The Potential Roles of Prophages in the Pathogenicity of Klebsiella pneumoniae Strains from Kenya
Abstract
1. Introduction
2. Results
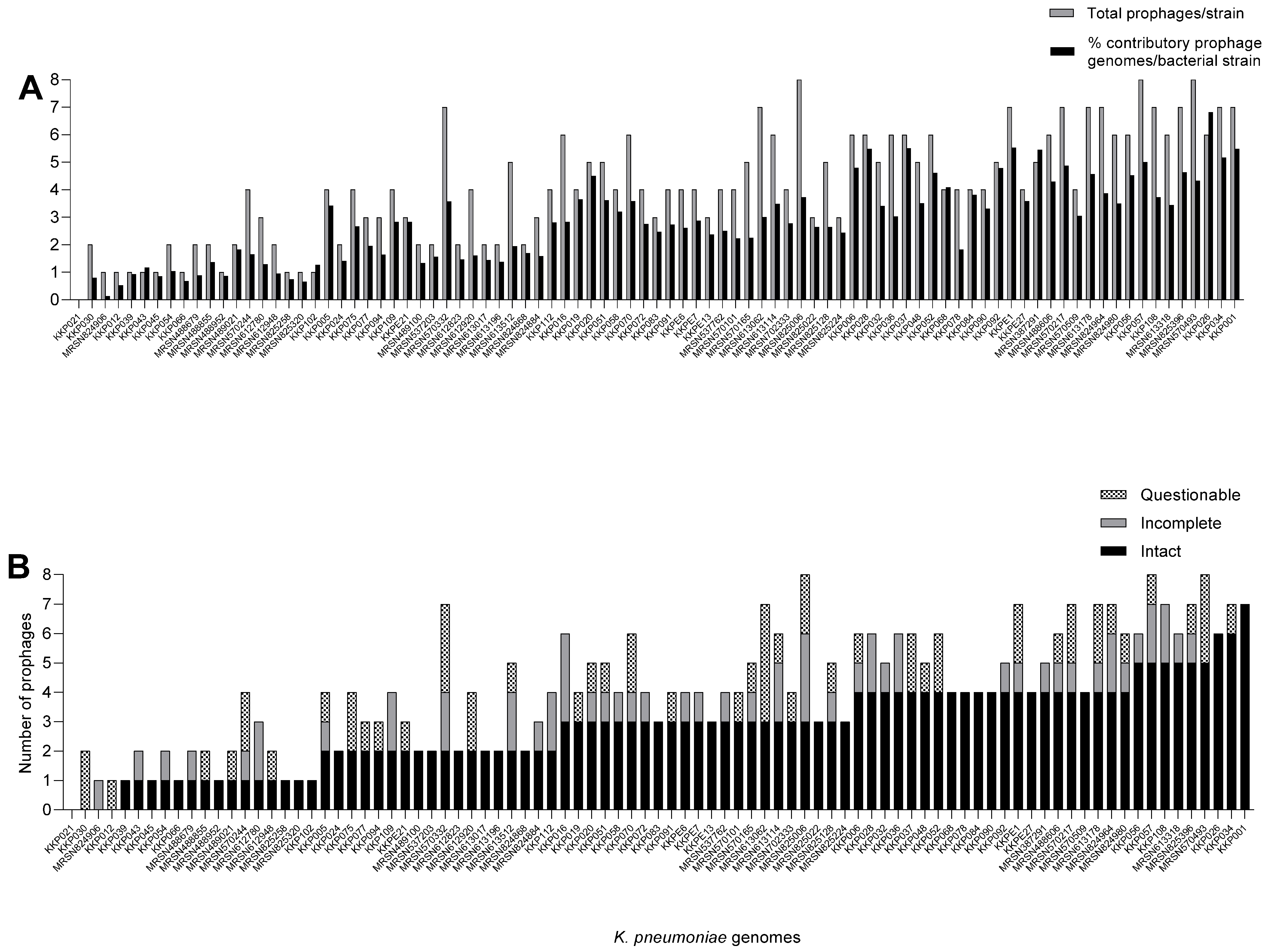
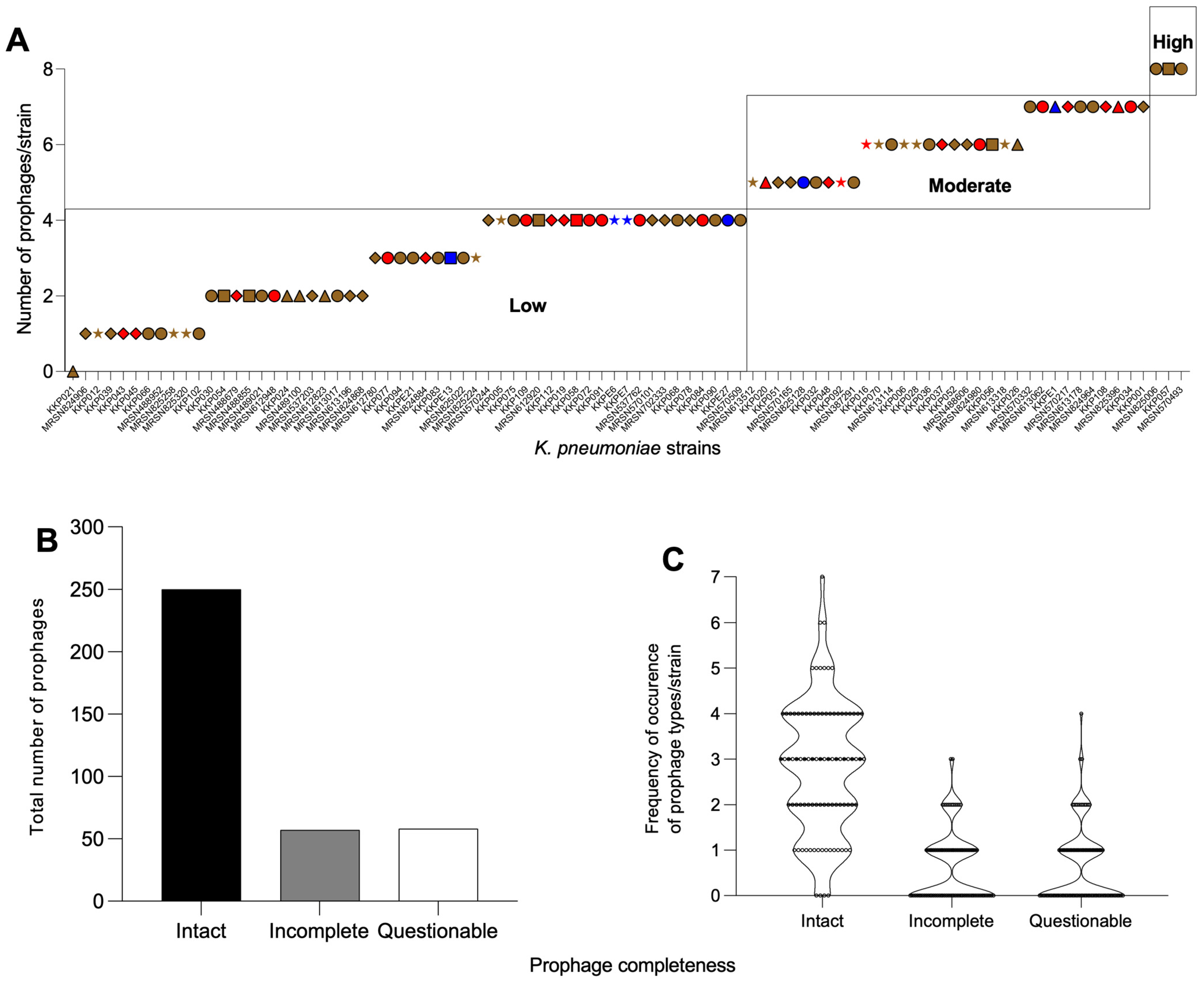
2.1. K. pneumoniae Strains from Kenya Carry Multiple and Diverse Prophages in Their Genomes
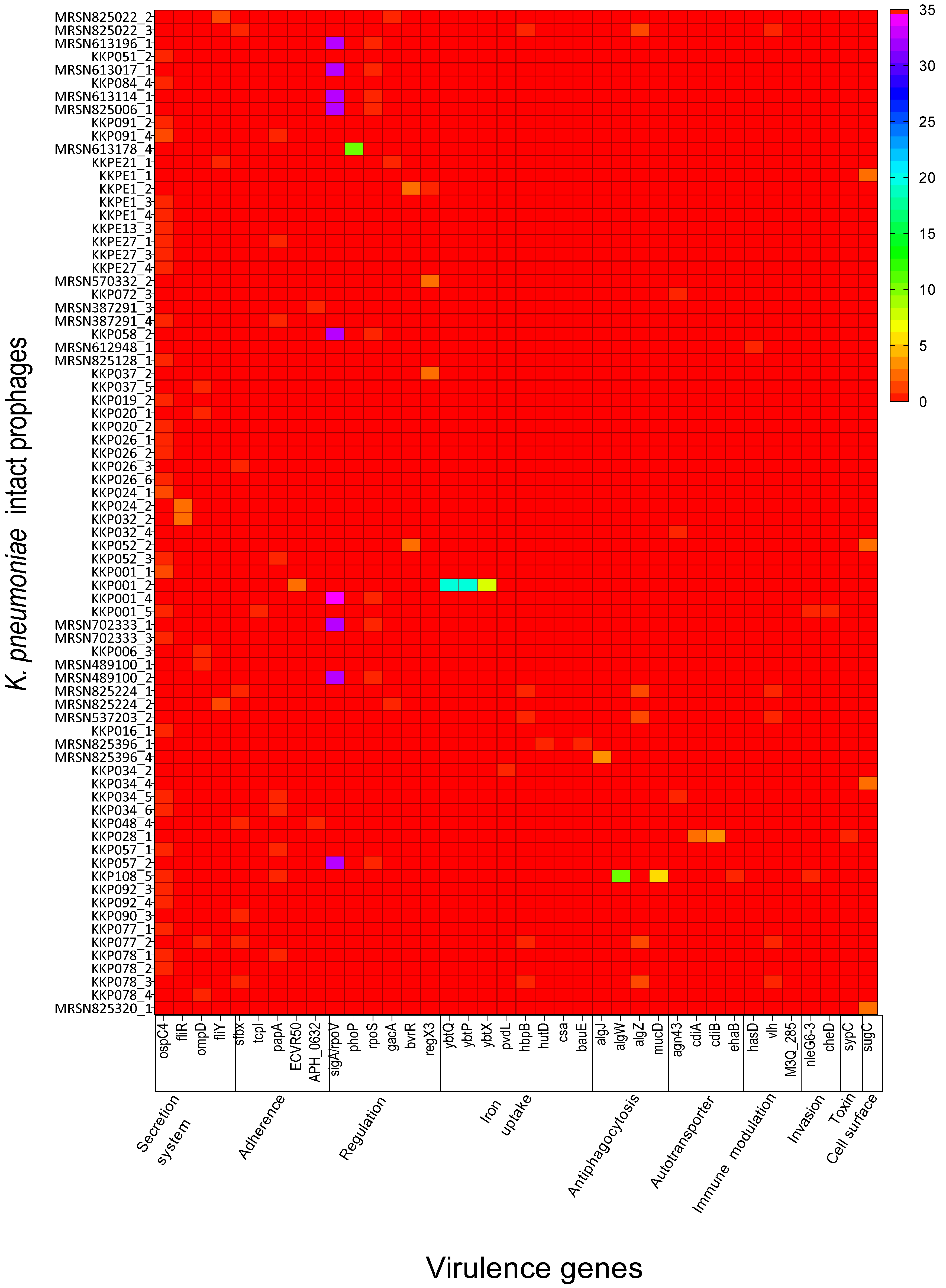
2.2. K. pneumoniae Intact Prophages Encode Several Virulence Genes That Are Related to Pathogenicity
2.3. K. pneumoniae Intact Prophages Are Genetically Diverse and Formed Three Distinct Lineages
3. Discussion
4. Materials and Methods
4.1. Collation of Bacterial Genomes
4.2. Identification and Extraction of Prophages from the Kenyan K. pneumoniae Genomes
4.3. Identification of Virulence Genes Within the Intact Prophages and Their Association with K. pneumoniae Pathogenicity
4.4. Phylogenetic Relationships of Intact Prophages of K. pneumoniae
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abbas, R.; Chakkour, M.; Zein El Dine, H.; Obaseki, E.F.; Obeid, S.T.; Jezzini, A.; Ghssein, G.; Ezzeddine, Z. General Overview of Klebsiella pneumonia: Epidemiology and the Role of Siderophores in Its Pathogenicity. Biology 2024, 13, 78. [Google Scholar] [CrossRef]
- Karimi, K.; Zarei, O.; Sedighi, P.; Taheri, M.; Doosti-Irani, A.; Shokoohizadeh, L. Investigation of Antibiotic Resistance and Biofilm Formation in Clinical Isolates of Klebsiella pneumoniae. Int. J. Microbiol. 2021, 2021, 5573388. [Google Scholar] [CrossRef] [PubMed]
- Guerra, M.E.S.; Destro, G.; Vieira, B.; Lima, A.S.; Ferraz, L.F.C.; Hakansson, A.P.; Darrieux, M.; Converso, T.R. Klebsiella pneumoniae Biofilms and Their Role in Disease Pathogenesis. Front. Cell Infect. Microbiol. 2022, 12, 877995. [Google Scholar] [CrossRef]
- Rawat, D.; Nair, D. Extended-spectrum β-lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2010, 2, 263–274. [Google Scholar] [CrossRef]
- Henson, S.P.; Boinett, C.J.; Ellington, M.J.; Kagia, N.; Mwarumba, S.; Nyongesa, S.; Mturi, N.; Kariuki, S.; Scott, J.A.G.; Thomson, N.R.; et al. Molecular epidemiology of Klebsiella pneumoniae invasive infections over a decade at Kilifi County Hospital in Kenya. Int. J. Med. Microbiol. 2017, 307, 422–429. [Google Scholar] [CrossRef]
- Wairimu, C.; Kariuki, S. Antimicrobial susceptibility and genetic basis of resistance of Klebsiella spp isolated from diarrheic and non-diarrheic children at health facilities in Mukuru informal settlement, Nairobi, Kenya. Int. J. Infect. Dis. 2023, 130, S131. [Google Scholar] [CrossRef]
- Pomakova, D.K.; Hsiao, C.B.; Beanan, J.M.; Olson, R.; MacDonald, U.; Keynan, Y.; Russo, T.A. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: An emerging and under-recognized pathogenic variant. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 981–989. [Google Scholar] [CrossRef]
- Das, M. Global update on hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 2024, 24, e621. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Sun, G.; Yu, Y.; Li, N.; Chen, M.; Jin, R.; Jiao, Y.; Wu, H. Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin. Infect. Dis. 2014, 58, 225–232. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, G.; Chao, X.; Xie, L.; Wang, H. The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae. Int. J. Environ. Res. Public Health 2020, 17, 6278. [Google Scholar] [CrossRef]
- Mendes, G.; Santos, M.L.; Ramalho, J.F.; Duarte, A.; Caneiras, C. Virulence factors in carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front. Microbiol. 2023, 14, 1325077. [Google Scholar] [CrossRef]
- Asokan, S.; Jacob, T.; Jacob, J.; AlSosowaa, A.A.; Cherian, T.; Peijnenburg, W.J.G.M.; Vijayan, S. Klebsiella pneumoniae: A growing threat in the era of antimicrobial resistance. Microbe 2025, 7, 100333. [Google Scholar] [CrossRef]
- Russo, A.; Fusco, P.; Morrone, H.L.; Trecarichi, E.M.; Torti, C. New advances in management and treatment of multidrug-resistant Klebsiella pneumoniae. Expert Rev. Anti Infect. Ther. 2023, 21, 41–55. [Google Scholar] [CrossRef]
- Ndlovu, T.; Kgosietsile, L.; Motshwarakgole, P.; Ndlovu, S.I. Evaluation of Potential Factors Influencing the Dissemination of Multidrug-Resistant Klebsiella pneumoniae and Alternative Treatment Strategies. Trop. Med. Infect. Dis. 2023, 8, 381. [Google Scholar] [CrossRef]
- O’Neill, J. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2015, 370, 20140082. [Google Scholar]
- Nale, J.Y.; Chan, B.; Nnadi, N.E.; Cheng, J.K.J.; Matts, S.; Nezam-Abadi, N.; Turkington, C.J.R.; Charreton, L.M.; Bola, H.; Nazir, R.; et al. Novel Escherichia coli-Infecting Bacteriophages Isolated from Uganda That Target Human Clinical Isolates. Phage 2023, 4, 141–149. [Google Scholar] [CrossRef]
- IHME. The Burden of Antimicrobial Resistance (AMR) in Kenya; Institute for Health Metrics and Evaluation: Seattle, WA, USA, 2019; Available online: https://www.healthdata.org/sites/default/files/2023-09/Kenya.pdf (accessed on 20 August 2025).
- Rwigi, D.; Nyerere, A.K.; Diakhate, M.M.; Kariuki, K.; Tickell, K.D.; Mutuma, T.; Tornberg, S.N.; Soge, O.O.; Walson, J.L.; Singa, B.; et al. Phenotypic and molecular characterization of β-lactamase-producing Klebsiella species among children discharged from hospital in Western Kenya. BMC Microbiol. 2024, 24, 135. [Google Scholar] [CrossRef]
- Muraya, A.; Kyany’a, C.; Kiyaga, S.; Smith, H.J.; Kibet, C.; Martin, M.J.; Kimani, J.; Musila, L. Antimicrobial Resistance and Virulence Characteristics of Klebsiella pneumoniae Isolates in Kenya by Whole-Genome Sequencing. Pathogens 2022, 11, 545. [Google Scholar] [CrossRef]
- Poirel, L.; Revathi, G.; Bernabeu, S.; Nordmann, P. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob. Agents Chemother. 2011, 55, 934–936. [Google Scholar] [CrossRef]
- Dieppa-Colón, E.; Martin, C.; Kosmopoulos, J.C.; Anantharaman, K. Prophage-DB: A comprehensive database to explore diversity, distribution, and ecology of prophages. Environ. Microbiome 2025, 20, 5. [Google Scholar] [CrossRef]
- Nadeem, A.; Wahl, L.M. Prophage as a genetic reservoir: Promoting diversity and driving innovation in the host community. Evolution 2017, 71, 2080–2089. [Google Scholar] [CrossRef]
- Andrews, K.; Landeryou, T.; Sicheritz-Pontén, T.; Nale, J.Y. Diverse Prophage Elements of Salmonella enterica Serovars Show Potential Roles in Bacterial Pathogenicity. Cells 2024, 13, 514. [Google Scholar] [CrossRef]
- Howard-Varona, C.; Hargreaves, K.R.; Abedon, S.T.; Sullivan, M.B. Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 2017, 11, 1511–1520. [Google Scholar] [CrossRef]
- Marques, A.T.; Tanoeiro, L.; Duarte, A.; Gonçalves, L.; Vítor, J.M.B.; Vale, F.F. Genomic Analysis of Prophages from Klebsiella pneumoniae Clinical Isolates. Microorganisms 2021, 9, 2252. [Google Scholar] [CrossRef]
- Wang, F.; Wang, D.; Hou, W.; Jin, Q.; Feng, J.; Zhou, D. Evolutionary Diversity of Prophage DNA in Klebsiella pneumoniae Chromosomes. Front. Microbiol. 2019, 10, 2840. [Google Scholar] [CrossRef]
- Bleriot, I.; Trastoy, R.; Blasco, L.; Fernández-Cuenca, F.; Ambroa, A.; Fernández-García, L.; Pacios, O.; Perez-Nadales, E.; Torre-Cisneros, J.; Oteo-Iglesias, J.; et al. Genomic analysis of 40 prophages located in the genomes of 16 carbapenemase-producing clinical strains of Klebsiella pneumoniae. Microb. Genom. 2020, 6, e000369. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Liu, P.; Jian, Z.; Yan, Q.; Tang, B.; Yang, A.; Liu, W. Genomic and clinical characterization of Klebsiella pneumoniae carrying the pks island. Front. Microbiol. 2023, 14, 1189120. [Google Scholar] [CrossRef]
- Fu, Y.; Wu, Y.; Yuan, Y.; Gao, M. Prevalence and Diversity Analysis of Candidate Prophages to Provide An Understanding on Their Roles in Bacillus thuringiensis. Viruses 2019, 11, 388. [Google Scholar] [CrossRef]
- Kang, F.; Chai, Z.; Li, B.; Hu, M.; Yang, Z.; Wang, X.; Liu, W.; Ren, H.; Jin, Y.; Yue, J. Characterization and Diversity of Klebsiella pneumoniae Prophages. Int. J. Mol. Sci. 2023, 24, 9116. [Google Scholar] [CrossRef]
- Li, W.; Liu, X.; Tsui, W.; Xu, A.; Li, D.; Zhang, X.; Li, P.; Bian, X.; Zhang, J. Identification and Comparative Genomic Analysis of Type VI Secretion Systems and Effectors in Klebsiella pneumoniae. Front. Microbiol. 2022, 13, 853744. [Google Scholar] [CrossRef]
- Perry, R.D.; Fetherston, J.D. Yersiniabactin iron uptake: Mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect. 2011, 13, 808–817. [Google Scholar] [CrossRef]
- Vollenweider, V.; Rehm, K.; Chepkirui, C.; Pérez-Berlanga, M.; Polymenidou, M.; Piel, J.; Bigler, L.; Kümmerli, R. Antimicrobial activity of iron-depriving pyoverdines against human opportunistic pathogens. Elife 2024, 13, RP92493. [Google Scholar] [CrossRef]
- Schalk, I.J.; Perraud, Q. Pseudomonas aeruginosa and its multiple strategies to access iron. Environ. Microbiol. 2023, 25, 811–831. [Google Scholar] [CrossRef]
- Xu, X.; Holt, S.C.; Kolodrubetz, D. Cloning and expression of two novel hemin binding protein genes from Treponema denticola. Infect. Immun. 2001, 69, 4465–4472. [Google Scholar] [CrossRef]
- Cook-Libin, S.; Sykes, E.M.E.; Kornelsen, V.; Kumar, A. Iron Acquisition Mechanisms and Their Role in the Virulence of Acinetobacter baumannii. Infect. Immun. 2022, 90, e0022322. [Google Scholar] [CrossRef]
- Litwin, C.M.; Calderwood, S.B. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 1993, 6, 137–149. [Google Scholar] [CrossRef]
- Peterson, J.W. Bacterial Pathogenesis. In Medical Microbiology; Baron, S., Ed.; University of Texas Medical Branch at Galveston Copyright © 1996; The University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Stones, D.H.; Krachler, A.M. Fatal attraction: How bacterial adhesins affect host signaling and what we can learn from them. Int. J. Mol. Sci. 2015, 16, 2626–2640. [Google Scholar] [CrossRef]
- Lane, M.C.; Mobley, H.L. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int. 2007, 72, 19–25. [Google Scholar] [CrossRef]
- Murphy, C.N.; Mortensen, M.S.; Krogfelt, K.A.; Clegg, S. Role of Klebsiella pneumoniae type 1 and type 3 fimbriae in colonizing silicone tubes implanted into the bladders of mice as a model of catheter-associated urinary tract infections. Infect. Immun. 2013, 81, 3009–3017. [Google Scholar] [CrossRef]
- Wang, H.; Guo, Y.; Liu, Z.; Chang, Z. The Type VI Secretion System Contributes to the Invasiveness of Liver Abscess Caused by Klebsiella pneumoniae. J. Infect. Dis. 2023, 228, 1127–1136. [Google Scholar] [CrossRef]
- Sarantis, H.; Grinstein, S. Subversion of phagocytosis for pathogen survival. Cell Host Microbe 2012, 12, 419–431. [Google Scholar] [CrossRef]
- Parrott, A.M.; Shi, J.; Aaron, J.; Green, D.A.; Whittier, S.; Wu, F. Detection of multiple hypervirulent Klebsiella pneumoniae strains in a New York City hospital through screening of virulence genes. Clin. Microbiol. Infect. 2021, 27, 583–589. [Google Scholar] [CrossRef]
- Pokharel, P.; Habouria, H.; Bessaiah, H.; Dozois, C.M. Serine Protease Autotransporters of the Enterobacteriaceae (SPATEs): Out and About and Chopping It Up. Microorganisms 2019, 7, 594. [Google Scholar] [CrossRef]
- Leseigneur, C.; Lê-Bury, P.; Pizarro-Cerdá, J.; Dussurget, O. Emerging Evasion Mechanisms of Macrophage Defenses by Pathogenic Bacteria. Front. Cell. Infect. Microbiol. 2020, 10, 577559. [Google Scholar] [CrossRef]
- Kristich, C.J.; Ordal, G.W. Bacillus subtilis CheD is a chemoreceptor modification enzyme required for chemotaxis. J. Biol. Chem. 2002, 277, 25356–25362. [Google Scholar] [CrossRef]
- Martin-Creuzburg, D.; Kowarik, C.; Straile, D. Cross-ecosystem fluxes: Export of polyunsaturated fatty acids from aquatic to terrestrial ecosystems via emerging insects. Sci. Total Environ. 2017, 577, 174–182. [Google Scholar] [CrossRef]
- Scholz-Schroeder, B.K.; Soule, J.D.; Gross, D.C. The sypA, sypB, and sypC Synthetase Genes Encode Twenty-Two Modules Involved in the Nonribosomal Peptide Synthesis of Syringopeptin by Pseudomonas syringae pv. syringae B301D. Mol. Plant-Microbe Interact.® 2003, 16, 271–280. [Google Scholar] [CrossRef]
- Kalscheuer, R.; Weinrick, B.; Veeraraghavan, U.; Besra, G.S.; Jacobs, W.R., Jr. Trehalose-recycling ABC transporter LpqY-SugA-SugB-SugC is essential for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 21761–21766. [Google Scholar] [CrossRef] [PubMed]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Ehsan, H. Antibiotic Resistance in Developing Countries: Emerging Threats and Policy Responses. Public Health Chall. 2025, 4, e70034. [Google Scholar] [CrossRef]
- Berry, S.K.; Rust, S.; Irving, L.; Bartholdson Scott, J.; Weinert, L.A.; Dougan, G.; Christie, G.; Warrener, P.; Minter, R.; Grant, A.J. Characterization of mAbs against Klebsiella pneumoniae type 3 fimbriae isolated in a target-independent phage display campaign. Microbiol. Spectr. 2024, 12, e0040024. [Google Scholar] [CrossRef]
- Sabat, A.J.; Budimir, A.; Nashev, D.; Sá-Leão, R.; van Dijl, J.; Laurent, F.; Grundmann, H.; Friedrich, A.W. Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Eurosurveillance 2013, 18, 20380. [Google Scholar] [CrossRef]
- Boeckaerts, D.; Stock, M.; Ferriol-González, C.; Oteo-Iglesias, J.; Sanjuán, R.; Domingo-Calap, P.; De Baets, B.; Briers, Y. Prediction of Klebsiella phage-host specificity at the strain level. Nat. Commun. 2024, 15, 4355. [Google Scholar] [CrossRef]
- Singh, R.P.; Kapoor, A.; Sinha, A.; Ma, Y.; Shankar, M. Virulence factors of Klebsiella pneumoniae: Insights into canonical and emerging mechanisms driving pathogenicity and drug resistance. Microbe 2025, 7, 100289. [Google Scholar] [CrossRef]
- Nale, J.Y.; Thanki, A.M.; Rashid, S.J.; Shan, J.; Vinner, G.K.; Dowah, A.S.A.; Cheng, J.K.J.; Sicheritz-Pontén, T.; Clokie, M.R.J. Diversity, Dynamics and Therapeutic Application of Clostridioides difficile Bacteriophages. Viruses 2022, 14, 2772. [Google Scholar] [CrossRef] [PubMed]
- Satta, G.; Pruzzo, C.; Debbia, E.; Calegari, L. Lysogenic conversion in Klebsiella pneumoniae: System which requires active immunity regulation for expression of the conversion phenomenon. J. Virol. 1978, 28, 786–794. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, J.A.M.; Buffet, A.; Haudiquet, M.; Rocha, E.P.C.; Rendueles, O. Modular prophage interactions driven by capsule serotype select for capsule loss under phage predation. ISME J. 2020, 14, 2980–2996. [Google Scholar] [CrossRef]
- Nirmal Kumar, G.P.; Sundarrajan, S.; Paul, V.D.; Nandini, S.; Saravanan, R.S.; Hariharan, S.; Sriram, B.; Padmanabhan, S. Use of prophage free host for achieving homogenous population of bacteriophages: New findings. Virus Res. 2012, 169, 182–187. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Guo, J. Prophage induction by non-antibiotic compounds promotes transformation of released antibiotic resistance genes from cell lysis. Water Res. 2024, 263, 122200. [Google Scholar] [CrossRef]
- Hu, J.; Ye, H.; Wang, S.; Wang, J.; Han, D. Prophage Activation in the Intestine: Insights Into Functions and Possible Applications. Front. Microbiol. 2021, 12, 785634. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Huang, L.; Yang, Y.; Xiang, Y.; Liu, J. Essential phage component induces resistance of bacterial community. Sci. Adv. 2024, 10, eadp5057. [Google Scholar] [CrossRef]
- Egido, J.E.; Toner-Bartelds, C.; Costa, A.R.; Brouns, S.J.J.; Rooijakkers, S.H.M.; Bardoel, B.W.; Haas, P.-J. Monitoring phage-induced lysis of gram-negatives in real time using a fluorescent DNA dye. Sci. Rep. 2023, 13, 856. [Google Scholar] [CrossRef]
- Lapras, B.; Marchand, C.; Merienne, C.; Medina, M.; Kolenda, C.; Laurent, F.; Pirot, F. Rationalisation of the purification process for a phage active pharmaceutical ingredient. Eur. J. Pharm. Biopharm. 2024, 203, 114438. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Calzada, A.; Fronzes, R.; Savva, C.G.; Chandran, V.; Lian, P.W.; Laeremans, T.; Pardon, E.; Steyaert, J.; Remaut, H.; Waksman, G.; et al. Structure of a bacterial type IV secretion core complex at subnanometre resolution. EMBO J. 2013, 32, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Vasanthrao, R.; Nidhin, I.K.; Taj, Z.; Chattopadhyay, I. Comprehensive whole metagenomics analysis uncovers microbial community and resistome variability across anthropogenically contaminated soils in urban and suburban areas of Tamil Nadu, India. Front. Microbiol. 2025, 16, 1649872. [Google Scholar] [CrossRef]
- Schwartz, D.A.; Lehmkuhl, B.K.; Lennon, J.T. Phage-Encoded Sigma Factors Alter Bacterial Dormancy. mSphere 2022, 7, e0029722. [Google Scholar] [CrossRef]
- Zhang, Y.; Liao, Y.-T.; Salvador, A.; Wu, V.C.H. Genomic Characterization of Two Shiga Toxin–Converting Bacteriophages Induced From Environmental Shiga Toxin–Producing Escherichia coli. Front. Microbiol. 2021, 12, 587696. [Google Scholar] [CrossRef]
- Khalil, R.K.S.; Skinner, C.; Patfield, S.; He, X. Phage-mediated Shiga toxin (Stx) horizontal gene transfer and expression in non-Shiga toxigenic Enterobacter and Escherichia coli strains. Pathog. Dis. 2016, 74, ftw037. [Google Scholar] [CrossRef]
- Shen, J.; Zhou, J.; Xu, Y.; Xiu, Z. Prophages contribute to genome plasticity of Klebsiella pneumoniae and may involve the chromosomal integration of ARGs in CG258. Genomics 2020, 112, 998–1010. [Google Scholar] [CrossRef]
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: Faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023, 51, W443–W450. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akhwale, J.K.; Mutai, I.J.; Nale, J.Y. The Potential Roles of Prophages in the Pathogenicity of Klebsiella pneumoniae Strains from Kenya. Antibiotics 2025, 14, 1145. https://doi.org/10.3390/antibiotics14111145
Akhwale JK, Mutai IJ, Nale JY. The Potential Roles of Prophages in the Pathogenicity of Klebsiella pneumoniae Strains from Kenya. Antibiotics. 2025; 14(11):1145. https://doi.org/10.3390/antibiotics14111145
Chicago/Turabian StyleAkhwale, Juliah K., Ivy J. Mutai, and Janet Y. Nale. 2025. "The Potential Roles of Prophages in the Pathogenicity of Klebsiella pneumoniae Strains from Kenya" Antibiotics 14, no. 11: 1145. https://doi.org/10.3390/antibiotics14111145
APA StyleAkhwale, J. K., Mutai, I. J., & Nale, J. Y. (2025). The Potential Roles of Prophages in the Pathogenicity of Klebsiella pneumoniae Strains from Kenya. Antibiotics, 14(11), 1145. https://doi.org/10.3390/antibiotics14111145





