Engineering of a Novel Amphibian Skin Peptide Isolated from Agua Rica Leaf Frog (Callimedusa ecuatoriana) into Active Antimicrobial Agents
Abstract
1. Introduction
2. Results
2.1. Molecular Cloning of cDNA Encoding PTR-CE1 Precursor
2.2. Peptide Design, Predicted Physicochemical Characteristics, and 3D Models
2.3. In Silico Bioactivity Predictions and Molecular Docking
2.4. Synthesis and Characterization of Peptides
2.5. Antimicrobial Activity of PTR-CE1 and Its Analogs
2.6. Hemolytic Activity of Peptides
3. Discussion
4. Materials and Methods
4.1. Collection of Callimedusa ecuatoriana Skin Secretions
4.2. “Shotgun” Cloning of the Novel Peptide Precursor from C. ecuatoriana Skin Secretion-Derived cDNA Library
4.3. Identification of the Novel Peptide from C. ecuatoriana Skin Secretion
4.4. Computer-Aided Peptide Design, In Silico Predictions, and Molecular Docking
4.5. Solid-Phase Peptide Synthesis (SPPS)
4.6. Minimum Inhibitory Concentration (MIC), Minimum Bactericidal Concentration (MBC), and Minimum Fungicidal Concentration (MFC)
4.7. Hemolytic Activity
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Minarini, L.A.D.R.; Andrade, L.N.D.; De Gregorio, E.; Grosso, F.; Naas, T.; Zarrilli, R.; Camargo, I.L.B.C. Editorial: Antimicrobial Resistance as a Global Public Health Problem: How Can We Address It? Front. Public Health 2020, 8, 612844. [Google Scholar] [CrossRef]
- O’Neill, J. Tacking Drug Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016. [Google Scholar]
- Bucataru, C.; Ciobanasu, C. Antimicrobial Peptides: Opportunities and Challenges in Overcoming Resistance. Microbiol. Res. 2024, 286, 127822. [Google Scholar] [CrossRef]
- Demori, I.; El Rashed, Z.; Corradino, V.; Catalano, A.; Rovegno, L.; Queirolo, L.; Salvidio, S.; Biggi, E.; Zanotti-Russo, M.; Canesi, L.; et al. Peptides for Skin Protection and Healing in Amphibians. Molecules 2019, 24, 347. [Google Scholar] [CrossRef]
- Xu, X.; Lai, R. The Chemistry and Biological Activities of Peptides from Amphibian Skin Secretions. Chem. Rev. 2015, 115, 1760–1846. [Google Scholar] [CrossRef] [PubMed]
- Modi, A.; Uniyal, A.; Akhilesh Tiwari, V.; Chouhan, D.; Agrawal, S.; Ummadisetty, O.; Tiwari, V. Dermorphin [D-Arg2, Lys4] (1–4) Amide Attenuates Burn Pain by Inhibiting TRPV1/NR2B Mediated Neuroinflammatory Signalling. Mol. Neurobiol. 2025, 62, 12668–12687. [Google Scholar] [CrossRef] [PubMed]
- Pantic, J.M.; Jovanovic, I.P.; Radosavljevic, G.D.; Arsenijevic, N.N.; Conlon, J.M.; Lukic, M.L. The Potential of Frog Skin-Derived Peptides for Development into Therapeutically-Valuable Immunomodulatory Agents. Molecules 2017, 22, 2071. [Google Scholar] [CrossRef]
- Conlon, J.M.; Owolabi, B.O.; Flatt, P.R.; Abdel-Wahab, Y.H.A. Amphibian Host-Defense Peptides with Potential for Type 2 Diabetes Therapy—An Updated Review. Peptides 2024, 175, 171180. [Google Scholar] [CrossRef] [PubMed]
- Tolos, A.M.; Moisa, C.; Dochia, M.; Popa, C.; Copolovici, L.; Copolovici, D.M. Anticancer Potential of Antimicrobial Peptides: Focus on Buforins. Polymers 2024, 16, 728. [Google Scholar] [CrossRef]
- Chen, D.; Zhou, X.; Chen, X.; Huang, L.; Xi, X.; Ma, C.; Zhou, M.; Wang, L.; Chen, T. Evaluating the Bioactivity of a Novel Antimicrobial and Anticancer Peptide, Dermaseptin-PS4 (Der-PS4), from the Skin Secretion of Phyllomedusa sauvagii. Molecules 2019, 24, 2974. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Zou, Z.; Yang, M.; Wu, C.; Su, Y.; Tang, J.; Yang, X. OM-LV20, a Novel Peptide from Odorous Frog Skin, Accelerates Wound Healing In Vitro and In Vivo. Chem. Biol. Drug Des. 2018, 91, 126–136. [Google Scholar] [CrossRef]
- Liu, S.; Long, Q.; Xu, Y.; Wang, J.; Xu, Z.; Wang, L.; Zhou, M.; Wu, Y.; Chen, T.; Shaw, C. Assessment of Antimicrobial and Wound Healing Effects of Brevinin-2Ta against the Bacterium Klebsiella pneumoniae in Dermally-Wounded Rats. Oncotarget 2017, 8, 111369–111385. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, X.; Fu, Z.; Wang, S.; Li, X.; Liu, N.; Feng, Z.; Yang, M.; Tang, J.; Yang, X. Identification and Characterization of a Novel Gene-Encoded Antioxidant Peptide Obtained from Amphibian Skin Secretions. Nat. Prod. Res. 2020, 34, 754–758. [Google Scholar] [CrossRef]
- Udovychenko, I.; Oliynyk, D.; Dudkina, J.; Halenova, T.; Savchuk, O. Analysis of the Common Spadefoot Toad (Pelobates fuscus) Skin Secretions on the Presence of the Potential Hemostasis System Effectors. Bull. Taras Shevchenko Natl. Univ. Kyiv. Ser. Biol. 2019, 77, 38–44. [Google Scholar] [CrossRef]
- Chen, X.; Liu, S.; Fang, J.; Zheng, S.; Wang, Z.; Jiao, Y.; Xia, P.; Wu, H.; Ma, Z.; Hao, L. Peptides Isolated from Amphibian Skin Secretions with Emphasis on Antimicrobial Peptides. Toxins 2022, 14, 722. [Google Scholar] [CrossRef]
- Prates, M.V.; Sforça, M.L.; Regis, W.C.B.; Leite, J.R.S.A.; Silva, L.P.; Pertinhez, T.A.; Araújo, A.L.T.; Azevedo, R.B.; Spisni, A.; Bloch, C. The NMR-Derived Solution Structure of a New Cationic Antimicrobial Peptide from the Skin Secretion of the Anuran Hyla punctata. J. Biol. Chem. 2004, 279, 13018–13026. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Kosikowska, P.; Lesner, A. Antimicrobial Peptides (AMPs) as Drug Candidates: A Patent Review (2003–2015). Expert. Opin. Ther. Pat. 2016, 26, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Erspamer, V.; Melchiorri, P.; Falconieri Erspamer, G.; Montecucchi, P.C.; de Castiglione, R. Phyllomedusa Skin: A Huge Factory and Store-House of a Variety of Active Peptides. Peptides 1985, 6, 7–12. [Google Scholar] [CrossRef]
- Ge, L.; Chen, X.; Ma, C.; Zhou, M.; Xi, X.; Wang, L.; Ding, A.; Duan, J.; Chen, T.; Shaw, C. Balteatide: A Novel Antimicrobial Decapeptide from the Skin Secretion of the Purple-Sided Leaf Frog. Phyllomedusa baltea. Sci. World J. 2014, 2014, 176214. [Google Scholar] [CrossRef]
- Shi, D.; Xi, X.; Wang, L.; Gao, Y.; Ma, C.; Chen, H.; Zhou, M.; Chen, T.; Shaw, C. Baltikinin: A New Myotropic Tryptophyllin-3 Peptide Isolated from the Skin Secretion of the Purple-Sided Leaf Frog, Phyllomedusa baltea. Toxins 2016, 8, 213. [Google Scholar] [CrossRef]
- Zhu, H.; Ding, X.; Li, W.; Lu, T.; Ma, C.; Xi, X.; Wang, L.; Zhou, M.; Burden, R.; Chen, T. Discovery of Two Skin-Derived Dermaseptins and Design of a TAT-Fusion Analogue with Broad-Spectrum Antimicrobial Activity and Low Cytotoxicity on Healthy Cells. PeerJ 2018, 2018, e5635. [Google Scholar] [CrossRef]
- Yin, W.; Yao, J.; Leng, X.; Ma, C.; Chen, X.; Jiang, Y.; Wang, T.; Chen, T.; Shaw, C.; Zhou, M.; et al. Enhancement of Antimicrobial Function by L/D-Lysine Substitution on a Novel Broad-Spectrum Antimicrobial Peptide, Phylloseptin-TO2: A Structure-Related Activity Research Study. Pharmaceutics 2024, 16, 1098. [Google Scholar] [CrossRef]
- Chen, Z.; Xi, X.; Lu, Y.; Hu, H.; Dong, Z.; Ma, C.; Wang, L.; Zhou, M.; Chen, T.; Du, S.; et al. In Vitro Activities of a Novel Antimicrobial Peptide Isolated from Phyllomedusa tomopterna. Microb. Pathog. 2021, 153, 104795. [Google Scholar] [CrossRef]
- Ron, S.R.; Read, M. Phyllomedusa ecuatoriana. Available online: https://bioweb.bio/faunaweb/amphibiaweb/FichaEspecie/Phyllomedusa%20ecuatoriana (accessed on 13 August 2025).
- Xuan, J.; Feng, W.; Wang, J.; Wang, R.; Zhang, B.; Bo, L.; Chen, Z.S.; Yang, H.; Sun, L. Antimicrobial Peptides for Combating Drug-Resistant Bacterial Infections. Drug Resist. Updates 2023, 68, 100954. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zuo, S.; Wang, B.; Zhang, K.; Wang, Y. Antimicrobial Mechanisms and Clinical Application Prospects of Antimicrobial Peptides. Molecules 2022, 27, 2675. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, L.; Dong, C. Antimicrobial Peptides: An Overview of Their Structure, Function and Mechanism of Action. Protein Pept. Lett. 2022, 29, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial Peptides: Application Informed by Evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef]
- Fjell, C.D.; Hiss, J.A.; Hancock, R.E.W.; Schneider, G. Designing Antimicrobial Peptides: Form Follows Function. Nat. Rev. Drug Discov. 2012, 11, 37–51. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.C.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q.Y. The Antimicrobial Peptides and Their Potential Clinical Applications. Am. J. Transl. Res. 2019, 11, 3919. [Google Scholar] [PubMed]
- Hancock, R.E.W.; Sahl, H.G. Antimicrobial and Host-Defense Peptides as New Anti-Infective Therapeutic Strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef]
- Otvos, L.; Wade, J.D. Current Challenges in Peptide-Based Drug Discovery. Front. Chem. 2014, 2, 62. [Google Scholar] [CrossRef]
- Mwangi, J.; Kamau, P.M.; Thuku, R.C.; Lai, R. Design Methods for Antimicrobial Peptides with Improved Performance. Zool. Res. 2023, 44, 1095–1114. [Google Scholar] [CrossRef]
- Bermúdez-Puga, S.; Morán-Marcillo, G.; Espinosa de los Monteros-Silva, N.; Naranjo, R.E.; Toscano, F.; Vizuete, K.; Torres Arias, M.; Almeida, J.R.; Proaño-Bolaños, C. Inspiration from Cruzioseptin-1: Membranolytic Analogue with Improved Antibacterial Properties. Amino Acids 2023, 55, 113–124. [Google Scholar] [CrossRef]
- Bermúdez-Puga, S.; Dias, M.; Lima Reis, I.; Freire de Oliveira, T.; Yokomizo de Almeida, S.R.; Mendes, M.A.; Moore, S.J.; Almeida, J.R.; Proaño-Bolaños, C.; Pinheiro de Souza Oliveira, R. Microscopic and Metabolomics Analysis of the Anti-Listeria Activity of Natural and Engineered Cruzioseptins. Biochimie 2024, 225, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Morán-Marcillo, G.; Sánchez Hinojosa, V.; de los Monteros-Silva, N.E.; Blasco-Zúñiga, A.; Rivera, M.; Naranjo, R.E.; Almeida, J.R.; Wang, L.; Zhou, M.; Chen, T.; et al. Picturins and Pictuseptins, Two Novel Antimicrobial Peptide Families from the Skin Secretions of the Chachi Treefrog, Boana picturata. J. Proteom. 2022, 264, 104633. [Google Scholar] [CrossRef]
- Pál, T.; Sonnevend, Á.; Galadari, S.; Conlon, J.M. Design of Potent, Non-Toxic Antimicrobial Agents Based upon the Structure of the Frog Skin Peptide, Pseudin-2. Regul. Pept. 2005, 129, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Kang, H.K.; Park, E.; Kim, M.K.; Park, Y. Bactericidal Activities and Action Mechanism of the Novel Antimicrobial Peptide Hylin A1 and Its Analog Peptides against Acinetobacter baumannii Infection. Eur. J. Pharm. Sci. 2022, 175, 106205. [Google Scholar] [CrossRef]
- Pál, T.; Abraham, B.; Sonnevend, Á.; Jumaa, P.; Conlon, J.M. Brevinin-1BYa: A Naturally Occurring Peptide from Frog Skin with Broad-Spectrum Antibacterial and Antifungal Properties. Int. J. Antimicrob. Agents 2006, 27, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Mechkarska, M.; Arafat, K.; Attoub, S.; Sonnevend, A. Analogues of the Frog Skin Peptide Alyteserin-2a with Enhanced Antimicrobial Activities against Gram-Negative Bacteria. J. Pept. Sci. 2012, 18, 270–275. [Google Scholar] [CrossRef]
- Kim, J.B.; Conlon, J.M.; Iwamuro, S.; Knoop, F.C. Antimicrobial Peptides from the Skin of the Japanese Mountain Brown Frog, Rana ornativentris. J. Pept. Res. 2001, 58, 349–356. [Google Scholar] [CrossRef]
- Proaño-Bolaños, C.; Zhou, M.; Wang, L.; Coloma, L.A.; Chen, T.; Shaw, C. Peptidomic Approach Identifies Cruzioseptins, a New Family of Potent Antimicrobial Peptides in the Splendid Leaf Frog, Cruziohyla calcarifer. J. Proteom. 2016, 146, 1–13. [Google Scholar] [CrossRef]
- Funk, C.; Caminer, M.; Ron, S.R. High Levels of Cryptic Species Diversity Uncovered in Amazonian Frogs. Proc. R. Soc. B Biol. Sci. 2012, 279, 1806–1814. [Google Scholar] [CrossRef] [PubMed]
- Brožek, R.; Kabelka, I.; Vácha, R. Effect of Helical Kink on Peptide Translocation across Phospholipid Membranes. J. Phys. Chem. B 2020, 124, 5940–5947. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Liang, X.; Liu, C.; Cheng, Y.; Zhou, L.; Wang, K.; Zhao, L. Influence of Proline Substitution on the Bioactivity of Mammalian-Derived Antimicrobial Peptide NK-2. Probiotics Antimicrob. Proteins 2018, 10, 118–127. [Google Scholar] [CrossRef]
- Park, S.H.; Kim, H.E.; Kim, C.M.; Yun, H.J.; Choi, E.C.; Lee, B.J. Role of Proline, Cysteine and a Disulphide Bridge in the Structure and Activity of the Anti-Microbial Peptide Gaegurin 5. Biochem. J. 2002, 368, 171–182. [Google Scholar] [CrossRef]
- Chen, G.; Miao, Y.; Ma, C.; Zhou, M.; Shi, Z.; Chen, X.; Burrows, J.F.; Xi, X.; Chen, T.; Wang, L. Brevinin-2GHK from Sylvirana guentheri and the Design of Truncated Analogs Exhibiting the Enhancement of Antimicrobial Activity. Antibiotics 2020, 9, 85. [Google Scholar] [CrossRef]
- Zelezetsky, I.; Tossi, A. Alpha-Helical Antimicrobial Peptides-Using a Sequence Template to Guide Structure-Activity Relationship Studies. Biochim. Biophys. Acta Biomembr. 2006, 1758, 1436–1449. [Google Scholar] [CrossRef]
- Eliseev, I.E.; Terterov, I.N.; Yudenko, A.N.; Shamova, O.V. Linking Sequence Patterns and Functionality of Alpha-Helical Antimicrobial Peptides. Bioinformatics 2019, 35, 2713–2717. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, J.; Chen, Y. Alpha-Helical Cationic Antimicrobial Peptides: Relationships of Structure and Function. Protein Cell 2010, 1, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Guilhaudis, L.; Cunning, T.S.; Delaney, J.J.; Ternan, N.G.; Mechkarska, M.; Attoub, S.; Conlon, J.M. Substitution of a Proline Residue in the Frog Skin Host-Defense Peptide, Figainin-2PL by D-Lysine Generates an Analog with Potent Activity against Antibiotic-Resistant ESKAPE Pathogens. Peptides 2025, 192, 171430. [Google Scholar] [CrossRef]
- Chaudhary, K.; Kumar, R.; Singh, S.; Tuknait, A.; Gautam, A.; Mathur, D.; Anand, P.; Varshney, G.C.; Raghava, G.P.S. A Web Server and Mobile App for Computing Hemolytic Potency of Peptides. Sci. Rep. 2016, 6, 22843. [Google Scholar] [CrossRef] [PubMed]
- Conlon, J.M.; Abraham, B.; Galadari, S.; Knoop, F.C.; Sonnevend, A.; Pál, T. Antimicrobial and Cytolytic Properties of the Frog Skin Peptide, Kassinatuerin-1 and Its L- and D-Lysine-Substituted Derivatives. Peptides 2005, 26, 2104–2110. [Google Scholar] [CrossRef]
- Tan, Y.; Chen, X.; Ma, C.; Xi, X.; Wang, L.; Zhou, M.; Burrows, J.F.; Kwok, H.F.; Chen, T. Biological Activities of Cationicity-Enhanced and Hydrophobicity-Optimized Analogues of an Antimicrobial Peptide, Dermaseptin-PS3, from the Skin Secretion of Phyllomedusa sauvagii. Toxins 2018, 10, 320. [Google Scholar] [CrossRef]
- Gong, Z.; Pei, X.; Ren, S.; Chen, X.; Wang, L.; Ma, C.; Xi, X.; Chen, T.; Shaw, C.; Zhou, M. Identification and Rational Design of a Novel Antibacterial Peptide Dermaseptin-Ac from the Skin Secretion of the Red-Eyed Tree Frog Agalychnis callidryas. Antibiotics 2020, 9, 243. [Google Scholar] [CrossRef]
- Juretić, D.; Simunić, J. Design of α-Helical Antimicrobial Peptides with a High Selectivity Index. Expert. Opin. Drug Discov. 2019, 14, 1053–1063. [Google Scholar] [CrossRef]
- Tamargo, J.; Le Heuzey, J.Y.; Mabo, P. Narrow Therapeutic Index Drugs: A Clinical Pharmacological Consideration to Flecainide. Eur. J. Clin. Pharmacol. 2015, 71, 549–567. [Google Scholar] [CrossRef] [PubMed]
- Azuma, E.; Choda, N.; Odaki, M.; Yano, Y.; Matsuzaki, K. Improvement of Therapeutic Index by the Combination of Enhanced Peptide Cationicity and Proline Introduction. ACS Infect. Dis. 2020, 6, 2271–2278. [Google Scholar] [CrossRef]
- Di, Y.P.; Lin, Q.; Chen, C.; Montelaro, R.C.; Doi, Y.; Deslouches, B. Enhanced Therapeutic Index of an Antimicrobial Peptide in Mice by Increasing Safety and Activity against Multidrug-Resistant Bacteria. Health Med. 2020, 6, eaay6817. [Google Scholar] [CrossRef]
- Clark, V.C. Collecting Arthropod and Amphibian Secretions for Chemical Analysis. In Behavioral Chemical Ecology; Zhiang, W., Liu, H., Eds.; Nova Science Publishers, Inc.: Hauppauge, NY, USA, 2009; ISBN 978-1-60741-099-7. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A Database on Sequences, Structures and Signatures of Antimicrobial Peptides. Nucleic Acids Res. 2016, 44, D1094–D1097. [Google Scholar] [CrossRef] [PubMed]
- Geourjon, C.; Deléage, G. Sopma: Significant Improvements in Protein Secondary Structure Prediction by Consensus Prediction from Multiple Alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A Web Server to Screen Sequences with Specific α-Helical Properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Rozenski, J. Peptide Mass Calculator, v3.2; Rega Institute for Medical Research: Leuven, Belgium, 1999. [Google Scholar]
- Yang, J.; Zhang, Y. I-TASSER Server: New Development for Protein Structure and Function Predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Schrödinger, L. The PyMol Molecular Graphics System, Versión 1.8; Schrdinger, LLC: New York, NY, USA, 2015.
- Feng, S.; Park, S.; Choi, Y.K.; Im, W. CHARMM-GUI Membrane Builder: Past, Current, and Future Developments and Applications. J. Chem. Theory Comput. 2023, 19, 2161–2185. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Proaño-Bolaños, C.; Blasco-Zúñiga, A.; Almeida, J.R.; Wang, L.; Llumiquinga, M.A.; Rivera, M.; Zhou, M.; Chen, T.; Shaw, C. Unravelling the Skin Secretion Peptides of the Gliding Leaf Frog, Agalychnis spurrelli (Hylidae). Biomolecules 2019, 9, 667. [Google Scholar] [CrossRef] [PubMed]

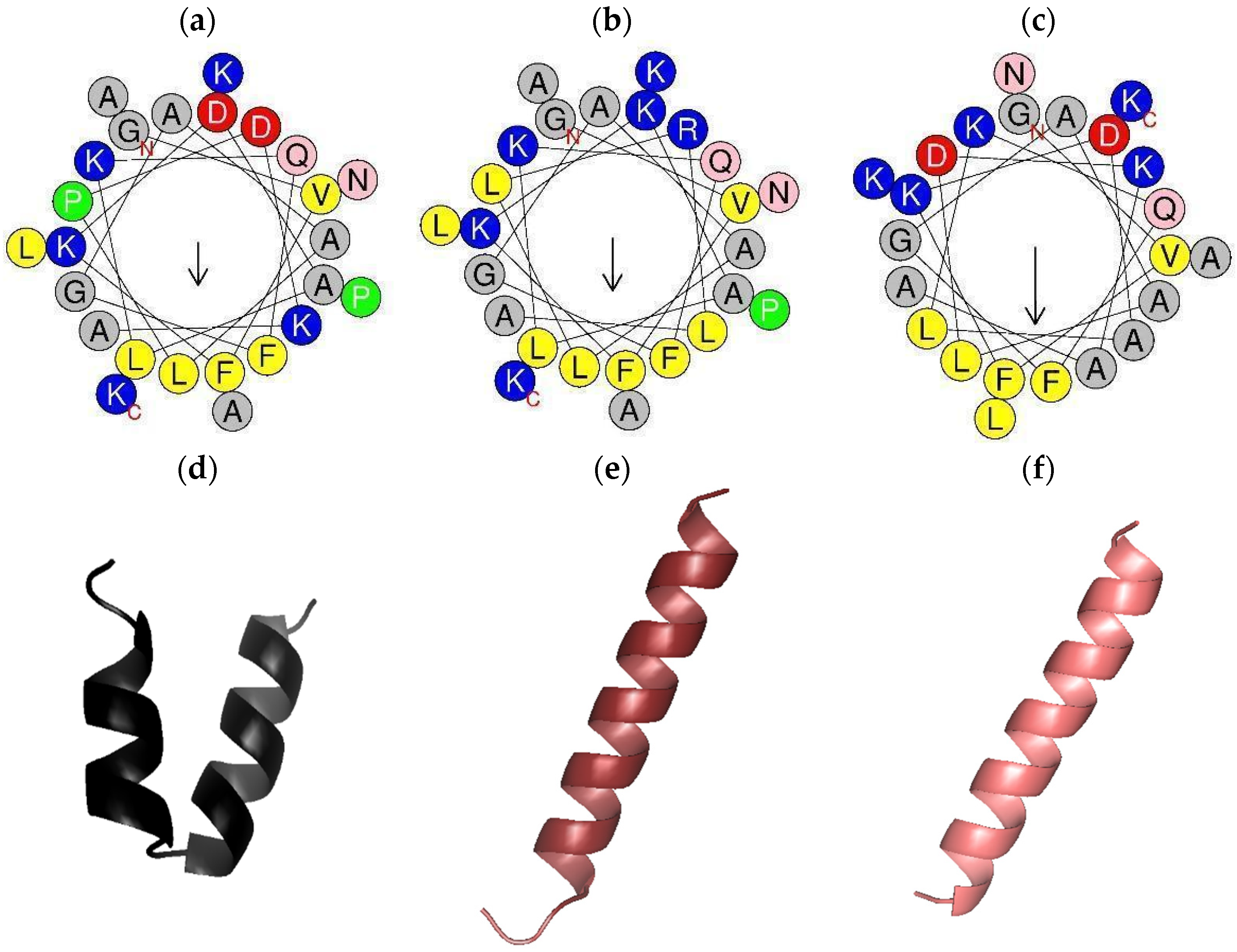
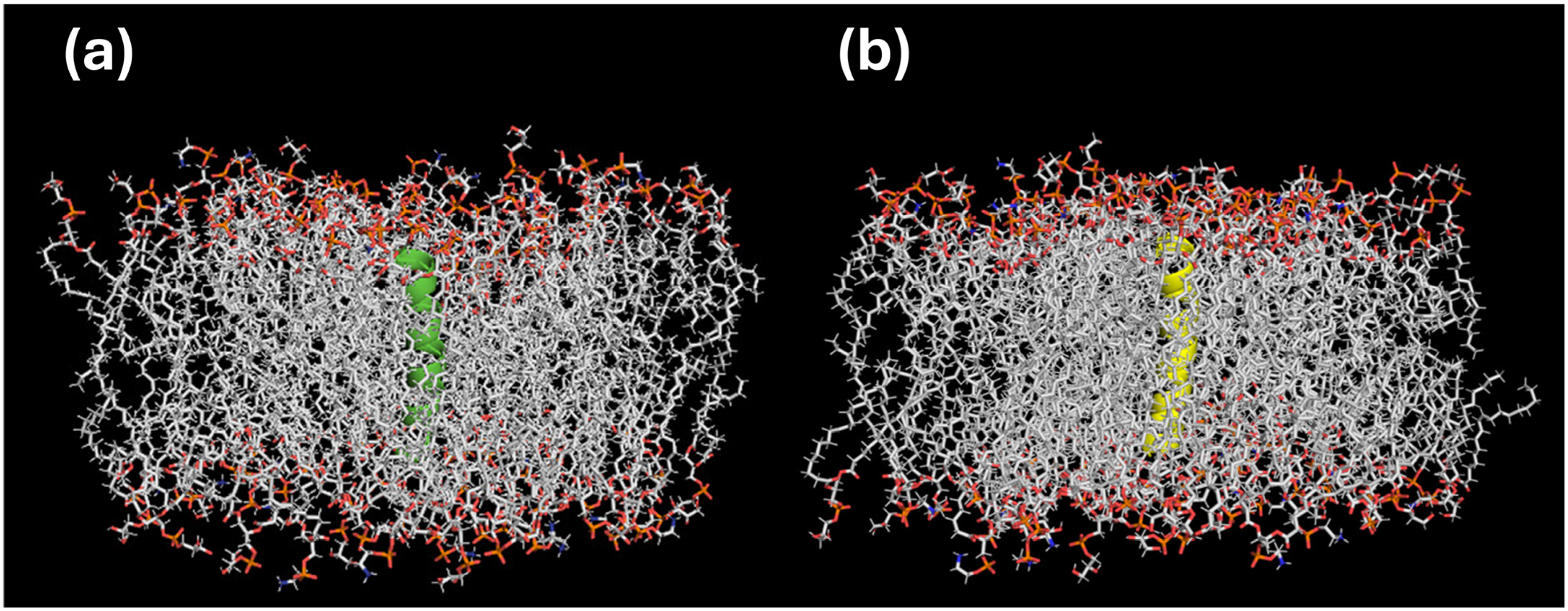

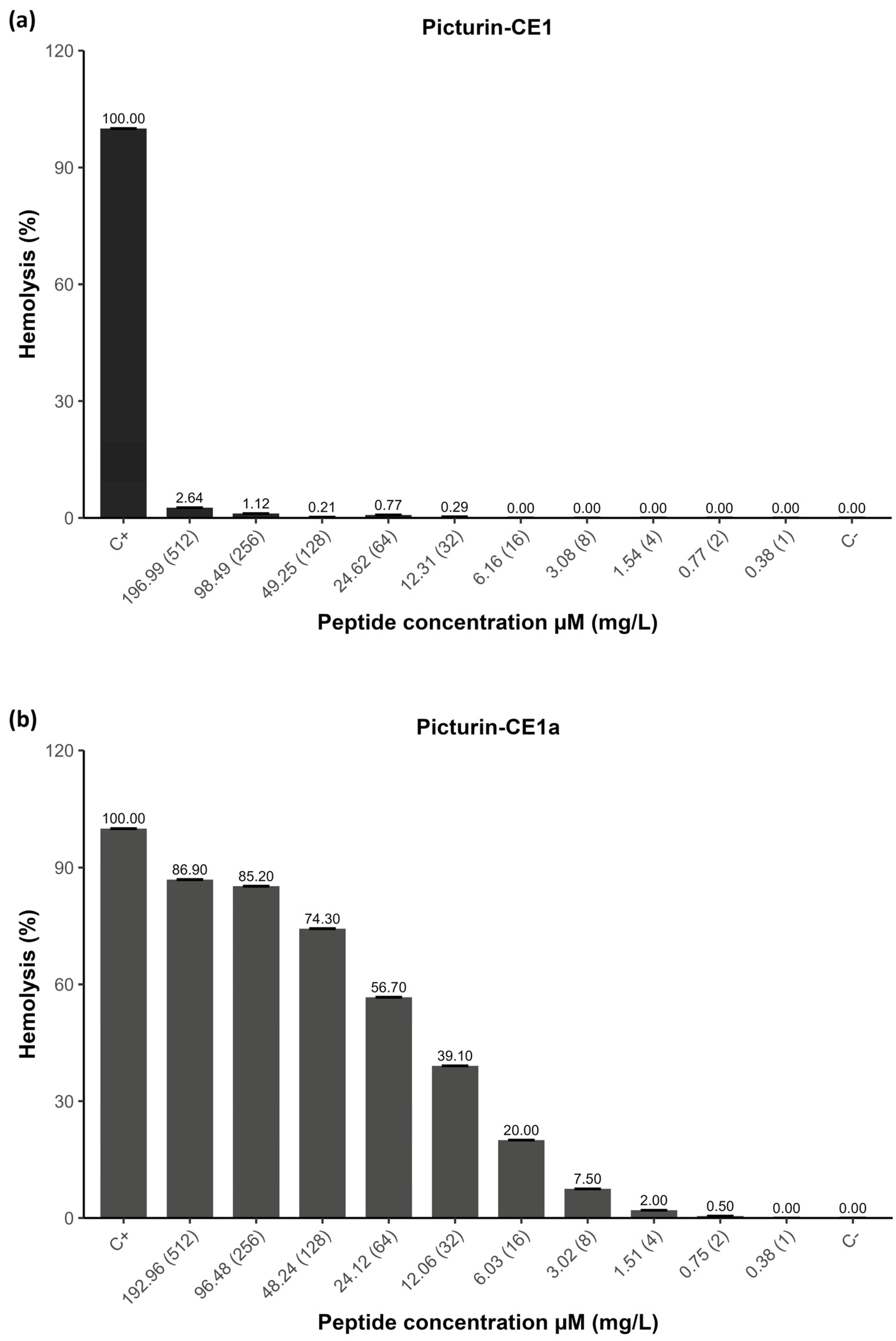
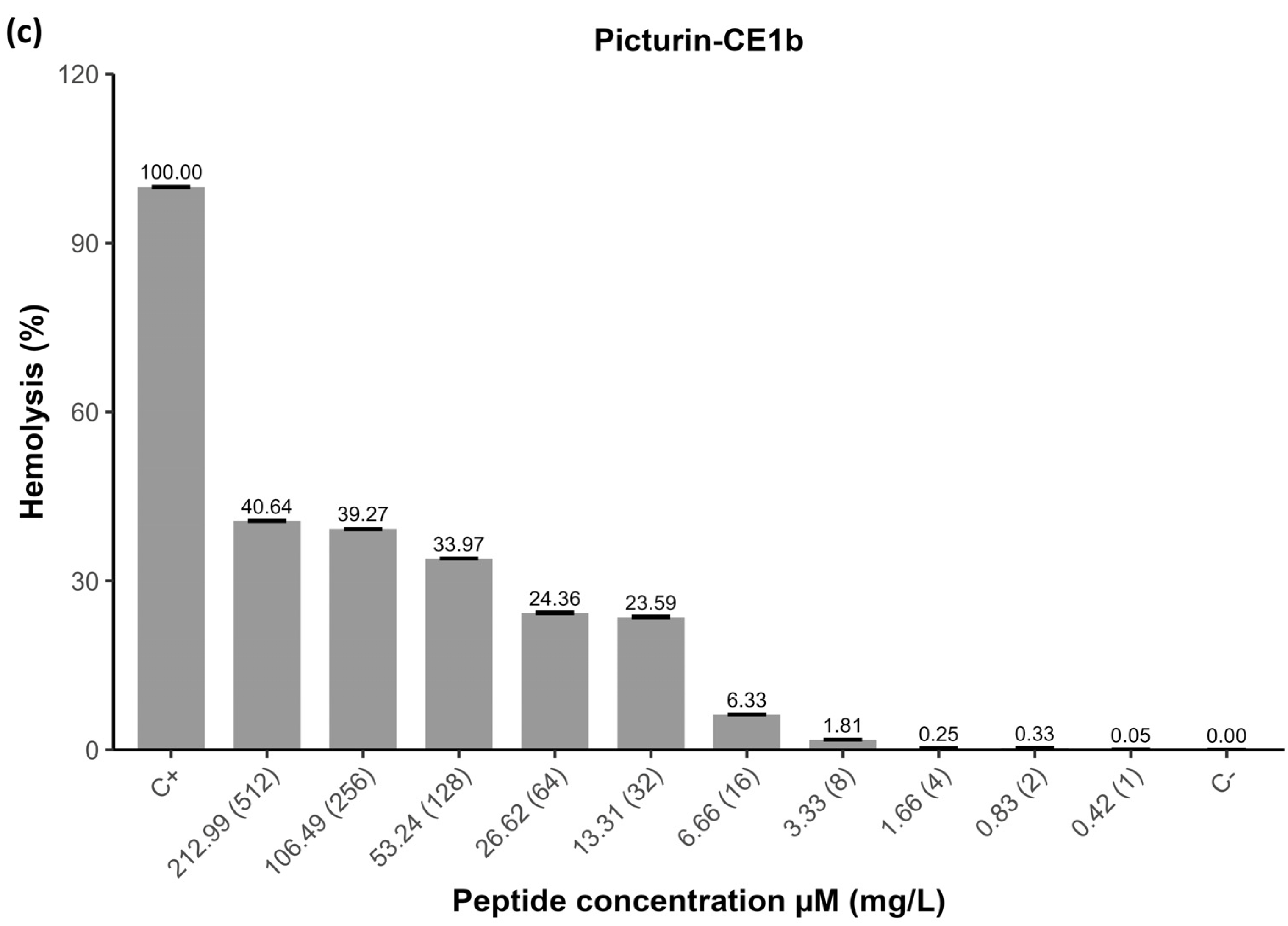
| Peptide | Signal Peptide | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ||||||||||||||||||||||||||||||||||||||||
| Picturin-CE1 | M | S | F | L | K | K | S | L | F | L | A | L | F | L | G | I | V | S | L | S | I | C | ||||||||||||||||||
| Picturin-2 | M | S | F | L | K | K | S | L | F | L | V | L | F | L | G | I | V | S | L | S | I | C | ||||||||||||||||||
| Picturin-3 | - | - | - | - | - | K | S | L | F | L | V | L | F | L | G | I | V | S | L | S | I | C | ||||||||||||||||||
| Picturin-1 | - | - | - | - | - | - | S | L | F | L | V | L | F | L | G | I | V | S | L | S | I | C | ||||||||||||||||||
| Boanin-3 | M | T | F | G | K | K | S | L | F | L | V | L | F | L | G | M | V | S | L | S | I | C | ||||||||||||||||||
| Pictuseptin-2 | M | S | F | L | K | K | S | L | F | L | V | L | F | L | G | I | V | S | L | S | I | C | ||||||||||||||||||
| Acidic spacer | ||||||||||||||||||||||||||||||||||||||||
| 23 | ||||||||||||||||||||||||||||||||||||||||
| Picturin-CE1 | E | E | E | K | R | - | - | Q | E | E | E | E | K | E | D | E | E | Y | E | E | G | Y | E | A | S | E | E | K | R | |||||||||||
| Picturin-2 | E | E | E | K | R | - | - | Q | E | E | E | E | K | E | D | E | E | Y | E | E | G | Y | E | A | S | E | E | K | R | |||||||||||
| Picturin-3 | E | E | E | K | R | - | - | Q | E | E | E | E | K | E | D | E | E | Y | E | E | G | Y | E | A | S | E | E | K | R | |||||||||||
| Picturin-1 | E | E | E | K | R | - | - | Q | E | E | E | E | K | E | D | E | E | Y | E | E | G | Y | E | A | S | E | E | K | R | |||||||||||
| Boanin-3 | Q | D | E | K | R | - | E | E | E | E | E | E | K | E | E | E | E | Y | E | E | G | N | E | E | H | K | E | K | R | |||||||||||
| Pictuseptin-2 | E | E | E | K | K | Q | A | E | E | E | E | E | K | Q | E | E | Q | Y | D | Q | E | N | E | E | Y | K | E | K | R | |||||||||||
| Mature peptide | Accession number | |||||||||||||||||||||||||||||||||||||||
| 52 | ||||||||||||||||||||||||||||||||||||||||
| Picturin-CE1 | - | - | - | G | V | F | K | D | A | L | K | Q | F | G | A | A | L | P | D | K | A | A | N | A | L | K | P | K | * | OQ438429.1 | ||||||||||
| Picturin-2 | - | - | - | G | V | F | K | D | A | L | K | Q | F | G | A | A | L | L | D | K | A | A | N | A | L | K | P | K | * | MN652614.1 | ||||||||||
| Picturin-3 | - | - | - | G | V | F | K | D | A | L | K | Q | F | G | A | A | L | L | D | Q | A | A | N | A | L | K | P | K | * | MN652615.1 | ||||||||||
| Picturin-1 | - | - | - | G | V | F | K | D | A | L | K | Q | L | G | A | A | L | L | D | K | A | A | N | A | L | K | P | K | * | MN652613.1 | ||||||||||
| Boanin-3 | - | F | L | G | A | L | F | A | I | G | K | A | I | G | K | A | I | L | P | L | A | V | K | A | F | N | P | Q | H | * | ON703100.1 | |||||||||
| Pictuseptin-2 | G | F | L | D | T | L | K | N | I | G | K | T | V | G | - | - | - | - | G | I | A | L | N | V | L | T | G | * | MW118451.1 | |||||||||||
| Peptide | Sequence | #Aas | Alpha Helix % | Hydrophobicity <H> | Hydrophobic Moment <mH> | Net Charge Z | Theoretical Mass | Ref. | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| h | h | h | h | h | h | h | h | h | h | h | h | h | c | c | h | h | h | h | h | h | h | c | t | t | |||||||||
| PTR-CE1 | G | V | F | K | D | A | L | K | Q | F | G | A | A | L | P | D | K | A | A | N | A | L | K | P | K | 25 | 80 | 0.236 | 0.506 | 3 | 2599.07 | ||
| h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | c | t | t | |||||||||
| PTR-CE1a | G | V | F | K | K | A | L | K | Q | F | G | A | A | L | L | R | L | A | A | N | A | L | K | P | K | a | 25 | 88 | 0.364 | 0.462 | 7 | 2653.30 | |
| h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | |||||||||||
| PTR-CE1b | G | V | F | K | D | A | L | K | Q | F | G | A | A | L | D | K | A | A | N | A | L | K | K | a | 23 | 100 | 0.193 | 0.468 | 4 | 2403.85 | |||
| h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | c | t | t | |||||||||
| PTR-1 | G | V | F | K | D | A | L | K | Q | L | G | A | A | L | L | D | K | A | A | N | A | L | K | P | K | 25 | 88 | 0.271 | 0.306 | 3 | 2581.10 | [37] | |
| h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | c | t | t | |||||||||
| PTR-2 | G | V | F | K | D | A | L | K | Q | F | G | A | A | L | L | D | K | A | A | N | A | L | K | P | K | 25 | 88 | 0.275 | 0.309 | 3 | 2615.12 | [37] | |
| h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | h | c | t | t | |||||||||
| PTR-3 | G | V | F | K | D | A | L | K | Q | F | G | A | A | L | L | D | Q | A | A | N | A | L | K | P | K | 25 | 88 | 0.306 | 0.326 | 2 | 2615.07 | [37] | |
| Peptide | CAMPR3 (SVM *) | HemoPI-2 | ToxinPred |
|---|---|---|---|
| Antimicrobial | |||
| PTR-CE1 | 0.915 | 0.52 | Non-toxin |
| PTR-CE1a | 0.994 | 0.58 | Non-toxin |
| PTR-CE1b | 0.908 | 0.54 | Non-toxin |
| MIC μM (mg/L) | MBC μM (mg/L) | Ref. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synthetic Peptide | E. coli | S. aureus | C. albicans | K. pneumoniae | P. aeruginosa | Bacillus subtilis | E. coli | S. aureus | C. albicans | K. pneumoniae | P. aeruginosa | Bacillus subtilis | |
| 25922 | 25923 | (AMP-RES) | (AMP-RES) | (AMP-RES) | 25922 | 25923 | (AMP-RES) | (AMP-RES) | (AMP-RES) | ||||
| Clinical Isolate | Clinical Isolate | Clinical Isolate | Clinical Isolate | Clinical Isolate | Clinical Isolate | ||||||||
| Picturin-CE1 | >196.99 | >196.99 | >196.99 | >196.99 | >196.99 | >196.99 | >196.99 | >196.99 | >196.99 | >196.99 | >196.99 | >196.99 | |
| (>512) | (>512) | (>512) | (>512) | (>512) | (>512) | (>512) | (>512) | (>512) | (>512) | (>512) | (>512) | ||
| Picturin-CE1a | 3.02 | 6.03 | 12.06 | 6.03 | 12.06 | 3.02 | 6.03 | 12.06 | >192.96 | 12.06 | 48.24 | 24.12 | |
| (8) | (16) | (32) | (16) | (32) | (8) | (16) | (32) | (>512) | (32) | (128) | (64) | ||
| Picturin-CE1b | 53.25 | >212.99 | 212.99 | 53.25 | 26.62 | 26.62 | 106.49 | >212.99 | >212.99 | 106.49 | >212.99 | 53.25 | |
| (128) | (>512) | (512) | (128) | (64) | (64) | (256) | (>512) | (>512) | (256) | (>512) | (128) | ||
| * Picturin-1 | 24.80 | 198.37 | >198.37 | ND | ND | ND | >198.37 | >198.37 | >198.37 | ND | ND | ND | [37] |
| (64) | (512) | (>512) | (>512) | (>512) | (>512) | ||||||||
| * Picturin-2 | 48.95 | >195.79 | >195.79 | ND | ND | ND | 48.95 | >195.79 | >195.79 | ND | ND | ND | [37] |
| (128) | (>512) | (>512) | (128) | (>512) | (>512) | ||||||||
| * Picturin-3 | 48.98 | 97.95 | >195.91 | ND | ND | ND | 48.98 | >195.91 | >195.91 | ND | ND | ND | [37] |
| (128) | (256) | (>512) | (128) | (>512) | (>512) | ||||||||
| * Pseudin-2 | 5 | 20 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | [38] |
| [Lys3,10,14,21] | |||||||||||||
| * Hylin a1-2A | 16 | 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | [39] |
| * Brevinin-1BYa | 20 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | [40] |
| [Ser18,Ser24] | |||||||||||||
| * Alysterin-2a | ND | 64 | 64 | ND | ND | ND | ND | ND | ND | ND | ND | ND | [41] |
| * Kassinatuerin-1 | 6.25 | 6.25 | 25 | ND | ND | ND | ND | ND | ND | ND | ND | ND | [42] |
| [Lys7,Lys18,Lys19] | |||||||||||||
| * Brevinin-2Ob | 4 | 9 | 40 | ||||||||||
| * Ampicillin | 46 | <11 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | [12] |
| * Fluconazole | ND | ND | 209 | ND | ND | ND | ND | ND | ND | ND | ND | ND | [16] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonilla-Jiménez, S.; Espinosa de los Monteros-Silva, N.; Morán-Marcillo, G.; Bermúdez-Puga, S.; Terán-Valdez, A.; Almeida, J.R.; Proaño-Bolaños, C. Engineering of a Novel Amphibian Skin Peptide Isolated from Agua Rica Leaf Frog (Callimedusa ecuatoriana) into Active Antimicrobial Agents. Antibiotics 2025, 14, 1186. https://doi.org/10.3390/antibiotics14121186
Bonilla-Jiménez S, Espinosa de los Monteros-Silva N, Morán-Marcillo G, Bermúdez-Puga S, Terán-Valdez A, Almeida JR, Proaño-Bolaños C. Engineering of a Novel Amphibian Skin Peptide Isolated from Agua Rica Leaf Frog (Callimedusa ecuatoriana) into Active Antimicrobial Agents. Antibiotics. 2025; 14(12):1186. https://doi.org/10.3390/antibiotics14121186
Chicago/Turabian StyleBonilla-Jiménez, Stefanny, Nina Espinosa de los Monteros-Silva, Giovanna Morán-Marcillo, Sebastián Bermúdez-Puga, Andrea Terán-Valdez, José R. Almeida, and Carolina Proaño-Bolaños. 2025. "Engineering of a Novel Amphibian Skin Peptide Isolated from Agua Rica Leaf Frog (Callimedusa ecuatoriana) into Active Antimicrobial Agents" Antibiotics 14, no. 12: 1186. https://doi.org/10.3390/antibiotics14121186
APA StyleBonilla-Jiménez, S., Espinosa de los Monteros-Silva, N., Morán-Marcillo, G., Bermúdez-Puga, S., Terán-Valdez, A., Almeida, J. R., & Proaño-Bolaños, C. (2025). Engineering of a Novel Amphibian Skin Peptide Isolated from Agua Rica Leaf Frog (Callimedusa ecuatoriana) into Active Antimicrobial Agents. Antibiotics, 14(12), 1186. https://doi.org/10.3390/antibiotics14121186









