) Enhances the Antibiotic Activity of Drugs by Cell Membrane Permeabilization Mechanism. Saudi J. Biol. Sci. 2017, 24, 59–64. [Google Scholar] [CrossRef]
- Dey, D.; Ray, R.; Hazra, B. Antitubercular and Antibacterial Activity of Quinonoid Natural Products Against Multi-Drug Resistant Clinical Isolates. Phytother. Res. 2014, 28, 1014–1021. [Google Scholar] [CrossRef]
- Mone, N.S.; Syed, S.; Ravichandiran, P.; Kamble, E.E.; Pardesi, K.R.; Salunke-Gawali, S.; Rai, M.; Vikram Singh, A.; Prasad Dakua, S.; Park, B.; et al. Synergistic and Additive Effects of Menadione in Combination with Antibiotics on Multidrug-Resistant Staphylococcus Aureus: Insights from Structure-Function Analysis of Naphthoquinones. ChemMedChem 2023. [Google Scholar] [CrossRef] [PubMed]
- Leitão, A.C.; Ferreira, T.L.; Gurgel do Amaral Valente Sá, L.; Rodrigues, D.S.; de Souza, B.O.; Barbosa, A.D.; Moreira, L.E.A.; de Andrade Neto, J.B.; Cabral, V.P.D.F.; Rios, M.E.F.; et al. Antibacterial Activity of Menadione Alone and in Combination with Oxacillin against Methicillin-Resistant Staphylococcus aureus and Its Impact on Biofilms. J. Med. Microbiol. 2023, 72, 001751. [Google Scholar] [CrossRef] [PubMed]
- Chiou, T.-J.; Tzeng, W.-F. The Roles of Glutathione and Antioxidant Enzymes in Menadione-Induced Oxidative Stress. Toxicology 2000, 154, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, J.P.; Martins, A.F.; Nunes, C.; Morais, C.M.; Lúcio, M.; Reis, S.; Pinheiro, T.J.T.; Geraldes, C.F.G.C.; Oliveira, P.J.; Jurado, A.S. A Biophysical Approach to Menadione Membrane Interactions: Relevance for Menadione-Induced Mitochondria Dysfunction and Related Deleterious/Therapeutic Effects. Biochim. Et Biophys. Acta (BBA)—Biomembr. 2013, 1828, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Shertzer, H.G.; Låstbom, L.; Sainsbury, M.; Moldéus, P. Menadione-Mediated Membrane Fluidity Alterations and Oxidative Damage in Rat Hepatocytes. Biochem. Pharmacol. 1992, 43, 2135–2141. [Google Scholar] [CrossRef] [PubMed]
- Yap, C.H.; Lim, S.K.; Chan, Y.L.; Chee, C.F.; Tay, S.T. Potential Application of Menadione for Antimicrobial Coating of Surgical Sutures. Biotechnol. Notes 2023, 4, 20–27. [Google Scholar] [CrossRef]
- Heidary, M.; Ebrahimi Samangani, A.; Kargari, A.; Kiani Nejad, A.; Yashmi, I.; Motahar, M.; Taki, E.; Khoshnood, S. Mechanism of Action, Resistance, Synergism, and Clinical Implications of Azithromycin. J. Clin. Lab. Anal. 2022, 36, e24427. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. Biotic Acts of Antibiotics. Front. Microbiol. 2013, 4, 241. [Google Scholar] [CrossRef] [PubMed]
- Steel, H.C.; Theron, A.J.; Cockeran, R.; Anderson, R.; Feldman, C. Pathogen- and Host-Directed Anti-Inflammatory Activities of Macrolide Antibiotics. Mediators Inflamm. 2012, 2012, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Fatsis-Kavalopoulos, N.; Sánchez-Hevia, D.L.; Andersson, D.I. Beyond the FIC Index: The Extended Information from Fractional Inhibitory Concentrations (FICs). J. Antimicrob. Chemother. 2024, 79, 2394–2396. [Google Scholar] [CrossRef] [PubMed]
- Odds, F.C. Synergy, Antagonism, and What the Chequerboard Puts between Them. J. Antimicrob. Chemother. 2003, 52, 1. [Google Scholar] [CrossRef]
- Kamal, M.A.M.; Bassil, J.; Loretz, B.; Hirsch, A.K.H.; Lee, S.; Lehr, C.-M. Arg-Biodynamers as Antibiotic Potentiators through Interacting with Gram-Negative Outer Membrane Lipopolysaccharides. Eur. J. Pharm. Biopharm. 2024, 200, 114336. [Google Scholar] [CrossRef]
- Gebreyohannes, G.; Nyerere, A.; Bii, C.; Sbhatu, D.B. Challenges of Intervention, Treatment, and Antibiotic Resistance of Biofilm-Forming Microorganisms. Heliyon 2019, 5, e02192. [Google Scholar] [CrossRef] [PubMed]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial Biofilms: From the Natural Environment to Infectious Diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Ceri, H.; Olson, M.E.; Stremick, C.; Read, R.R.; Morck, D.; Buret, A. The Calgary Biofilm Device: New Technology for Rapid Determination of Antibiotic Susceptibilities of Bacterial Biofilms. J. Clin. Microbiol. 1999, 37, 1771–1776. [Google Scholar] [CrossRef]
- Harrison, J.J.; Stremick, C.A.; Turner, R.J.; Allan, N.D.; Olson, M.E.; Ceri, H. Microtiter Susceptibility Testing of Microbes Growing on Peg Lids: A Miniaturized Biofilm Model for High-Throughput Screening. Nat. Protoc. 2010, 5, 1236–1254. [Google Scholar] [CrossRef] [PubMed]
- Mone, N.S.; Kamble, E.E.; Pardesi, K.R.; Satpute, S.K. Antibacterial and Antibiofilm Potency of Menadione Against Multidrug-Resistant, S. Aureus. Curr. Microbiol. 2022, 79, 282. [Google Scholar] [CrossRef] [PubMed]
- Ruhal, R.; Kataria, R. Biofilm Patterns in Gram-Positive and Gram-Negative Bacteria. Microbiol. Res. 2021, 251, 126829. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, D.; Sehgal, T. Biofilm Development in Gram-Positive and Gram-Negative Bacteria. In Focus on Bacterial Biofilms; IntechOpen: London, UK, 2022. [Google Scholar]
- San Mauro, A.J.S.; Høiby, N.; Ciofu, O. Increased Susceptibility to Azithromycin of Pseudomonas Aeruginosa Biofilms Using RPMI 1640 Testing Media. APMIS 2024, 132, 1086–1095. [Google Scholar] [CrossRef]
- Loor, G.; Kondapalli, J.; Schriewer, J.M.; Chandel, N.S.; Vanden Hoek, T.L.; Schumacker, P.T. Menadione Triggers Cell Death through ROS-Dependent Mechanisms Involving PARP Activation without Requiring Apoptosis. Free Radic. Biol. Med. 2010, 49, 1925–1936. [Google Scholar] [CrossRef]
- Schlievert, P.M.; Merriman, J.A.; Salgado-Pabón, W.; Mueller, E.A.; Spaulding, A.R.; Vu, B.G.; Chuang-Smith, O.N.; Kohler, P.L.; Kirby, J.R. Menaquinone Analogs Inhibit Growth of Bacterial Pathogens. Antimicrob. Agents Chemother. 2013, 57, 5432–5437. [Google Scholar] [CrossRef]
- Van Acker, H.; Coenye, T. The Role of Reactive Oxygen Species in Antibiotic-Mediated Killing of Bacteria. Trends Microbiol. 2017, 25, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Xiuhong Wang, J.Z.; Zhao, X. Post-Stress Bacterial Cell Death Mediated by Reactive Oxygen Species. Prog. Nucl. Energy 6 Biol. Sci. 2019, 116, 10064–10071. [Google Scholar] [CrossRef] [PubMed]
- Blanch-Asensio, M.; Dey, S.; Sankaran, S. In Vitro Assembly of Plasmid DNA for Direct Cloning in Lactiplantibacillus Plantarum WCSF1. PLoS ONE 2023, 18, e0281625. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.G.; Palmer, A.E.; Tsien, R.Y. Improved Monomeric Red, Orange and Yellow Fluorescent Proteins Derived from Discosoma Sp. Red Fluorescent Protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef]
- Nakano, H.; Murai, S.; Moriwaki, K. Regulation of the Release of Damage-Associated Molecular Patterns from Necroptotic Cells. Biochem. J. 2022, 479, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Scheffer, L.L.; Sreetama, S.C.; Sharma, N.; Medikayala, S.; Brown, K.J.; Defour, A.; Jaiswal, J.K. Mechanism of Ca2+-Triggered ESCRT Assembly and Regulation of Cell Membrane Repair. Nat. Commun. 2014, 5, 5646. [Google Scholar] [CrossRef] [PubMed]
- Sankaran, S.; del Campo, A. Optoregulated Protein Release from an Engineered Living Material. Adv. Biosyst. 2019, 3, 1800312. [Google Scholar] [CrossRef] [PubMed]
- Criddle, D.N.; Gillies, S.; Baumgartner-Wilson, H.K.; Jaffar, M.; Chinje, E.C.; Passmore, S.; Chvanov, M.; Barrow, S.; Gerasimenko, O.V.; Tepikin, A.V.; et al. Menadione-Induced Reactive Oxygen Species Generation via Redox Cycling Promotes Apoptosis of Murine Pancreatic Acinar Cells. J. Biol. Chem. 2006, 281, 40485–40492. [Google Scholar] [CrossRef] [PubMed]
- Negri, L.B.; Mannaa, Y.; Korupolu, S.; Farinelli, W.A.; Anderson, R.R.; Gelfand, J.A. Vitamin K3 (Menadione) Is a Multifunctional Microbicide Acting as a Photosensitizer and Synergizing with Blue Light to Kill Drug-Resistant Bacteria in Biofilms. J. Photochem. Photobiol. B 2023, 244, 112720. [Google Scholar] [CrossRef] [PubMed]
- FDA FDA-Drug Approvals and Databases. Available online: https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&ApplNo=002139 (accessed on 3 February 2025).
- Chivé, C.; Mc Cord, C.; Sanchez-Guzman, D.; Brookes, O.; Joseph, P.; Lai Kuen, R.; Phan, G.; Baeza-Squiban, A.; Devineau, S.; Boland, S. 3D Model of the Bronchial Epithelial Barrier to Study Repeated Exposure to Xenobiotics: Application to Silver Nanoparticles. Environ. Toxicol. Pharmacol. 2023, 103, 104281. [Google Scholar] [CrossRef] [PubMed]
- Meziu, E.; Shehu, K.; Koch, M.; Schneider, M.; Kraegeloh, A. Impact of Mucus Modulation by N-Acetylcysteine on Nanoparticle Toxicity. Int. J. Pharm. X 2023, 6, 100212. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, E. Comparison of Conventional and Advanced in Vitro Models in the Toxicity Testing of Nanoparticles. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- López-García, J.; Lehocký, M.; Humpolíček, P.; Sáha, P. HaCaT Keratinocytes Response on Antimicrobial Atelocollagen Substrates: Extent of Cytotoxicity, Cell Viability and Proliferation. J. Funct. Biomater. 2014, 5, 43–57. [Google Scholar] [CrossRef]
- McCormick, M.L.; Denning, G.M.; Reszka, K.J.; Bilski, P.; Buettner, G.R.; Rasmussen, G.T.; Railsback, M.A.; Britigan, B.E. Biological Effects of Menadione Photochemistry: Effects of Menadione on Biological Systems May Not Involve Classical Oxidant Production. Biochem. J. 2000, 350, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Gerasimenko, J.V.; Gerasimenko, O.V.; Palejwala, A.; Tepikin, A.V.; Petersen, O.H.; Watson, A.J.M. Menadione-Induced Apoptosis: Roles of Cytosolic Ca2+elevations and the Mitochondrial Permeability Transition Pore. J. Cell Sci. 2002, 115, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Murgia, X.; Loretz, B.; Hartwig, O.; Hittinger, M.; Lehr, C.-M. The Role of Mucus on Drug Transport and Its Potential to Affect Therapeutic Outcomes. Adv. Drug Deliv. Rev. 2018, 124, 82–97. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on Transformation of Escherichia Coli with Plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Mathiesen, G.; Sveen, A.; Brurberg, M.; Fredriksen, L.; Axelsson, L.; Eijsink, V.G. Genome-Wide Analysis of Signal Peptide Functionality in Lactobacillus Plantarum WCFS1. BMC Genom. 2009, 10, 425. [Google Scholar] [CrossRef]
- Weisblum, B. Erythromycin Resistance by Ribosome Modification. Antimicrob. Agents Chemother. 1995, 39, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Jost, C.; Bidet, P.; Carrère, T.; Mariani-Kurkdjian, P.; Bonacorsi, S. Susceptibility of Enterohaemorrhagic Escherichia coli to Azithromycin in France and Analysis of Resistance Mechanisms. J. Antimicrob. Chemother. 2016, 71, 1183–1187. [Google Scholar] [CrossRef]
- Yang, X.; Liu, X.; Yang, C.; Chan, E.W.-C.; Zhang, R.; Chen, S. A Conjugative IncI1 Plasmid Carrying Erm(B) and BlaCTX-M-104 That Mediates Resistance to Azithromycin and Cephalosporins. Microbiol. Spectr. 2021, 9, e00286-21. [Google Scholar] [CrossRef] [PubMed]
- Oberhardt, M.A.; Puchałka, J.; Fryer, K.E.; Martins dos Santos, V.A.P.; Papin, J.A. Genome-Scale Metabolic Network Analysis of the Opportunistic Pathogen Pseudomonas aeruginosa PAO1. J. Bacteriol. 2008, 190, 2790–2803. [Google Scholar] [CrossRef] [PubMed]
- Klockgether, J.; Munder, A.; Neugebauer, J.; Davenport, C.F.; Stanke, F.; Larbig, K.D.; Heeb, S.; Schöck, U.; Pohl, T.M.; Wiehlmann, L.; et al. Genome Diversity of Pseudomonas aeruginosa PAO1 Laboratory Strains. J. Bacteriol. 2010, 192, 1113–1121. [Google Scholar] [CrossRef] [PubMed]
- Miller, H.J. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia Coli and Related Bacteria; Cold Spring Harbor Laboratory Press: Plainview, NY, USA, 1992. [Google Scholar]
- Cold Spring Harbor Protocols. Available online: https://cshprotocols.cshlp.org/content/2010/8/pdb.rec12295.short (accessed on 3 February 2025).
- Eucast. Eucast Determination of Minimum Inhibitory Concentrations (MICs) of Antibacterial Agents by Broth Dilution. Clin. Microbiol. Infect. 2003, 9, ix–xv. [Google Scholar] [CrossRef]
- Caballero, U.; Kim, S.; Eraso, E.; Quindós, G.; Vozmediano, V.; Schmidt, S.; Jauregizar, N. In Vitro Synergistic Interactions of Isavuconazole and Echinocandins against Candida Auris. Antibiotics 2021, 10, 355. [Google Scholar] [CrossRef] [PubMed]
- Eruslanov, E.; Kusmartsev, S. Identification of ROS Using Oxidized DCFDA and Flow-Cytometry. In Advanced Protocols in Oxidative Stress II; Humana Press: Totowa, NJ, USA, 2010; pp. 57–72. [Google Scholar]
- Kim, H.; Xue, X. Detection of Total Reactive Oxygen Species in Adherent Cells by 2′,7′-Dichlorodihydrofluorescein Diacetate Staining. J. Vis. Exp. 2020, 160, e60682. [Google Scholar] [CrossRef]
- Halliwell, B.; Whiteman, M. Measuring Reactive Species and Oxidative Damage in Vivo and in Cell Culture: How Should You Do It and What Do the Results Mean? Br. J. Pharmacol. 2004, 142, 231–255. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P.; Bayir, H.; Belousov, V.; Chang, C.J.; Davies, K.J.A.; Davies, M.J.; Dick, T.P.; Finkel, T.; Forman, H.J.; Janssen-Heininger, Y.; et al. Guidelines for Measuring Reactive Oxygen Species and Oxidative Damage in Cells and in Vivo. Nat. Metab. 2022, 4, 651–662. [Google Scholar] [CrossRef]
- Ravichandiran, P.; Sheet, S.; Premnath, D.; Kim, A.R.; Yoo, D.J. 1,4-Naphthoquinone Analogues: Potent Antibacterial Agents and Mode of Action Evaluation. Molecules 2019, 24, 1437. [Google Scholar] [CrossRef]
- Fogh, J.; Fogh, J.M.; Orfeo, T. One Hundred and Twenty-Seven Cultured Human Tumor Cell Lines Producing Tumors in Nude Mice23. J. Natl. Cancer Inst. 1977, 59, 221–226. [Google Scholar] [CrossRef]
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In Vitro Cultivation of Human Tumors: Establishment of Cell Lines Derived From a Series of Solid Tumors2. J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef]
- Loret, T.; Peyret, E.; Dubreuil, M.; Aguerre-Chariol, O.; Bressot, C.; le Bihan, O.; Amodeo, T.; Trouiller, B.; Braun, A.; Egles, C.; et al. Air-Liquid Interface Exposure to Aerosols of Poorly Soluble Nanomaterials Induces Different Biological Activation Levels Compared to Exposure to Suspensions. Part. Fibre Toxicol. 2016, 13, 58. [Google Scholar] [CrossRef]
- Montefusco-Pereira, C.V.; Horstmann, J.C.; Ebensen, T.; Beisswenger, C.; Bals, R.; Guzmán, C.A.; Schneider-Daum, N.; de Souza Carvalho-Wodarz, C.; Lehr, C.M. Aeruginosa Infected 3D Co-Culture of Bronchial Epithelial Cells and Macrophages at Air-Liquid Interface for Preclinical Evaluation of Anti-Infectives. J. Vis. Exp. 2020, 160, e61069. [Google Scholar] [CrossRef]
- Aliyazdi, S.; Frisch, S.; Hidalgo, A.; Frank, N.; Krug, D.; Müller, R.; Schaefer, U.F.; Vogt, T.; Loretz, B.; Lehr, C.-M. 3D Bioprinting of E. Coli MG1655 Biofilms on Human Lung Epithelial Cells for Building Complex in Vitro Infection Models. Biofabrication 2023, 15, 035019. [Google Scholar] [CrossRef] [PubMed]
- Crabbé, A.; Liu, Y.; Matthijs, N.; Rigole, P.; De La Fuente-Nùñez, C.; Davis, R.; Ledesma, M.A.; Sarker, S.; Van Houdt, R.; Hancock, R.E.W.; et al. Antimicrobial Efficacy against Pseudomonas Aeruginosa Biofilm Formation in a Three-Dimensional Lung Epithelial Model and the Influence of Fetal Bovine Serum. Sci. Rep. 2017, 7, 43321. [Google Scholar] [CrossRef]
- Mekontso, J.A.; Farrukh, U.; Trujillo, S.; del Campo, A. A Practical Workflow for Cytocompatibility Assessment of Living Therapeutic Materials. Biomater. Adv. 2025, 169, 214182. [Google Scholar] [CrossRef]
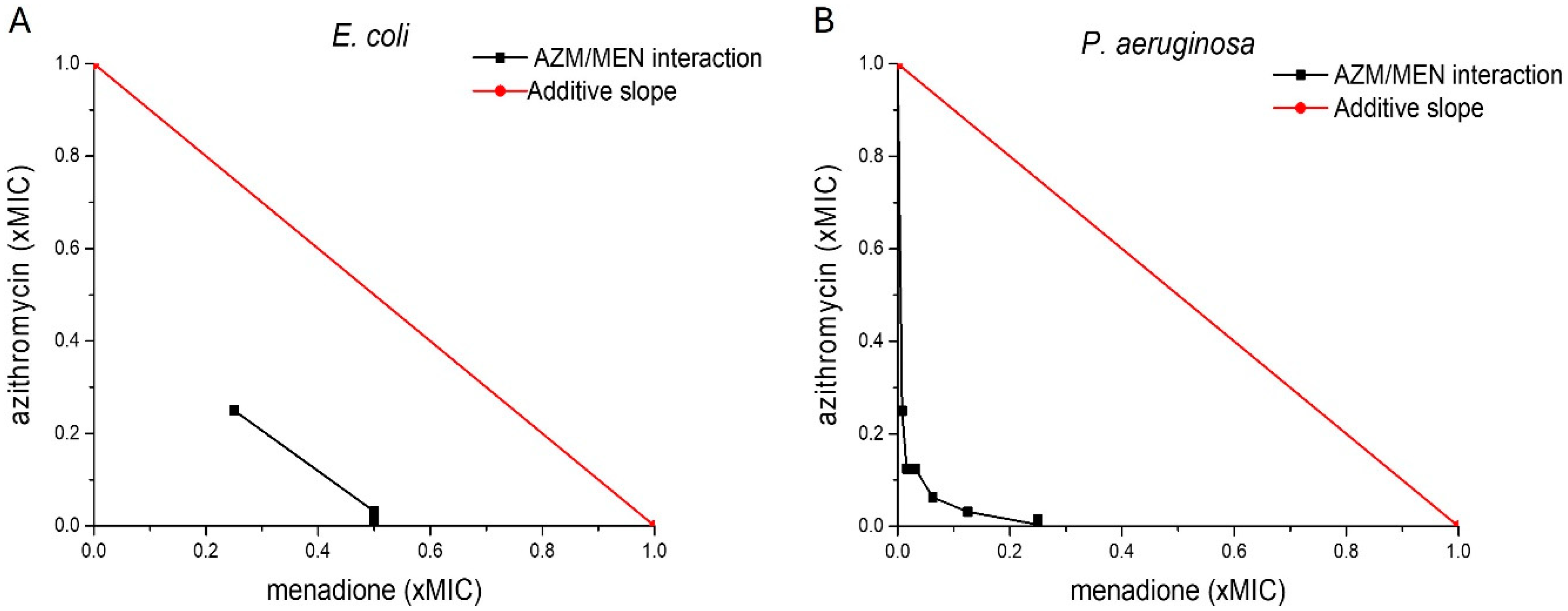
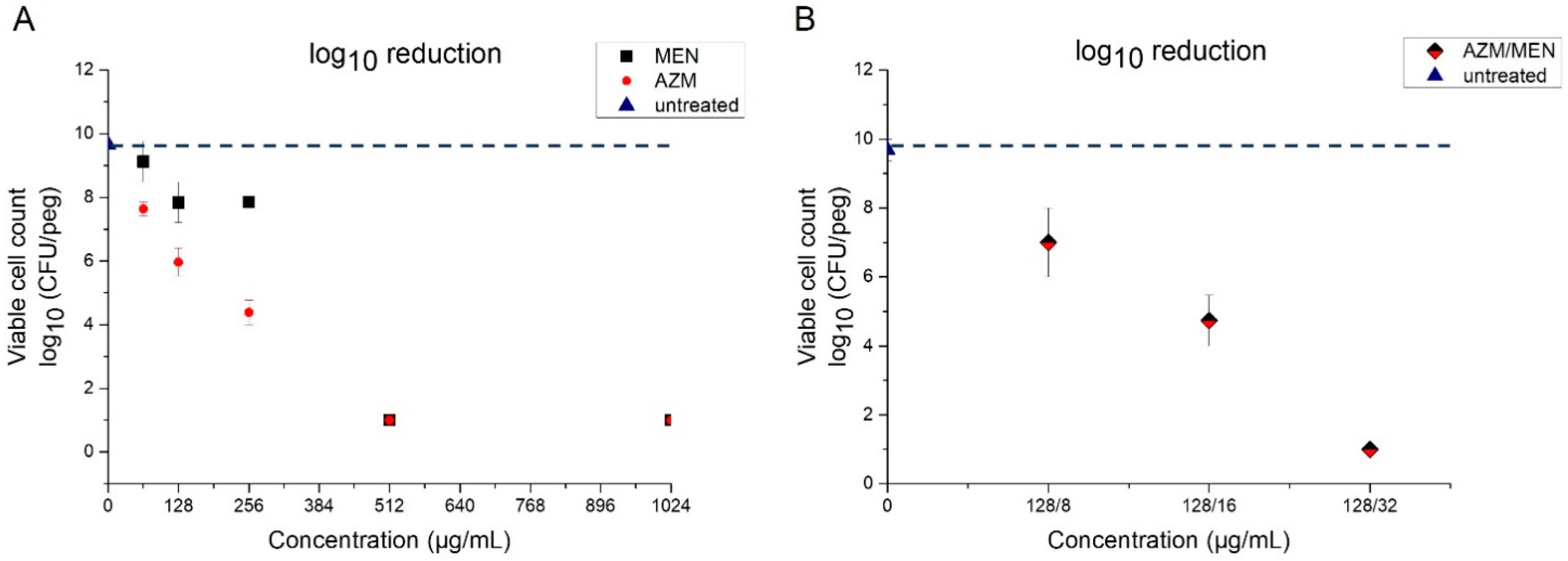


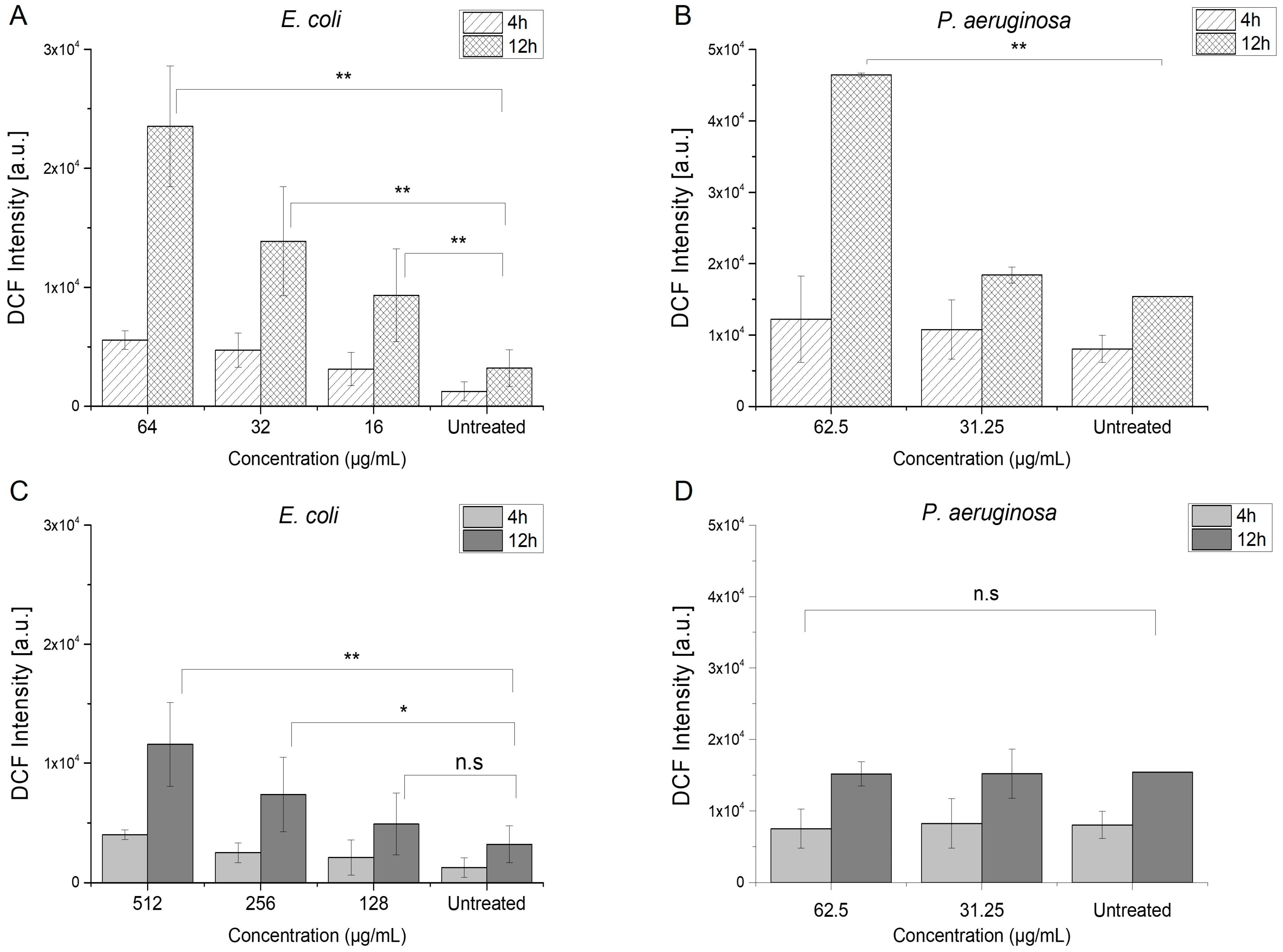
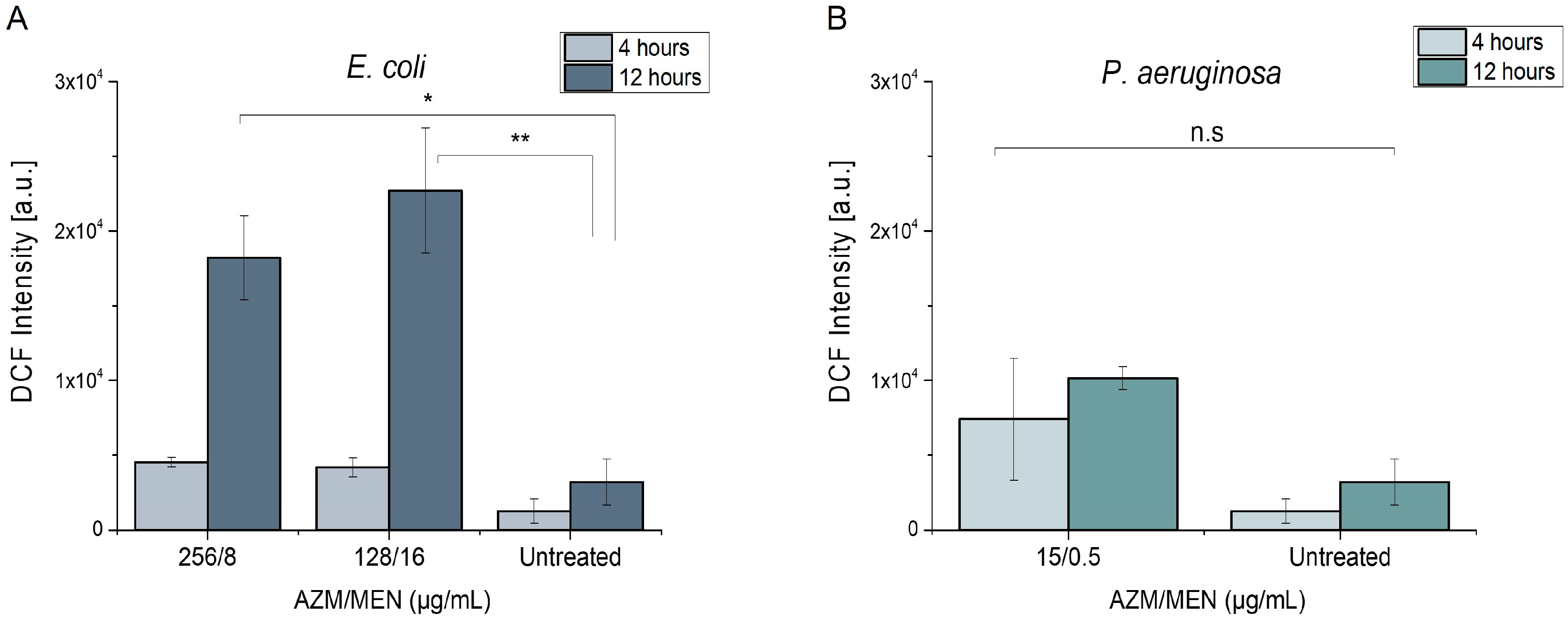


| Bacterial Strain | Azithromycin | Menadione | Azithromycin/Menadione | |
|---|---|---|---|---|
| MIC (µg/mL) | MIC (µg/mL) | MIC (µg/mL) | FICI | |
| AZMr-E. coli DH5α | 512 | 64 | 128/16 | 0.5 |
| P. aeruginosa | 62.5 | 62.5 | 15/0.5 | 0.248 |
| Bacterial Strain | MBEC (µg/mL) | ||
|---|---|---|---|
| Azithromycin | Menadione | Azithromycin/Menadione | |
| P. aeruginosa | 256 | 512 | 128/16 |
| Strain | Resistance | Plasmid | Fluorescence | Source | Abbreviation in Text |
|---|---|---|---|---|---|
| drug-resistant E. coli DH5α | ermB | pLp3050sNuc | mCherry | NEB, Art.No.C2987 | AZMr-E. coli DH5α |
| Pseudomonas aeruginosa PAO1 | - | - | - | DSM 22644 | P. aeruginosa |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shehu, K.; Schneider, M.; Kraegeloh, A. Menadione as Antibiotic Adjuvant Against P. aeruginosa: Mechanism of Action, Efficacy and Safety. Antibiotics 2025, 14, 163. https://doi.org/10.3390/antibiotics14020163
Shehu K, Schneider M, Kraegeloh A. Menadione as Antibiotic Adjuvant Against P. aeruginosa: Mechanism of Action, Efficacy and Safety. Antibiotics. 2025; 14(2):163. https://doi.org/10.3390/antibiotics14020163
Chicago/Turabian StyleShehu, Kristela, Marc Schneider, and Annette Kraegeloh. 2025. "Menadione as Antibiotic Adjuvant Against P. aeruginosa: Mechanism of Action, Efficacy and Safety" Antibiotics 14, no. 2: 163. https://doi.org/10.3390/antibiotics14020163
APA StyleShehu, K., Schneider, M., & Kraegeloh, A. (2025). Menadione as Antibiotic Adjuvant Against P. aeruginosa: Mechanism of Action, Efficacy and Safety. Antibiotics, 14(2), 163. https://doi.org/10.3390/antibiotics14020163








