Validation of Automated Bacterial Suspension Preparation by Colibri® and Plate Streaking by WASP® for Antibiotic Disk Diffusion Susceptibility Testing
Abstract
1. Introduction
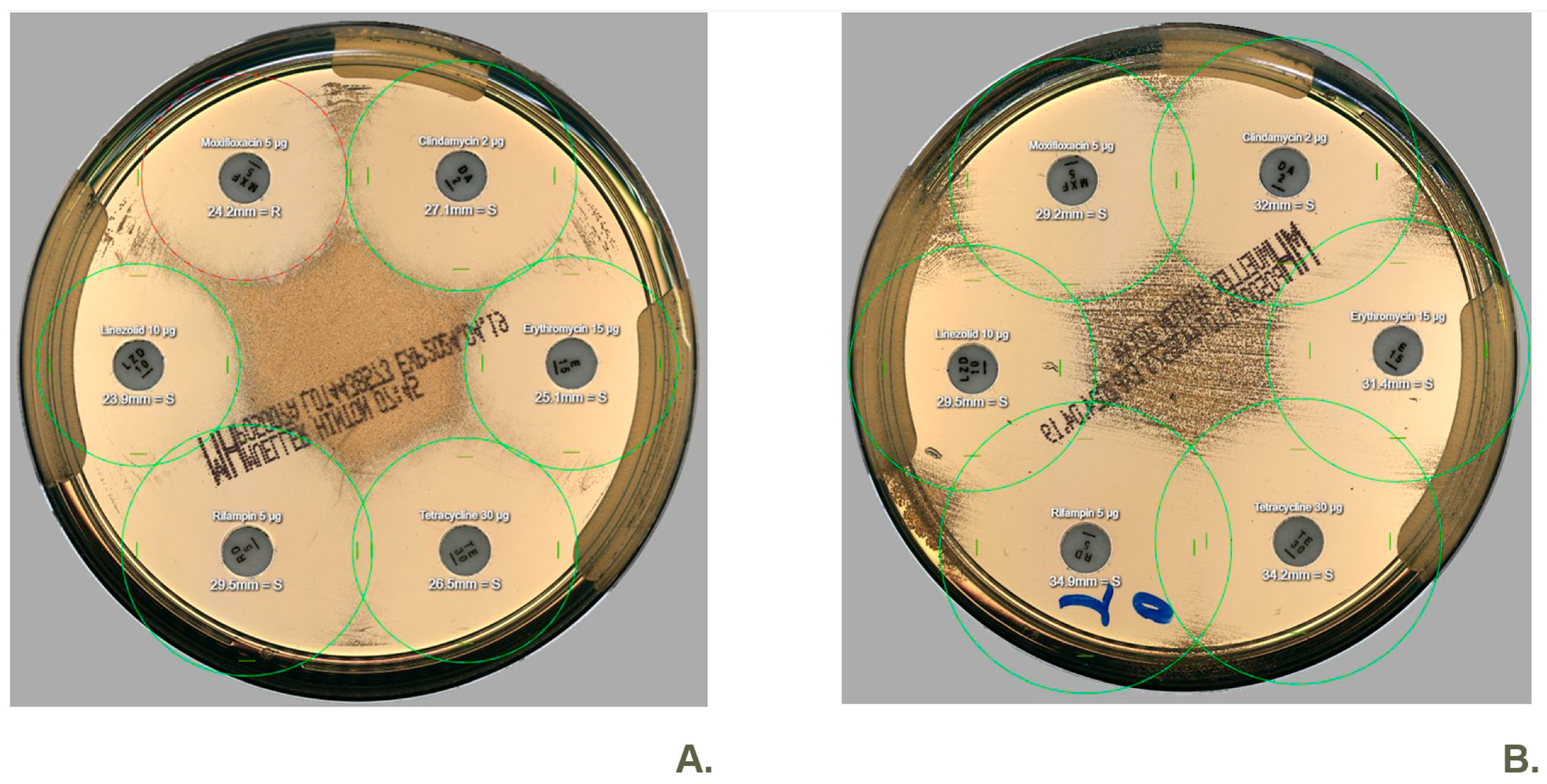
2. Results
3. Discussion
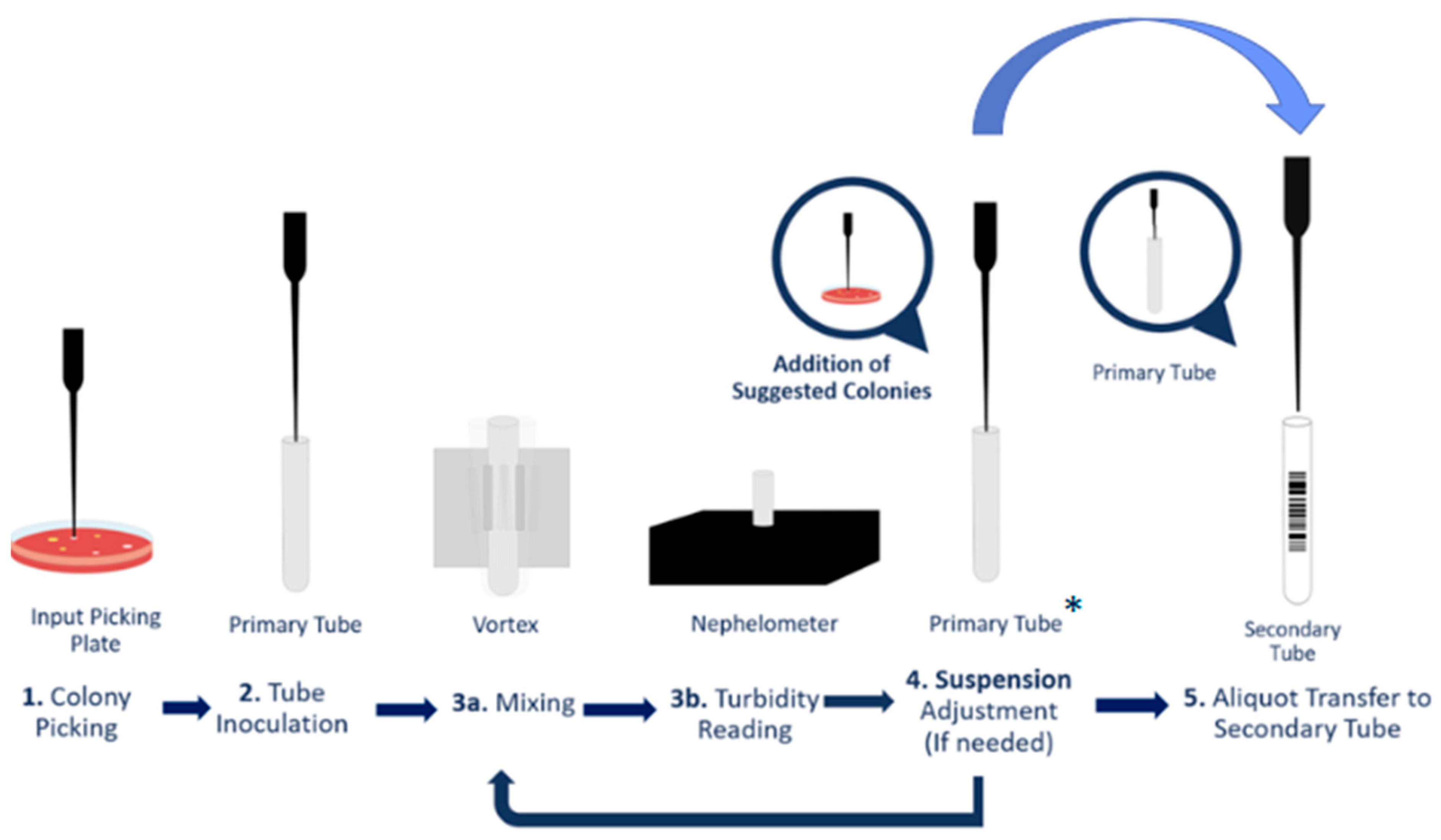
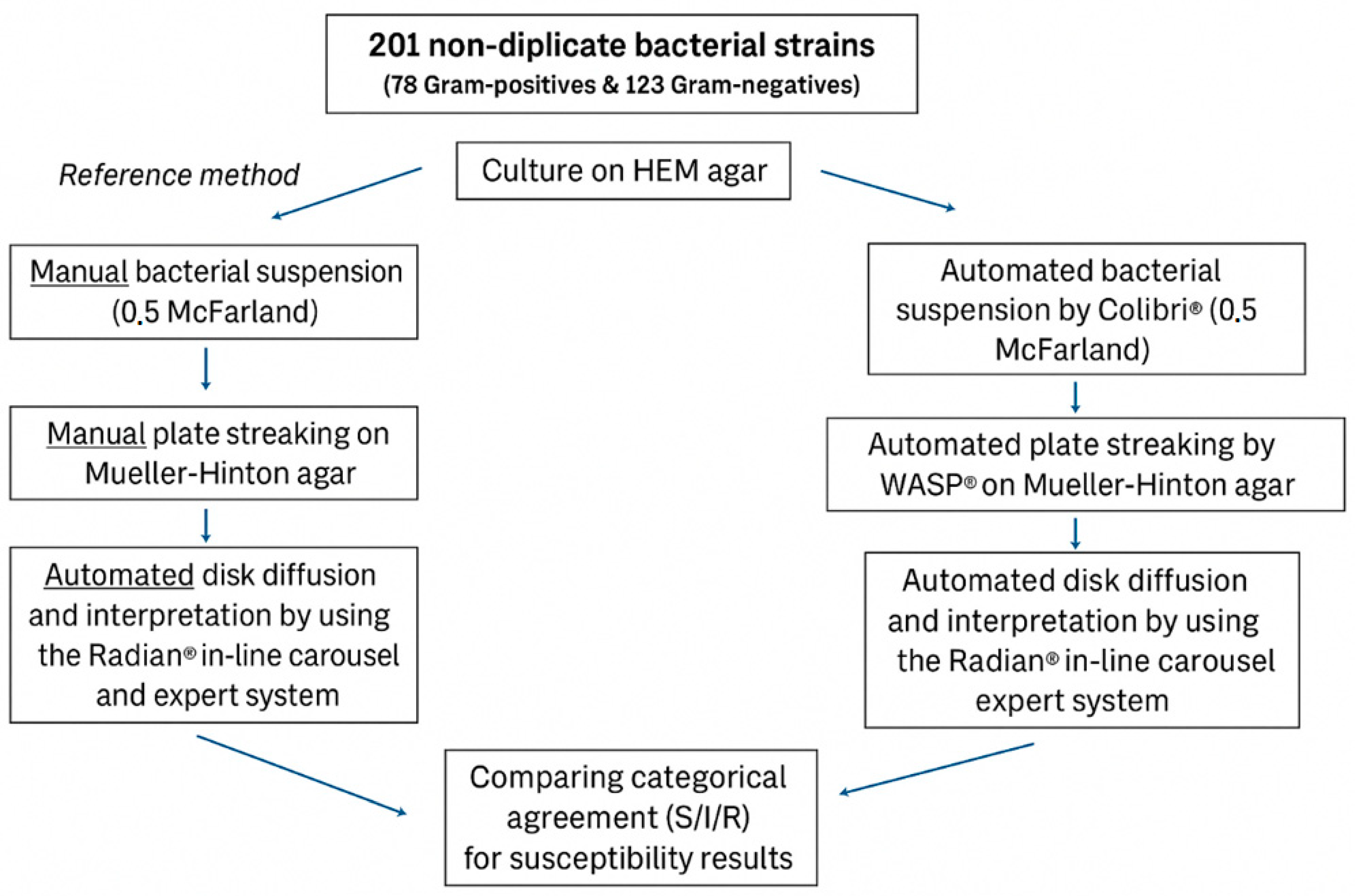
4. Materials and Methods
4.1. Bacterial Strains
4.2. Identification of Species
4.3. Identification of Resistance Mechanisms and Phenotypes
4.4. Antimicrobial Susceptibility Testing
4.5. Quality Control
4.6. Statistical Analysis and Discordant Results
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hombach, M.; Maurer, F.P.; Pfiffner, T.; Böttger, E.C.; Furrer, R. Standardization of Operator-Dependent Variables Affecting Precision and Accuracy of the Disk Diffusion Method for Antibiotic Susceptibility Testing. J. Clin. Microbiol. 2015, 53, 3864–3869. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, K.; Atas, C.; Simsek, E.; Coban, A.Y. The Effect of Inoculum Size on Antimicrobial Susceptibility Testing of Mycobacterium tuberculosis. Microbiol. Spectr. 2023, 11, e0031923. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.L.; Ledeboer, N.; Burnham, C.D. Clinical Microbiology is Growing Up: The Total Laboratory Automation Revolution. Clin. Chem. 2019, 65, 634–643. [Google Scholar] [CrossRef] [PubMed]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [PubMed]
- Croxatto, A.; Prod’hom, G.; Faverjon, F.; Rochais, Y.; Greub, G. Laboratory automation in clinical bacteriology: What system to choose? Clin. Microbiol. Infect. 2016, 22, 217–235. [Google Scholar] [CrossRef] [PubMed]
- Cherkaoui, A.; Renzi, G.; Vuilleumier, N.; Schrenzel, J. Copan WASPLab automation significantly reduces incubation times and allows earlier culture readings. Clin. Microbiol. Infect. 2019, 25, 1430.e5–1430.e12. [Google Scholar] [CrossRef] [PubMed]
- Pham, M.L.; Van Horn, K.; Zarate, E.; Pickering, E.; Murphy, C.; Bryant, K. A multicenter evaluation of Copan’s Colibrí™, an automated instrument for MALDI TOF MS target application for bacterial identification. Diagn. Microbiol. Infect. Dis. 2024, 108, 116098. [Google Scholar] [CrossRef] [PubMed]
- Heestermans, R.; Herroelen, P.; Emmerechts, K.; Vandoorslaer, K.; De Geyter, D.; Demuyser, T.; Wybo, I.; Piérard, D.; Muyldermans, A. Validation of the Colibrí Instrument for Automated Preparation of MALDI-TOF MS Targets for Yeast Identification. J. Clin. Microbiol. 2022, 60, e0023722. [Google Scholar] [CrossRef] [PubMed]
- Cherkaoui, A.; Renzi, G.; Vuilleumier, N.; Schrenzel, J. Performance of Fully Automated Antimicrobial Disk Diffusion Susceptibility Testing Using Copan WASP Colibri Coupled to the Radian In-Line Carousel and Expert System. J. Clin. Microbiol. 2021, 59, e0077721. [Google Scholar] [CrossRef] [PubMed]
- Callebaut, K.; Stoefs, A.; Emmerechts, K.; Vandoorslaer, K.; Wybo, I.; De Geyter, D.; Demuyser, T.; Piérard, D.; Muyldermans, A. Evaluation of Automated Disk Diffusion Antimicrobial Susceptibility Testing Using Radian® In-Line Carousel. Curr. Microbiol. 2024, 81, 196. [Google Scholar] [CrossRef] [PubMed]
- Herroelen, P.H.; Heestermans, R.; Emmerechts, K.; Vandoorslaer, K.; Wybo, I.; Piérard, D.; Muyldermans, A. alidation of Rapid Antimicrobial Susceptibility Testing directly from blood cultures using WASPLab®, including Colibrí™ and Radian® in-Line Carousel. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, T.; Weiss, D.S. Heteroresistance is a cause of discrepant antibiotic susceptibility testing results. Lancet Microbe 2024, 5, e312. [Google Scholar] [CrossRef] [PubMed]
- Lenhard, J.R.; Bulman, Z.P. Inoculum effect of β-lactam antibiotics. J. Antimicrob. Chemother. 2019, 74, 2825–2843. [Google Scholar] [CrossRef] [PubMed]
- Quiblier, C.; Jetter, M.; Rominski, M.; Mouttet, F.; Böttger, E.C.; Keller, P.M.; Hombach, M. Performance of Copan WASP for Routine Urine Microbiology. J. Clin. Microbiol. 2016, 54, 585–592. [Google Scholar] [CrossRef]
- Van Honacker, E.; Vandendriessche, S.; Coorevits, L.; Verhasselt, B.; Boelens, J. Impact of the introduction of EUCAST’s concept of “area of technical uncertainty”. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 203–207. [Google Scholar] [CrossRef]
- Andersson, D.I.; Nicoloff, H.; Hjort, K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat. Rev. Microbiol. 2019, 17, 479–496. [Google Scholar] [CrossRef]
- ISO 20776-2:2021; Evaluation of Performance of Antimicrobial Susceptibility Test Devices Against Reference Broth Micro-Dilution. Clinical Laboratory Testing and In Vitro Diagnostic Test Systems—Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices. ISO: Geneva, Switzerland, 2021.
- CLSI Document M7; Institute, Clinical and Laboratory Standards. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard, 11th ed. CLSI: Berwyn, PA, USA, 2018.
- EUCAST Breakpoints Table Version 13.1, Published on 29 June 2023. Clinical Breakpoints and Dosing of Antibiotics. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 13 March 2025).
- Boutiba-Ben Boubaker, I.; Ben Abbes, R.; Ben Abdallah, H.; Mamlouk, K.; Mahjoubi, F.; Kammoun, A.; Hammami, A.; Ben Redjeb, S. Evaluation of a cefoxitin disk diffusion test for the routine detection of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2004, 10, 762–765. [Google Scholar] [CrossRef]
- CLSI Document M02-Ed11; Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard. CLSI: Berwyn, PA, USA, 2012.
- EUCAST Disk Diffusion Method, Version 9.0. Antimicrobial Susceptibility Testing. 2021. Available online: https://www.eucast.org/ast_of_bacteria/disk_diffusion_methodology (accessed on 13 March 2025).
- Matuschek, E.; Brown, D.F.; Kahlmeter, G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin. Microbiol. Infect. 2014, 20, O255–O266. [Google Scholar] [CrossRef]
- Copan. Radian; Pushing Antimicrobial Susceptibility Testing at Full Speed. Product Focus. 2022. Available online: https://mediadelivery.copangroup.com/wp-content/uploads/2022/11/MMWB-GEN01ENR06_DIGITAL.pdf (accessed on 13 March 2025).
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K. CLSI Methods Development and Standardization Working Group of the Subcommittee on AST. CLSI Methods Development and Standardization Working Group Best Practices for Evaluation of AST. J. Clin. Microbiol. 2018, 56, e01934-17. [Google Scholar] [CrossRef] [PubMed]
- CLSI M52; Verification of Commercial Microbial Identification and Antimicrobial Susceptibility Testing Systems. CLSI: Berwyn, PA, USA, 2015; revised on January 2020.
| S/I/R Agreement | ||||||
|---|---|---|---|---|---|---|
| CA | mE | ME | VME | NA | Total | |
| Amikacin 30 | 123 | 0 | 0 | 0 | 0 | 123 |
| Amoxicillin/clavulanic acid 30 | 93 | 0 | 0 | 0 | 207 | 93 |
| Ampicillin 2 | 18 | 0 | 0 | 0 | 14 | 18 |
| Ampicillin 10 | 94 | 0 | 0 | 0 | 2 | 94 |
| Aztreonam 30 | 110 | 9 | 0 | 0 | 0 | 119 |
| Cefadroxil 30 | 94 | 0 | 0 | 0 | 2 | 94 |
| Cefepime 30 | 101 | 18 | 0 | 0 | 0 | 119 |
| Cefoxitin 30 | 43 | 0 | 0 | 0 | 2 | 43 |
| Ceftazidime 10 | 111 | 8 | 0 | 0 | 0 | 119 |
| Ceftriaxone 30 | 91 | 3 | 0 | 0 | 2 | 94 |
| Cefuroxime 30 | 73 | 0 | 0 | 0 | 17 | 73 |
| Ciprofloxacin 5 | 173 | 10 | 0 | 0 | 1 | 183 |
| Clindamycin 2 | 54 | 0 | 0 | 0 | 2 | 54 |
| Erythromycin 15 | 54 | 0 | 0 | 0 | 2 | 54 |
| Fosfomycin 200 | 34 | 0 | 0 | 0 | 30 | 34 |
| Gentamicin 10 | 140 | 0 | 1 | 0 | 4 | 141 |
| Gentamicin 30 | 0 | 0 | 0 | 0 | 16 | 0 |
| Linezolid 10 | 72 | 0 | 0 | 0 | 0 | 72 |
| Meropenem 10 | 109 | 14 | 0 | 0 | 0 | 123 |
| Moxifloxacin 5 | 145 | 0 | 2 | 0 | 4 | 147 |
| Nitrofurantoin 100 | 43 | 0 | 0 | 0 | 46 | 43 |
| Oxacillin 1 | 0 | 0 | 0 | 0 | 16 | 0 |
| Penicillin G 1 | 38 | 0 | 0 | 0 | 3 | 38 |
| Piperacillin/tazobactam 36 | 30 | 1 | 1 | 0 | 0 | 32 |
| Rifampicin 5 | 54 | 0 | 0 | 0 | 2 | 54 |
| Temocillin 30 | 72 | 0 | 0 | 0 | 18 | 72 |
| Tetracycline 30 | 54 | 0 | 0 | 0 | 2 | 54 |
| Trimethoprim/sulfamethoxazole 25 | 150 | 0 | 0 | 0 | 4 | 150 |
| Vancomycin 5 | 29 | 0 | 0 | 0 | 14 | 29 |
| Total | 2202 | 63 | 4 | 0 | 410 | 2269 |
| % | 97.0% | 2.8% | 0.4% | 0.0% | / | / |
| Strain Reference | Species | Type of Error | Discrepant Antibiotic(s) | Results Before Rerun | Results After Rerun |
|---|---|---|---|---|---|
| Q23/023 | S. epidermidis | VME | cefoxitin | Colibri S, manual R | Colibri S, manual S |
| Q23/064 | S. aureus | VME | moxifloxacin | Colibri S, manual R | Colibri S, manual S |
| REF-814 | E. faecalis | VME | linezolid, vancomycin | Colibri S, manual R | Colibri S, manual S |
| Q21/003 | S. aureus | ME | moxifloxacin | Colibri R, manual S | Colibri R, manual S |
| Q22/035 | E. faecium | ME | linezolid | Colibri R, manual S | Colibri S, manual S |
| REF-739 | S. pyogenes | ME | penicillin | Colibri R, manual S | Colibri S, manual S |
| Q23/013 | E. cloacae | ME | moxifloxacin | Colibri R, manual S | Colibri R, manual S |
| REF-727 | E. cloacae | ME | moxifloxacin | Colibri R, manual S | Colibri R, manual R |
| REF-793 | K. aerogenes | ME | amikacin*, gentamicin | Colibri R, manual S | Colibri R, manual S |
| 23/0645 | K. pneumoniae | ME | amikacin | Colibri R, manual S | Colibri S, manual S |
| 23/1560 | E. coli | ME | nitrofurantoin | Colibri R, manual S | Colibri S, manual S |
| Q09/035 | P. aeruginosa | ME | meropenem | Colibri R, manual S | Colibri S, manual S |
| REF-708 | K. pneumoniae | ME | cefepime | Colibri R, manual S | Colibri R, manual R |
| REF-716 | K. oxytoca | ME | piperacillin/tazobactam | Colibri R, manual S | Colibri R, manual S |
| REF-717 | K. pneumoniae | ME | amikacin | Colibri R, manual S | Colibri R, manual R |
| Q09/028 | K. pneumoniae | ME | cefuroxime | Colibri R, manual S | Colibri S, manual S |
| Q18/011 | K. pneumoniae | ME | cefuroxime | Colibri R, manual S | Colibri S, manual S |
| Q22/033 | K. oxytoca | ME | amikacin | Colibri R, manual S | Colibri R, manual R |
| Q23/030 | E. coli | ME | amikacin | Colibri R, manual S | Colibri R, manual R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanstokstraeten, R.; Van Honacker, E.; Emmerechts, K.; Janssen, Z.; Michel, C.; Van Der Kelen, G.; Vandoorslaer, K.; De Geyter, D.; Vanmechelen, B. Validation of Automated Bacterial Suspension Preparation by Colibri® and Plate Streaking by WASP® for Antibiotic Disk Diffusion Susceptibility Testing. Antibiotics 2025, 14, 1178. https://doi.org/10.3390/antibiotics14121178
Vanstokstraeten R, Van Honacker E, Emmerechts K, Janssen Z, Michel C, Van Der Kelen G, Vandoorslaer K, De Geyter D, Vanmechelen B. Validation of Automated Bacterial Suspension Preparation by Colibri® and Plate Streaking by WASP® for Antibiotic Disk Diffusion Susceptibility Testing. Antibiotics. 2025; 14(12):1178. https://doi.org/10.3390/antibiotics14121178
Chicago/Turabian StyleVanstokstraeten, Robin, Eveline Van Honacker, Kristof Emmerechts, Zan Janssen, Charlotte Michel, Goran Van Der Kelen, Kristof Vandoorslaer, Deborah De Geyter, and Bram Vanmechelen. 2025. "Validation of Automated Bacterial Suspension Preparation by Colibri® and Plate Streaking by WASP® for Antibiotic Disk Diffusion Susceptibility Testing" Antibiotics 14, no. 12: 1178. https://doi.org/10.3390/antibiotics14121178
APA StyleVanstokstraeten, R., Van Honacker, E., Emmerechts, K., Janssen, Z., Michel, C., Van Der Kelen, G., Vandoorslaer, K., De Geyter, D., & Vanmechelen, B. (2025). Validation of Automated Bacterial Suspension Preparation by Colibri® and Plate Streaking by WASP® for Antibiotic Disk Diffusion Susceptibility Testing. Antibiotics, 14(12), 1178. https://doi.org/10.3390/antibiotics14121178






