Dynamic Spread of Antibiotic Resistance Determinants by Conjugation to a Human-Derived Gut Microbiota in a Transplanted Mouse Model
Abstract
1. Introduction
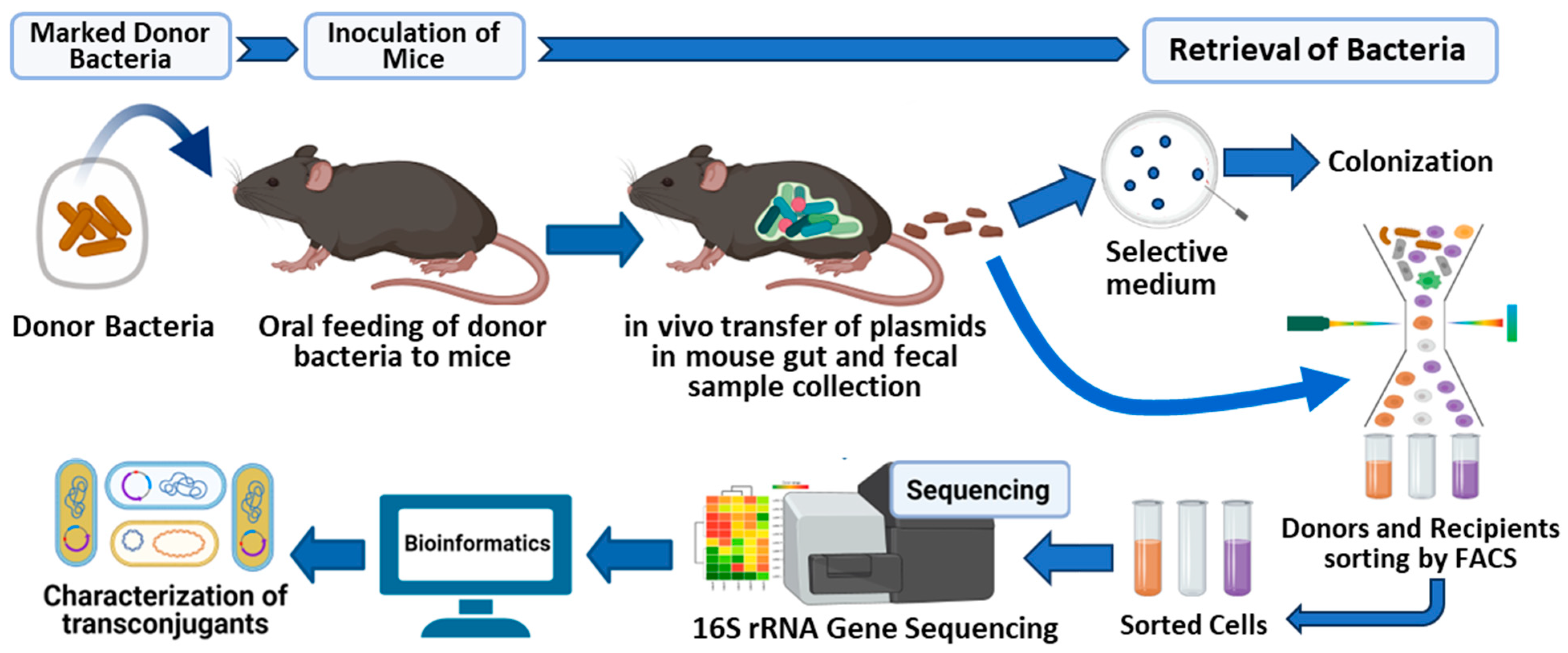
2. Results
2.1. Validation of Murine Model Carrying Human-Derived Gut Microbiota for These Experiments
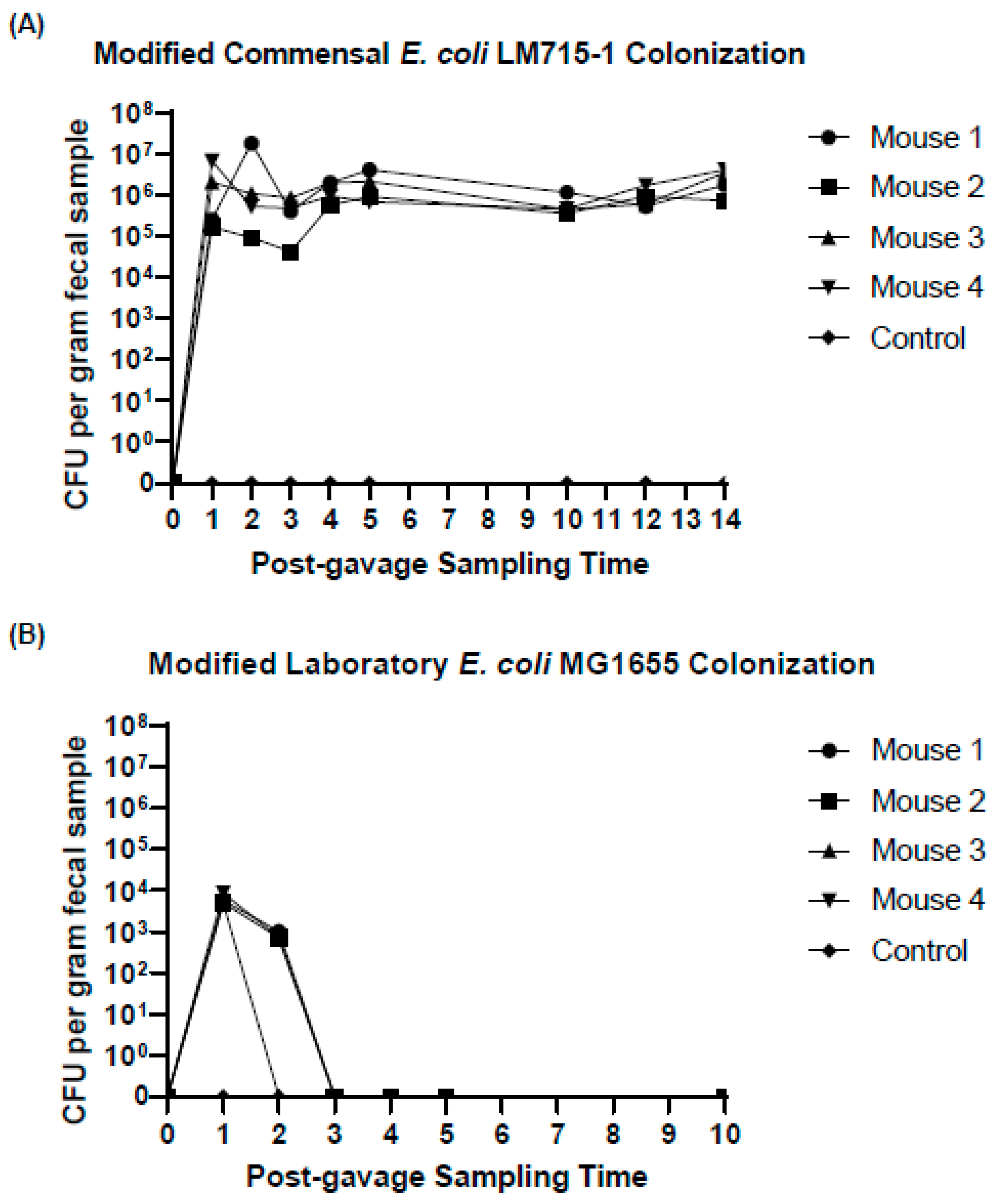
2.2. Fluorescently Labeled Commensal Donor E. coli Strain Colonized the Gut Longer than a Similarly Labeled Laboratory Strain of E. coli
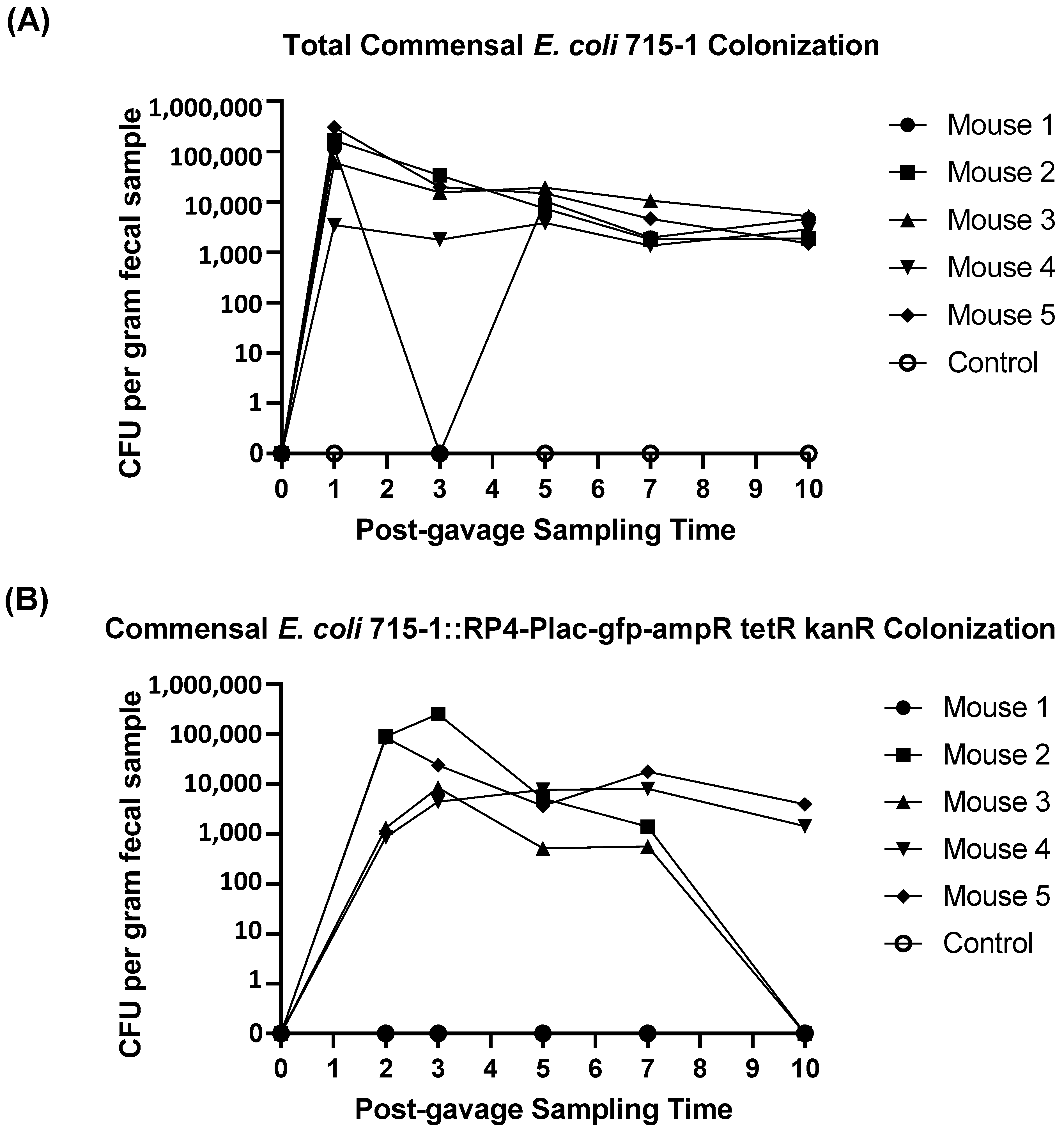
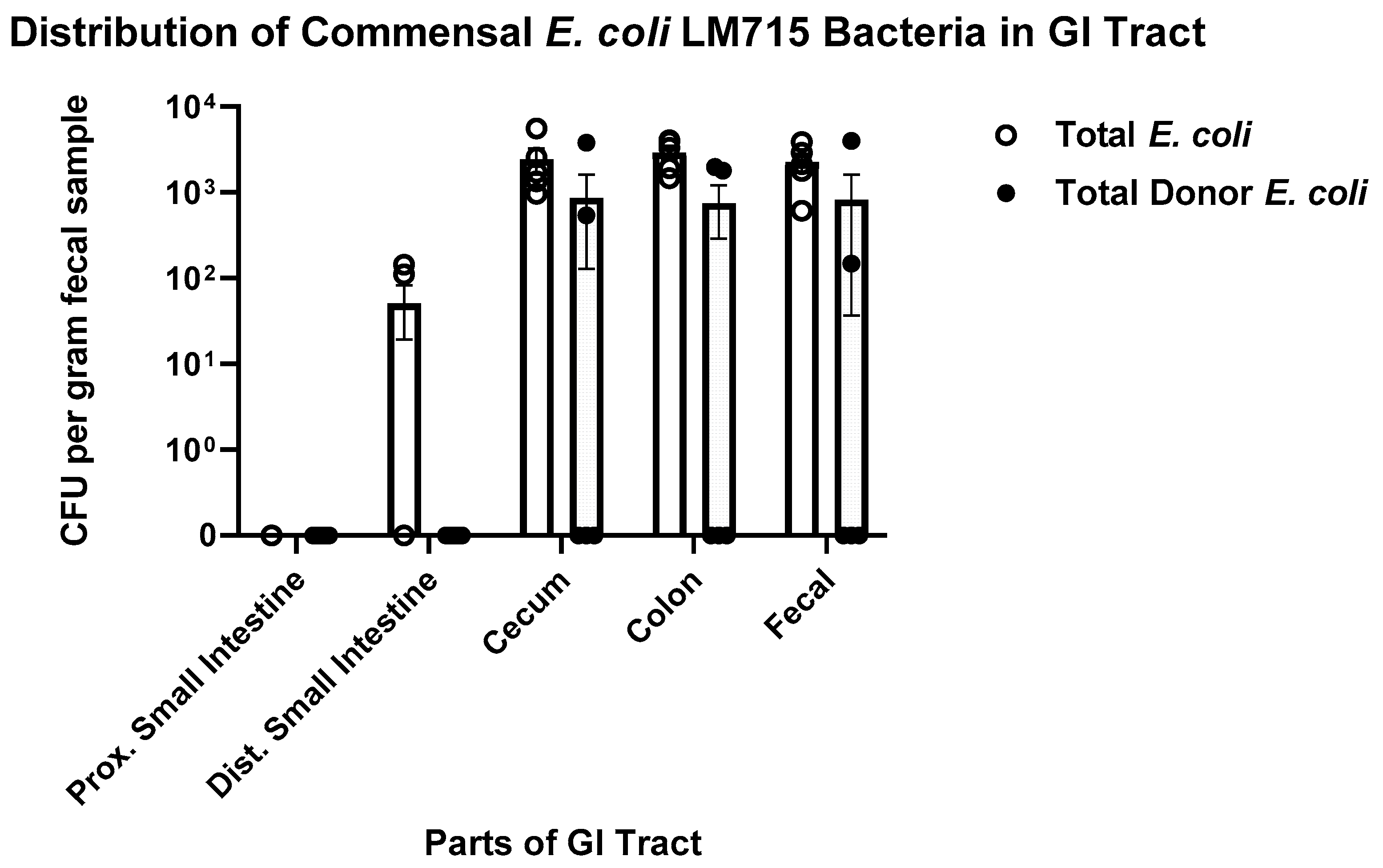
2.3. Commensal Donor E. coli LM715-1 Strain Carrying the RP4 Plasmid Persisted in the Gut Without Antibiotic Treatment
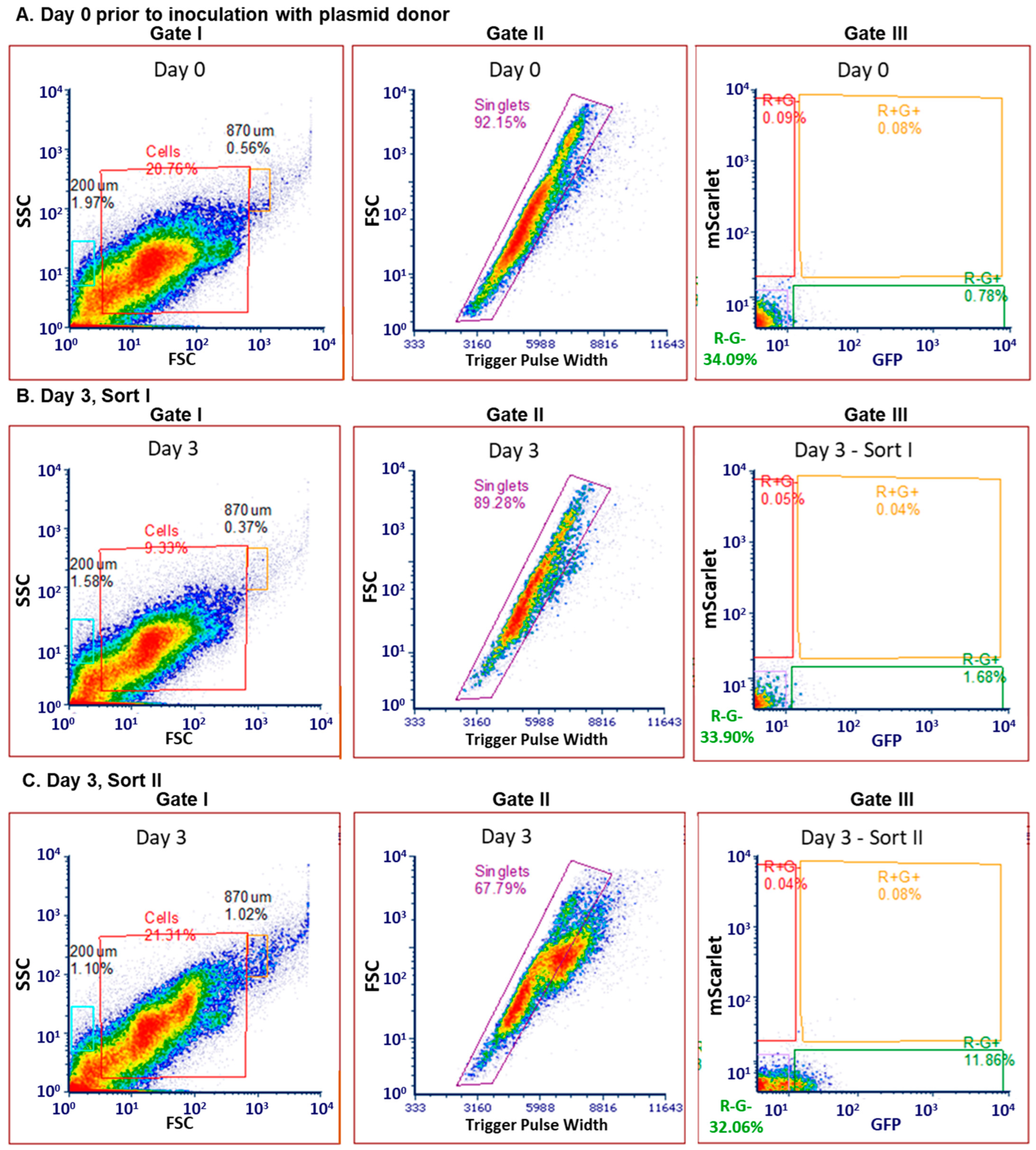
2.4. Determination of FACS Gating Strategy with Commensal Donor E. coli LM715-1 Before Inoculation into Mice
2.5. Sorting of Transconjugant Bacteria That Received Labeled RP4 Plasmid from Commensal Donor E. coli LM715-1
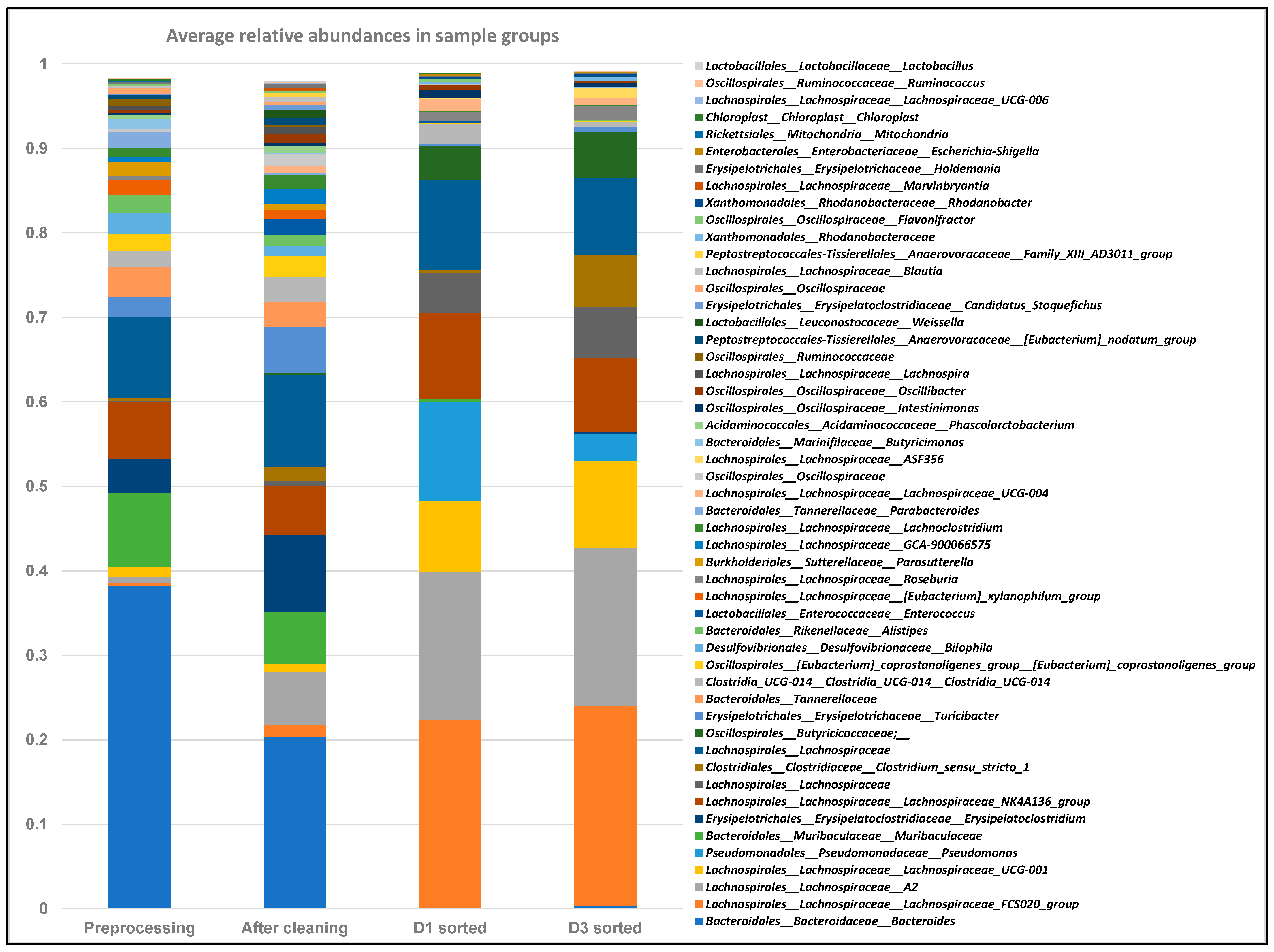
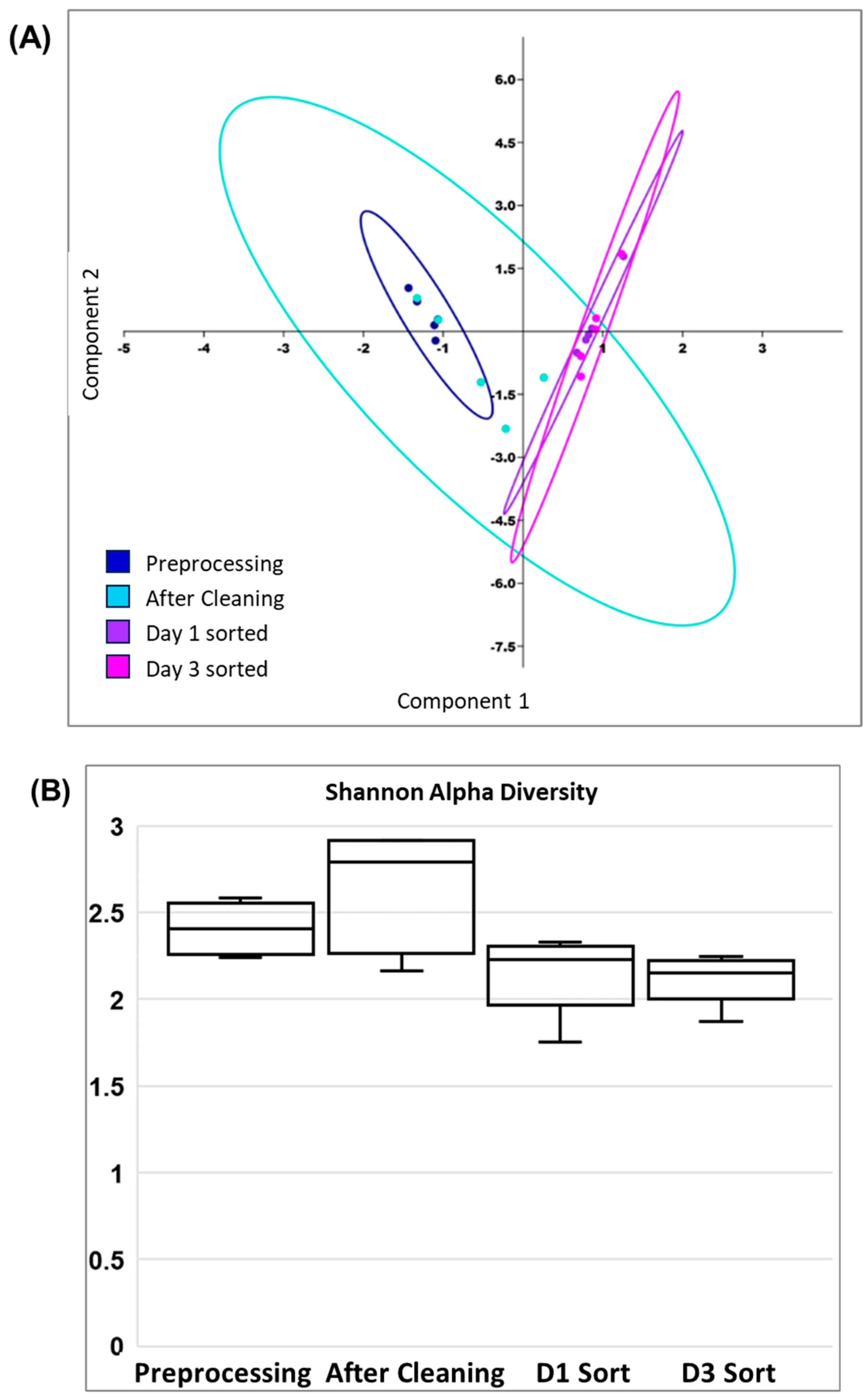
2.6. FACS Putative Transconjugants Comprised Diverse Population of Human Gut Microbiota Members
3. Discussion
4. Materials and Methods
4.1. Animal Ethics Statement
4.2. Media, Chemicals, and Reagents
4.3. Inoculation of Mice with Donor Strain
4.4. Assessing Colonization of Donor Strain in Fecal and GI Samples
4.5. Screening of Human Adult Fecal Microbiota Transplanted Mice for Presence of Bacteria with Antibiotic Resistance Phenotypes
4.6. Detection of Fluorescence in Donor and Transconjugant Bacteria
4.7. Validation of Antibiotic Resistance Markers and Fluorescence in Donor and Transconjugants
4.8. Fluorescence-Activated Cell Sorting of Donor and Transconjugant Bacteria
4.9. DNA Extraction from FACS Transconjugant Bacterial Cells
4.10. 16S rRNA Gene Sequencing and Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- CDC. Antibiotic Resistance Threats in the United States, 2019; US Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019; p. 114. Available online: www.cdc.gov/DrugResistance/Biggest-Threats.html (accessed on 8 January 2020).
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; The Review on Antimicrobial Resistance; Wellcome Trust: London, UK, 2016. [Google Scholar]
- CDC. Antibiotic Resistance Threats in the United States, 2013; CDC: Atlanta, GA, USA, 2013. [Google Scholar]
- Wallace, M.J.; Fishbein, S.R.S.; Dantas, G. Antimicrobial resistance in enteric bacteria: Current state and next-generation solutions. Gut Microbes 2020, 12, 1799654. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef]
- Mazel, D.; Davies, J. Antibiotic resistance in microbes. Cell. Mol. Life Sci. 1999, 56, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Barlow, M. What Antimicrobial Resistance Has Taught Us About Horizontal Gene Transfer; Humana Press: Totowa, NJ, USA, 2009; pp. 397–411. [Google Scholar]
- Alekshun, M.N.; Levy, S.B. Molecular Mechanisms of Antibacterial Multidrug Resistance. Cell 2007, 128, 1037–1050. [Google Scholar] [CrossRef]
- Dubnau, D. DNA Uptake in Bacteria. Annu. Rev. Microbiol. 1999, 53, 217–244. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.C.; Paul, J.H. Gene transfer by transduction in the marine environment. Appl. Environ. Microbiol. 1998, 64, 2780–2787. [Google Scholar] [CrossRef]
- Schicklmaier, P.; Schmieger, H. Frequency of generalized transducing phages in natural isolates of the Salmonella typhimurium complex. Appl. Environ. Microbiol. 1995, 61, 1637–1640. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V.; Passiatore, J.E.; Cannon, F. The mob and oriT mobilization functions of a bacterial plasmid promote its transfer to plants. Nature 1988, 328, 172–175. [Google Scholar] [CrossRef]
- Heinemann, J.A.; Sprague, G.F. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 1989, 340, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.A.; Diene, S.M.; Rolain, J.-M. Human microbiomes and antibiotic resistance. Hum. Microb. J. 2018, 10, 43–52. [Google Scholar] [CrossRef]
- Anthony, W.E.; Burnham, C.A.D.; Dantas, G.; Kwon, J.H. The Gut Microbiome as a Reservoir for Antimicrobial Resistance. J. Infect. Dis. 2021, 223, S209–S213. [Google Scholar] [CrossRef] [PubMed]
- Heuer, H.; Krãgerrecklenfort, E.; Wellington, E.M.H.; Egan, S.; Elsas, J.D.; Overbeek, L.; Collard, J.-M.; Guillaume, G.; Karagouni, A.D.; Nikolakopoulou, T.L.; et al. Gentamicin resistance genes in environmental bacteria: Prevalence and transfer. FEMS Microbiol. Ecol. 2002, 42, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Klümper, U.; Riber, L.; Dechesne, A.; Sannazzarro, A.; Hansen, L.H.; Sørensen, S.J.; Smets, B.F. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 2015, 9, 934–945. [Google Scholar] [CrossRef] [PubMed]
- Ott, L.C.; Stromberg, Z.R.; Redweik, G.A.J.; Wannemuehler, M.J.; Mellata, M. Mouse Genetic Background Affects Transfer of an Antibiotic Resistance Plasmid in the Gastrointestinal Tract. mSphere 2020, 5, e00847-19. [Google Scholar] [CrossRef]
- Gumpert, H.; Kubicek-Sutherland, J.Z.; Porse, A.; Karami, N.; Munck, C.; Linkevicius, M.; Adlerberth, I.; Wold, A.E.; Andersson, D.I.; Sommer, M.O.A. Transfer and Persistence of a Multi-Drug Resistance Plasmid in situ of the Infant Gut Microbiota in the Absence of Antibiotic Treatment. Front. Microbiol. 2017, 8, 1852. [Google Scholar] [CrossRef]
- Maeusli, M.; Lee, B.; Miller, S.; Reyna, Z.; Lu, P.; Yan, J.; Ulhaq, A.; Skandalis, N.; Spellberg, B.; Luna, B. Horizontal Gene Transfer of Antibiotic Resistance from Acinetobacter baylyi to Escherichia coli on Lettuce and Subsequent Antibiotic Resistance Transmission to the Gut Microbiome. mSphere 2020, 5, e00329-20. [Google Scholar] [CrossRef]
- Neil, K.; Allard, N.; Grenier, F.; Burrus, V.; Rodrigue, S. Highly efficient gene transfer in the mouse gut microbiota is enabled by the Incl2 conjugative plasmid TP114. Commun. Biol. 2020, 3, 523. [Google Scholar] [CrossRef]
- Ding, C.; Yang, D.; Ma, J.; Jin, M.; Shen, Z.; Shi, D.; Tian, Z.; Kang, M.; Li, J.; Qiu, Z. Effects of free antibiotic resistance genes in the environment on intestinal microecology of mice. Ecotoxicol. Environ. Saf. 2020, 204, 111119. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, J.; Wang, B.; Wang, W.; Sun, Y.; Li, L.; Xu, H.; Wang, M. Spatially and Temporally Confined Response of Gastrointestinal Antibiotic Resistance Gene Levels to Sulfadiazine and Extracellular Antibiotic Resistance Gene Exposure in Mice. Biology 2023, 12, 210. [Google Scholar] [CrossRef]
- Ott, L.C.; Mellata, M. Models for Gut-Mediated Horizontal Gene Transfer by Bacterial Plasmid Conjugation. Front. Microbiol. 2022, 13, 891548. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Chen, H.; Li, N.; Wang, T.; Liang, W. The Spread of Antibiotic Resistance Genes In Vivo Model. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 3348695. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, J.; Li, S.C. Understanding Horizontal Gene Transfer network in human gut microbiota. Gut Pathog. 2020, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Brito, I.L. Examining horizontal gene transfer in microbial communities. Nat. Rev. Microbiol. 2021, 19, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.; Auchtung, J.M.; Schaefer, L.; Eaton, K.A.; Britton, R.A. Humanized microbiota mice as a model of recurrent Clostridium difficile disease. Microbiome 2015, 3, 35. [Google Scholar] [CrossRef]
- Moya-Uribe, I.A.; Bell, J.A.; Zanetti, A.; Jantre, S.; Huebner, M.; Arshad, S.H.; Ewart, S.L.; Mansfield, L.S. Fecal Microbiota Transplants (FMT) of Three Distinct Human Communities to Germ-Free Mice Exacerbated Inflammation and Decreased Lung Function in Their Offspring mBio. 2025. [Google Scholar]
- Brooks, P.T.; Brakel, K.A.; Bell, J.A.; Bejcek, C.E.; Gilpin, T.; Brudvig, J.M.; Mansfield, L.S. Transplanted human fecal microbiota enhanced Guillain Barré syndrome autoantibody responses after Campylobacter jejuni infection in C57BL/6 mice. Microbiome 2017, 5, 92. [Google Scholar] [CrossRef]
- Sher, A.A.; Van Allen, M.E.; Ahmed, H.; Whitehead-Tillery, C.; Rafique, S.; Bell, J.A.; Zhang, L.; Mansfield, L.S. Conjugative RP4 Plasmid-Mediated Transfer of Antibiotic Resistance Genes to Commensal and Multidrug-Resistant Enteric Bacteria In Vitro. Microorganisms 2023, 11, 193. [Google Scholar] [CrossRef]
- Klümper, U.; Dechesne, A.; Riber, L.; Brandt, K.K.; Gülay, A.; Sørensen, S.J.; Smets, B.F. Metal stressors consistently modulate bacterial conjugal plasmid uptake potential in a phylogenetically conserved manner. ISME J. 2017, 11, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Ronda, C.; Chen, S.P.; Cabral, V.; Yaung, S.J.; Wang, H.H. Metagenomic engineering of the mammalian gut microbiome in situ. Nat. Methods 2019, 16, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, L.S.; Ethridge, A.D.; Brooks, P.T.; Brakel, K.A.; Bell, J.A. Effect of Host Genetic Background on Development of Autoimmunity in Transplanted and Vertically Transmitted Human Microbiota Mouse Models. J. Allergy Clin. Immunol. 2018, 141, AB184. [Google Scholar] [CrossRef]
- Jiang, X.; Ellabaan, M.M.H.; Charusanti, P.; Munck, C.; Blin, K.; Tong, Y.; Weber, T.; Sommer, M.O.A.; Lee, S.Y. Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat. Commun. 2017, 8, 15784. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, K.J.; Reyes, A.; Wang, B.; Selleck, E.M.; Sommer, M.O.A.; Dantas, G. The shared antibiotic resistome of soil bacteria and human pathogens. Science 2012, 337, 1107–1111. [Google Scholar] [CrossRef]
- Bakkeren, E.; Huisman, J.S.; Fattinger, S.A.; Hausmann, A.; Furter, M.; Egli, A.; Slack, E.; Sellin, M.E.; Bonhoeffer, S.; Regoes, R.R.; et al. Salmonella persisters promote the spread of antibiotic resistance plasmids in the gut. Nature 2019, 573, 276–280. [Google Scholar] [CrossRef]
- Kydd, L.N.; Alalhareth, F.; Mendez, A.; Hohn, M.; Radunskaya, A.; Kojouharov, H.; Jaworski, J. Introduction of Plasmid to the Murine Gut via Consumption of an Escherichia coli Carrier and Examining the Impact of Bacterial Dosing and Antibiotics on Persistence. Regen. Eng. Transl. Med. 2022, 8, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Hadziabdic, S.; Fischer, J.; Malorny, B.; Borowiak, M.; Guerra, B.; Kaesbohrer, A.; Gonzalez-Zorn, B.; Szabo, I. In vivo transfer and microevolution of avian native IncA/C 2 blaNDM-1-carrying plasmid pRH-1238 during a broiler chicken infection study. Antimicrob. Agents Chemother. 2018, 62, e02128-17. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Yang, D.; Jin, M.; Liu, W.; Zhao, X.; Li, C.; Zhao, T.; Wang, J.; Gao, Z.; Shen, Z.; et al. Aquatic animals promote antibiotic resistance gene dissemination in water via conjugation: Role of different regions within the zebra fish intestinal tract, and impact on fish intestinal microbiota. Mol. Ecol. 2017, 26, 5318–5333. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Singer, R.S.; Isaacson, R.E.; Danzeisen, J.L.; Lang, K.; Kobluk, K.; Rivet, B.; Borewicz, K.; Frye, J.G.; Englen, M.; et al. In Vivo transmission of an incA/C plasmid in Escherichia coli depends on tetracycline concentration, and acquisition of the plasmid results in a variable cost of fitness. Appl. Environ. Microbiol. 2015, 81, 3561–3570. [Google Scholar] [CrossRef]
- Kang, K.; Imamovic, L.; Misiakou, M.A.; Bornakke Sørensen, M.; Heshiki, Y.; Ni, Y.; Zheng, T.; Li, J.; Ellabaan, M.M.H.; Colomer-Lluch, M.; et al. Expansion and persistence of antibiotic-specific resistance genes following antibiotic treatment. Gut Microbes 2021, 13, 1900995. [Google Scholar] [CrossRef] [PubMed]
- Musovic, S.; Klümper, U.; Dechesne, A.; Magid, J.; Smets, B.F. Long-term manure exposure increases soil bacterial community potential for plasmid uptake. Environ. Microbiol. Rep. 2014, 6, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Klümper, U.; Liu, Y.; Yang, Y.; Wei, Q.; Lin, J.-G.; Gu, J.-D.; Li, M. Metagenomic and metatranscriptomic analyses reveal activity and hosts of antibiotic resistance genes in activated sludge. Environ. Int. 2019, 129, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Geisenberger, O.; Ammendola, A.; Christensen, B.B.; Molin, S.; Schleifer, K.-H.; Eberl, L. Monitoring the conjugal transfer of plasmid RP4 in activated sludge and in situ identification of the transconjugants. FEMS Microbiol. Lett. 1999, 174, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Soda, S.; Otsuki, H.; Inoue, D.; Tsutsui, H.; Sei, K.; Ike, M. Transfer of antibiotic multiresistant plasmid RP4 from escherichia coli to activated sludge bacteria. J. Biosci. Bioeng. 2008, 106, 292–296. [Google Scholar] [CrossRef]
- Schlechter, R.O.; Jun, H.; Bernach, M.; Oso, S.; Boyd, E.; Muñoz-Lintz, D.A.; Dobson, R.C.J.; Remus, D.M.; Remus-Emsermann, M.N.P. Chromatic Bacteria—A Broad Host-Range Plasmid and Chromosomal Insertion Toolbox for Fluorescent Protein Expression in Bacteria. Front. Microbiol. 2018, 9, 3052. [Google Scholar] [CrossRef] [PubMed]
- Zahavy, E.; Ber, R.; Gur, D.; Abramovich, H.; Freeman, E.; Maoz, S.; Yitzhaki, S.; Zahavy, E.; Abramovich, H.; Freeman, E.; et al. Application of Nanoparticles for the Detection and Sorting of Pathogenic Bacteria by Flow-Cytometry. Nano-Biotechnol. Biomed. Diagn. Res. 2012, 733, 23–36. [Google Scholar] [CrossRef]
- Spencer, S.J.; Tamminen, M.V.; Preheim, S.P.; Guo, M.T.; Briggs, A.W.; Brito, I.L.; Weitz, D.A.; Pitkänen, L.K.; Vigneault, F.; Virta, M.P.; et al. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J. 2016, 10, 427–436. [Google Scholar] [CrossRef]
- Gawad, C.; Koh, W.; Quake, S.R. Single-cell genome sequencing: Current state of the science. Nat. Rev. Genet. 2016, 17, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Mansfield, L.S.; Bell, J.A.; Wilson, D.L.; Murphy, A.J.; Elsheikha, H.M.; Rathinam, V.A.K.; Fierro, B.R.; Linz, J.E.; Young, V.B. C57BL/6 and congenic interleukin-10-deficient mice can serve as models of Campylobacter jejuni colonization and enteritis. Infect. Immun. 2007, 75, 1099–1115. [Google Scholar] [CrossRef] [PubMed]
- Skinner, S.O.; Sepúlveda, L.A.; Xu, H.; Golding, I. Measuring mRNA copy-number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization (smFISH). Nat. Protoc. 2013, 8, 1100. [Google Scholar] [CrossRef] [PubMed]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the miseq illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Weber, N.; Liou, D.; Dommer, J.; Macmenamin, P.; Quiñones, M.; Misner, I.; Oler, A.J.; Wan, J.; Kim, L.; Coakley McCarthy, M.; et al. Nephele: A cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics 2018, 34, 1411–1413. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Hammer, D.A.T.; Ryan, P.D.; Hammer, Ø.; Harper, D.A.T. Past: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1. [Google Scholar]

| Primer | Product Size (in bp) | Primer Sequence (5′-3′) | Target Gene | Gene Bank Accession No. References |
|---|---|---|---|---|
| gfpF | 182 | ggtgaaggtgaaggtgatgc | gfp | U73901.1 |
| gfpR | cttctggcatggcagacttg | |||
| mScarletF | 371 | cgcgtgatgaactttgaaga | mScarlet-I | KY021424.1 |
| mScarletR | tcgctgcgttcatactgttc |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sher, A.A.; Whitehead-Tillery, C.E.; Peer, A.M.; Bell, J.A.; Vocelle, D.B.; Dippel, J.T.; Zhang, L.; Mansfield, L.S. Dynamic Spread of Antibiotic Resistance Determinants by Conjugation to a Human-Derived Gut Microbiota in a Transplanted Mouse Model. Antibiotics 2025, 14, 152. https://doi.org/10.3390/antibiotics14020152
Sher AA, Whitehead-Tillery CE, Peer AM, Bell JA, Vocelle DB, Dippel JT, Zhang L, Mansfield LS. Dynamic Spread of Antibiotic Resistance Determinants by Conjugation to a Human-Derived Gut Microbiota in a Transplanted Mouse Model. Antibiotics. 2025; 14(2):152. https://doi.org/10.3390/antibiotics14020152
Chicago/Turabian StyleSher, Azam A., Charles E. Whitehead-Tillery, Ashley M. Peer, Julia A. Bell, Daniel B. Vocelle, Joshua T. Dippel, Lixin Zhang, and Linda S. Mansfield. 2025. "Dynamic Spread of Antibiotic Resistance Determinants by Conjugation to a Human-Derived Gut Microbiota in a Transplanted Mouse Model" Antibiotics 14, no. 2: 152. https://doi.org/10.3390/antibiotics14020152
APA StyleSher, A. A., Whitehead-Tillery, C. E., Peer, A. M., Bell, J. A., Vocelle, D. B., Dippel, J. T., Zhang, L., & Mansfield, L. S. (2025). Dynamic Spread of Antibiotic Resistance Determinants by Conjugation to a Human-Derived Gut Microbiota in a Transplanted Mouse Model. Antibiotics, 14(2), 152. https://doi.org/10.3390/antibiotics14020152







