Genomic Characterization of Multiple Antibiotic-Resistant Enterococcus in Farm Animals in Ningxia Province, China
Abstract
1. Introduction
2. Results
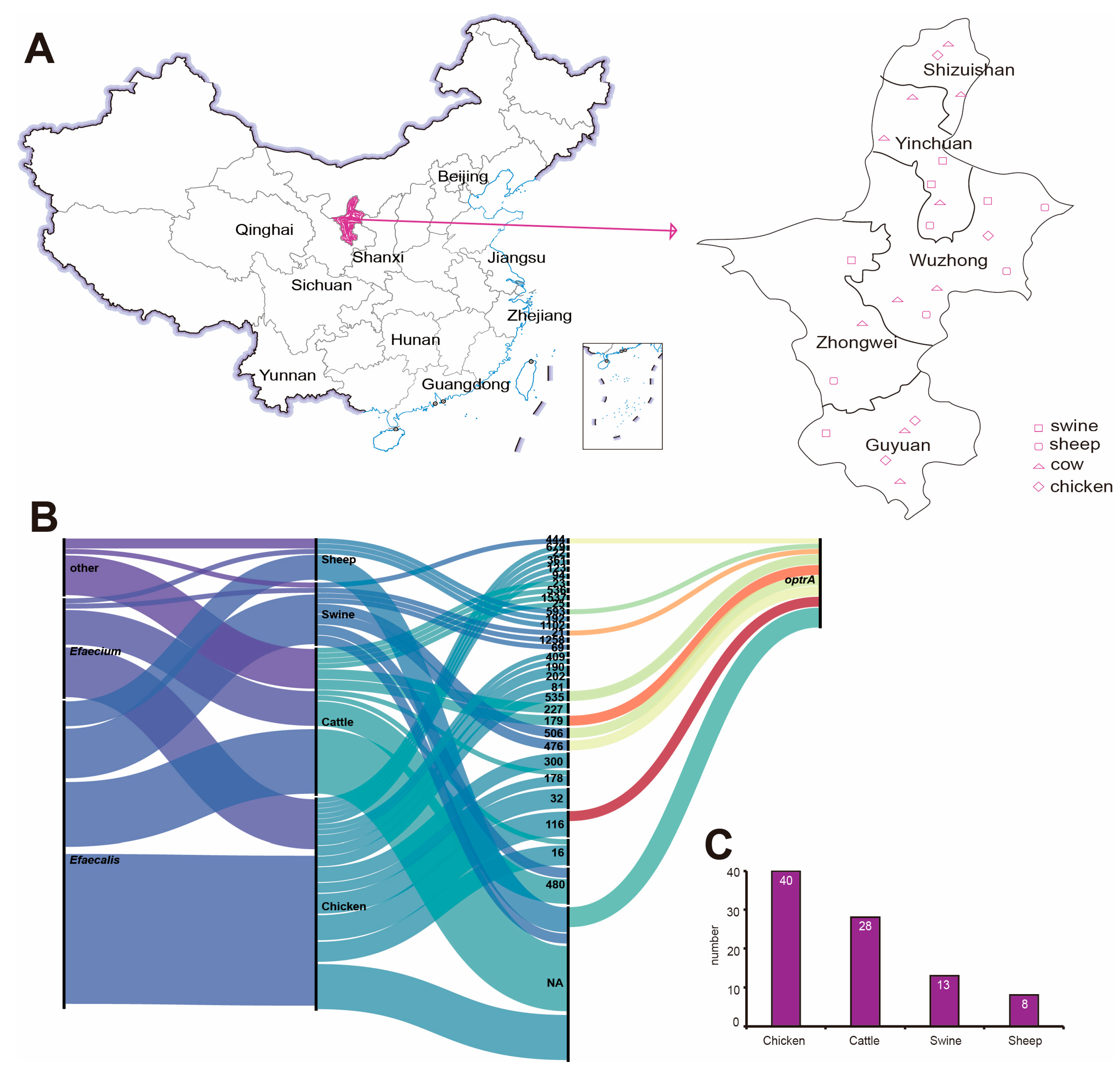
2.1. Strain Source
2.2. Results of Antibiotic Sensitivity Test
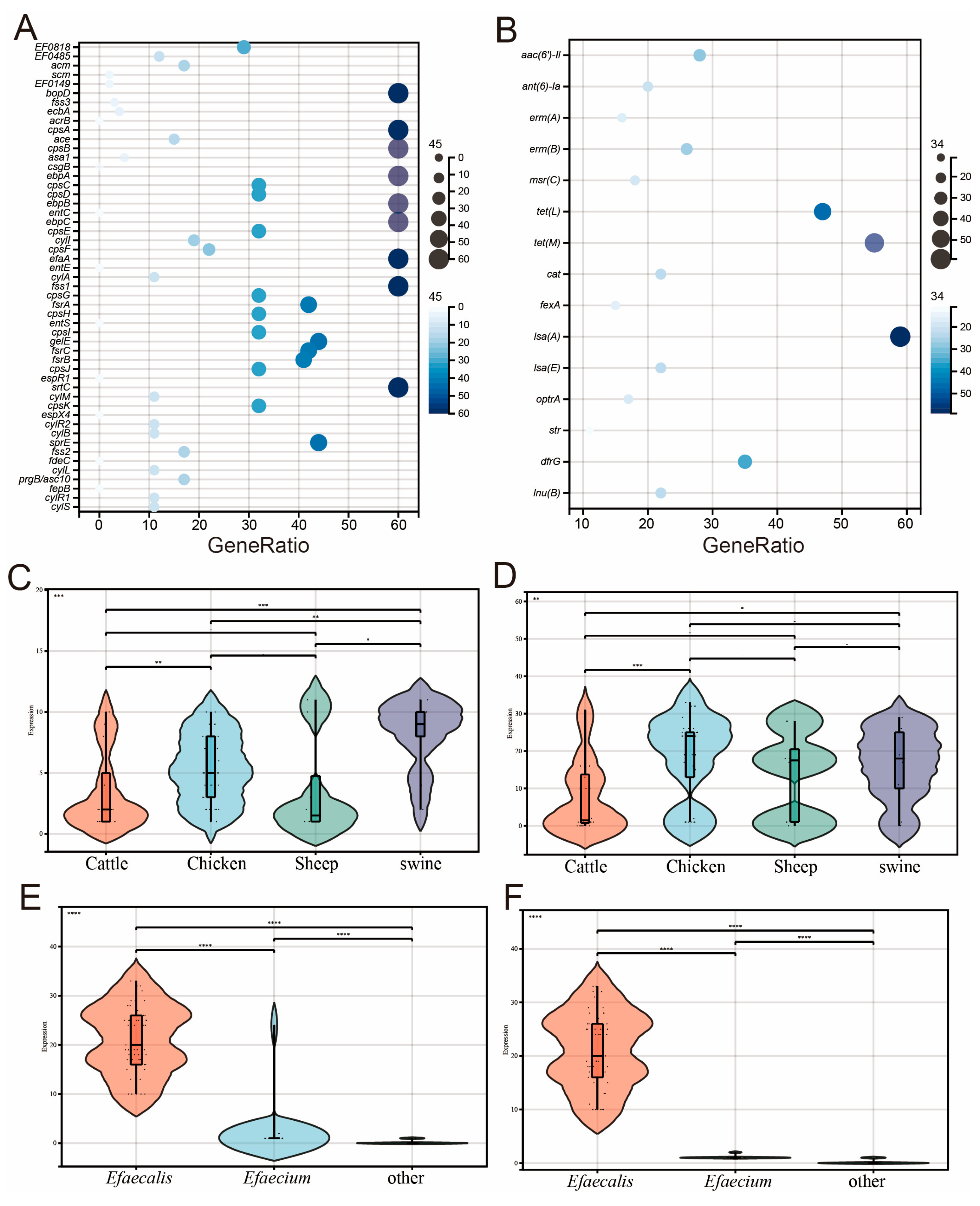
2.3. Results of Characteristic Genes
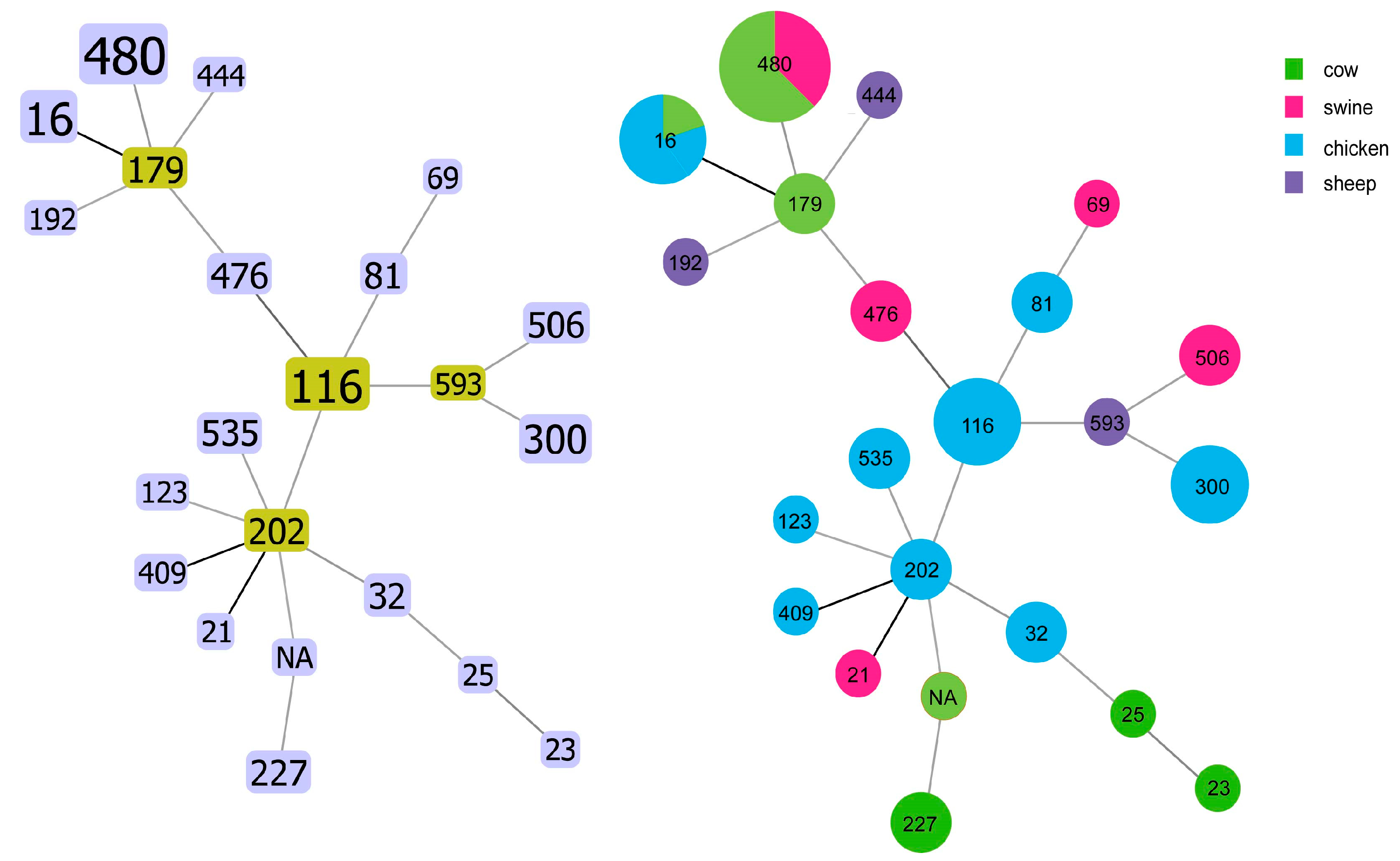
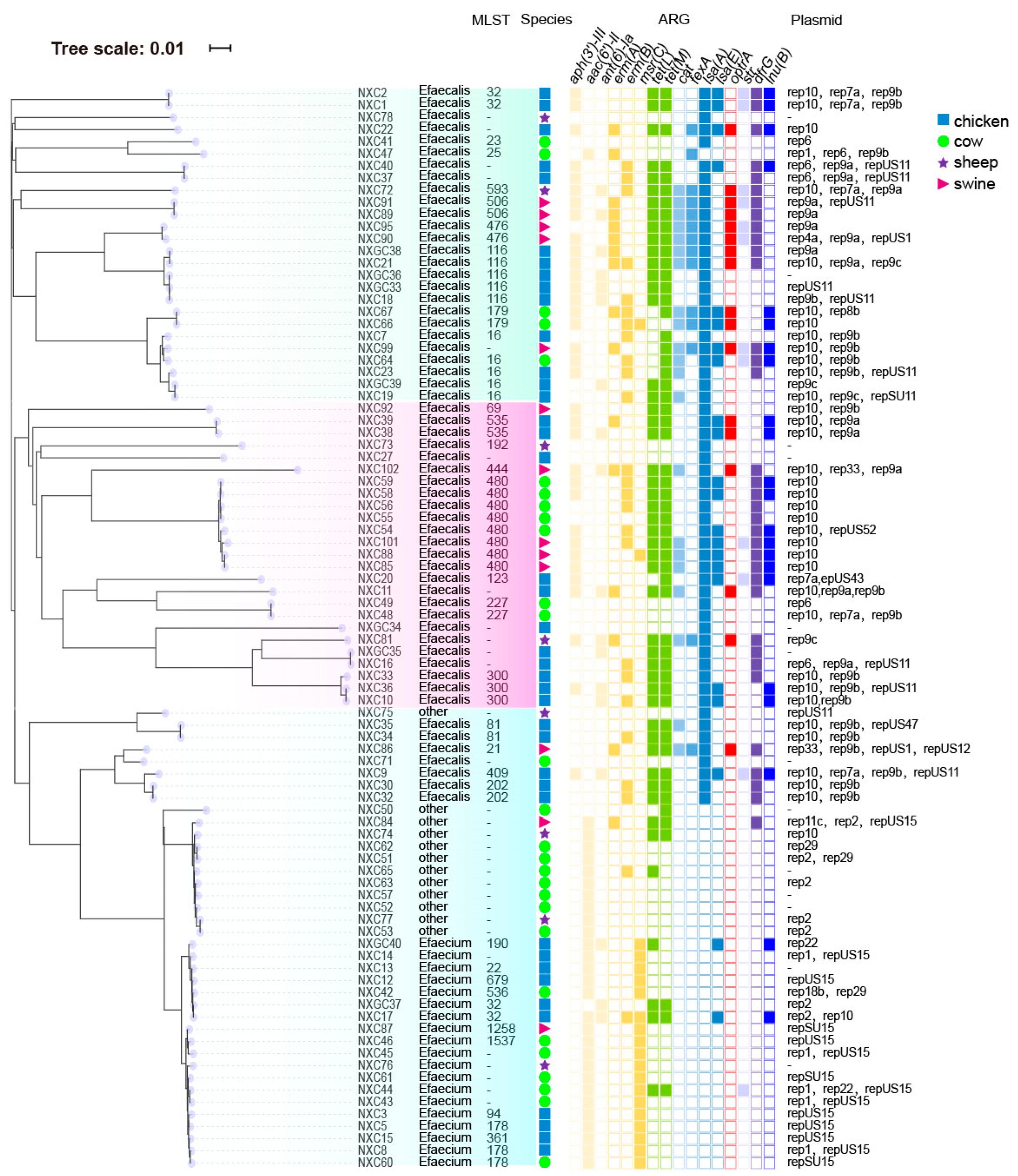
2.4. Results of MLST Sequence Typing
2.5. Phylogenetic Tree of SNP and Characteristics of Antimicrobial Resistance Genes and Plasmids Carried by Enterococcus
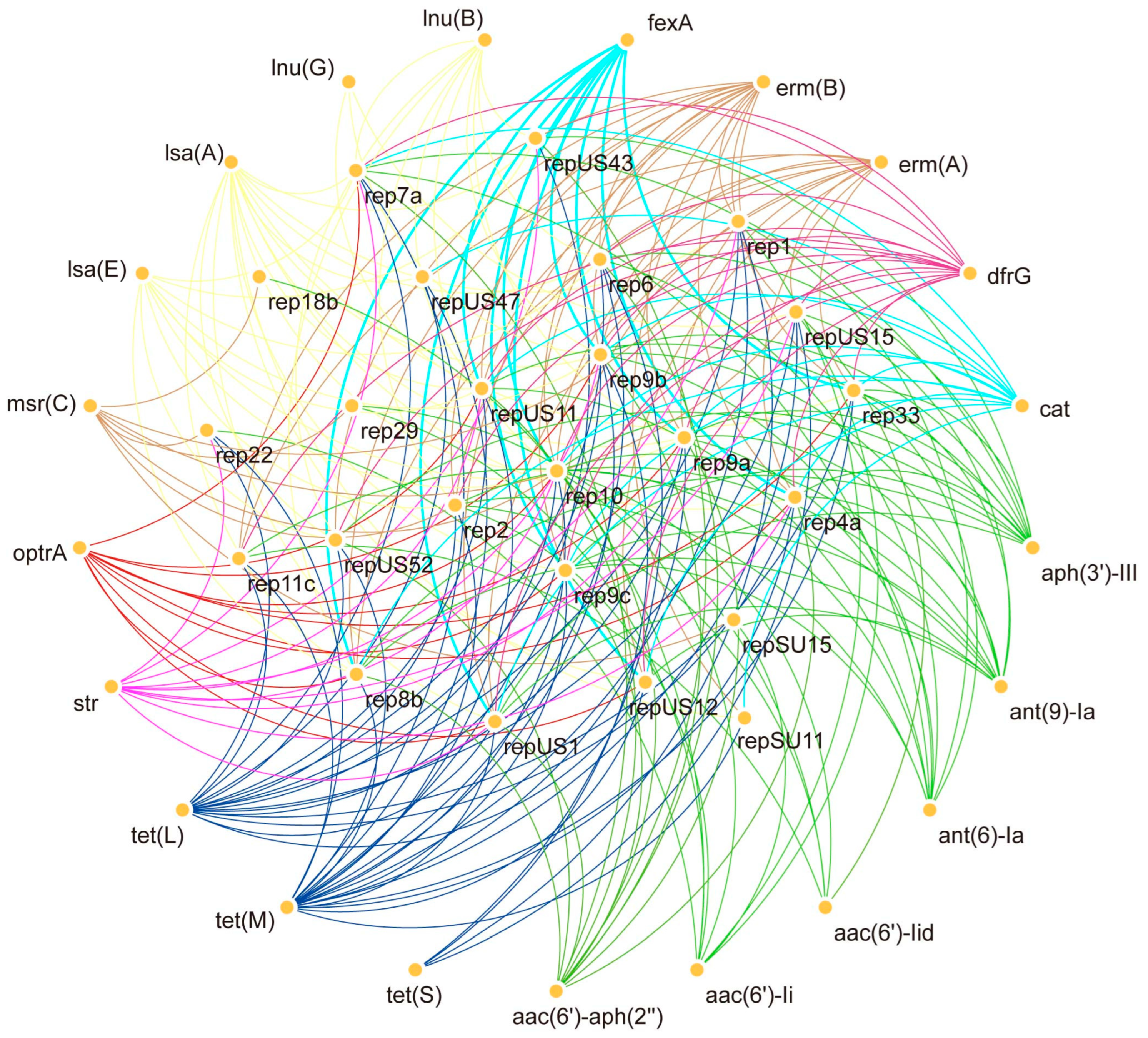
2.6. Co-Existence of Plasmids with Antibiotic-Resistant Genes
3. Discussion
4. Materials and Methods
4.1. Source of Samples
4.2. Antimicrobial Susceptibility Testing
4.3. Whole Genome Sequencing and Results Analysis
4.4. SNP Phylogenetic Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial Resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6, 185–227. [Google Scholar] [CrossRef]
- Farnleitner, A.H.; Ryzinska-Paier, G.; Reischer, G.H.; Burtscher, M.M.; Knetsch, S.; Kirschner, A.K.; Dirnböck, T.; Kuschnig, G.; Mach, R.L.; Sommer, R. Escherichia coli and enterococci are sensitive and reliable indicators for human, livestock and wildlife faecal pollution in alpine mountainous water resources. J. Appl. Microbiol. 2010, 109, 1599–1608. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control publishes Annual epidemiological report 2011. Euro Surveill. 2011, 16, 20012.
- Radhouani, H.; Igrejas, G.; Carvalho, C.; Pinto, L.; Gonçalves, A.; Lopez, M.; Sargo, R.; Cardoso, L.; Martinho, A.; Rego, V.; et al. Clonal lineages, antibiotic resistance and virulence factors in vancomycin-resistant enterococci isolated from fecal samples of red foxes (Vulpes vulpes). J. Wildl. Dis. 2011, 47, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Iweriebor, B.C.; Obi, L.C.; Okoh, A.I. Macrolide, glycopeptide resistance and virulence genes in Enterococcus species isolates from dairy cattle. J. Med. Microbiol. 2016, 65, 641–648. [Google Scholar] [CrossRef]
- Iweriebor, B.C.; Obi, L.C.; Okoh, A.I. Virulence and antimicrobial resistance factors of Enterococcus spp. isolated from fecal samples from piggery farms in Eastern Cape, South Africa. BMC Microbiol. 2015, 15, 136. [Google Scholar] [CrossRef]
- Todokoro, D.; Suzuki, T.; Kobayakawa, S.; Tomita, H.; Ohashi, Y.; Akiyama, H. Postoperative Enterococcus faecalis endophthalmitis: Virulence factors leading to poor visual outcome. Jpn. J. Ophthalmol. 2017, 61, 408–414. [Google Scholar] [CrossRef]
- Buetti, N.; Tabah, A.; Loiodice, A.; Ruckly, S.; Aslan, A.T.; Montrucchio, G.; Cortegiani, A.; Saltoglu, N.; Kayaaslan, B.; Aksoy, F.; et al. Different epidemiology of bloodstream infections in COVID-19 compared to non-COVID-19 critically ill patients: A descriptive analysis of the Eurobact II study. Crit. Care 2022, 26, 319. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; De Angelis, G.; Menchinelli, G.; D’Inzeo, T.; Fiori, B.; De Maio, F.; Cortazzo, V.; Sanguinetti, M.; Spanu, T. Risk Factors for Mortality in Adult COVID-19 Patients Who Develop Bloodstream Infections Mostly Caused by Antimicrobial-Resistant Organisms: Analysis at a Large Teaching Hospital in Italy. J. Clin. Med. 2021, 10, 1752. [Google Scholar] [CrossRef]
- Cultrera, R.; Barozzi, A.; Libanore, M.; Marangoni, E.; Pora, R.; Quarta, B.; Spadaro, S.; Ragazzi, R.; Marra, A.; Segala, D.; et al. Co-Infections in Critically Ill Patients with or without COVID-19: A Comparison of Clinical Microbial Culture Findings. Int. J. Environ. Res. Public Health 2021, 18, 4358. [Google Scholar] [CrossRef]
- Kojima, A.; Morioka, A.; Kijima, M.; Ishihara, K.; Asai, T.; Fujisawa, T.; Tamura, Y.; Takahashi, T. Classification and antimicrobial susceptibilities of Enterococcus species isolated from apparently healthy food-producing animals in Japan. Zoonoses Public Health 2010, 57, 137–141. [Google Scholar] [CrossRef]
- Xu, X.; Lin, D.; Yan, G.; Ye, X.; Wu, S.; Guo, Y.; Zhu, D.; Hu, F.; Zhang, Y.; Wang, F.; et al. vanM, a new glycopeptide resistance gene cluster found in Enterococcus faecium. Antimicrob. Agents Chemother. 2010, 54, 4643–4647. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Leclercq, R. Twenty-five years of shared life with vancomycin-resistant enterococci: Is it time to divorce? J. Antimicrob. Chemother. 2013, 68, 731–742. [Google Scholar] [CrossRef]
- Ahmed, M.O.; Baptiste, K.E. Vancomycin-Resistant Enterococci: A Review of Antimicrobial Resistance Mechanisms and Perspectives of Human and Animal Health. Microb. Drug Resist. 2018, 24, 590–606. [Google Scholar] [CrossRef]
- Hegstad, K.; Mikalsen, T.; Coque, T.M.; Werner, G.; Sundsfjord, A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin. Microbiol. Infect. 2010, 16, 541–554. [Google Scholar]
- Gordoncillo, M.J.; Donabedian, S.; Bartlett, P.C.; Perri, M.; Zervos, M.; Kirkwood, R.; Febvay, C. Isolation and molecular characterization of vancomycin-resistant Enterococcus faecium from swine in Michigan, USA. Zoonoses Public Health 2013, 60, 319–326. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, K.; Lai, J.; Wu, C.; Shen, J.; Wang, Y. Prevalence and antimicrobial resistance of Enterococcus species of food animal origin from Beijing and Shandong Province, China. J. Appl. Microbiol. 2013, 114, 555–563. [Google Scholar]
- Kiruthiga, A.; Padmavathy, K.; Shabana, P.; Naveenkumar, V.; Gnanadesikan, S.; Malaiyan, J. Improved detection of esp, hyl, asa1, gelE, cylA virulence genes among clinical isolates of Enterococci. BMC Res. Notes 2020, 13, 170. [Google Scholar] [CrossRef]
- Mikalsen, T.; Pedersen, T.; Willems, R.; Coque, T.M.; Werner, G.; Sadowy, E.; van Schaik, W.; Jensen, L.B.; Sundsfjord, A.; Hegstad, K. Investigating the mobilome in clinically important lineages of Enterococcus faecium and Enterococcus faecalis. BMC Genom. 2015, 16, 282. [Google Scholar]
- Zou, J.; Tang, Z.; Yan, J.; Liu, H.; Chen, Y.; Zhang, D.; Zhao, J.; Tang, Y.; Zhang, J.; Xia, Y. Dissemination of Linezolid Resistance Through Sex Pheromone Plasmid Transfer in Enterococcus faecalis. Front. Microbiol. 2020, 11, 1185. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Li, D.; Shan, X.; Schwarz, S.; Zhang, S.M.; Chen, Y.X.; Ouyang, W.; Du, X.D. Analysis of two pheromone-responsive conjugative multiresistance plasmids carrying the novel mobile optrA locus from Enterococcus faecalis. Infect. Drug Resist. 2019, 12, 2355–2362. [Google Scholar] [CrossRef]
- Dönhöfer, A.; Franckenberg, S.; Wickles, S.; Berninghausen, O.; Beckmann, R.; Wilson, D.N. Structural basis for TetM-mediated tetracycline resistance. Proc. Natl. Acad. Sci. USA 2012, 109, 16900–16905. [Google Scholar]
- Parent, R.; Roy, P.H. The chloramphenicol acetyltransferase gene of Tn2424: A new breed of cat. J. Bacteriol. 1992, 174, 2891–2897. [Google Scholar] [CrossRef]
- Morroni, G.; Brenciani, A.; Antonelli, A.; D’Andrea, M.M.; Di Pilato, V.; Fioriti, S.; Mingoia, M.; Vignaroli, C.; Cirioni, O.; Biavasco, F.; et al. Characterization of a Multiresistance Plasmid Carrying the optrA and cfr Resistance Genes From an Enterococcus faecium Clinical Isolate. Front. Microbiol. 2018, 9, 2189. [Google Scholar] [CrossRef] [PubMed]
- Cavaco, L.M.; Bernal, J.F.; Zankari, E.; Léon, M.; Hendriksen, R.S.; Perez-Gutierrez, E.; Aarestrup, F.M.; Donado-Godoy, P. Detection of linezolid resistance due to the optrA gene in Enterococcus faecalis from poultry meat from the American continent (Colombia). J. Antimicrob. Chemother. 2017, 72, 678–683. [Google Scholar]
- Mendes, R.E.; Deshpande, L.M.; Jones, R.N. Linezolid update: Stable in vitro activity following more than a decade of clinical use and summary of associated resistance mechanisms. Drug Resist. Updates 2014, 17, 1–12. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef]
- Nüesch-Inderbinen, M.; Raschle, S.; Stevens, M.J.A.; Schmitt, K.; Stephan, R. Linezolid-resistant Enterococcus faecalis ST16 harbouring optrA on a Tn6674-like element isolated from surface water. J. Glob. Antimicrob. Resist. 2021, 25, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Nawaz, M.S.; Khan, S.A.; Steele, R. Detection and characterization of erythromycin-resistant methylase genes in Gram-positive bacteria isolated from poultry litter. Appl. Microbiol. Biotechnol. 2002, 59, 377–381. [Google Scholar] [PubMed]
- Ben Said, L.; Dziri, R.; Sassi, N.; Lozano, C.; Ben Slama, K.; Ouzari, I.; Torres, C.; Klibi, N. Species distribution, antibiotic resistance and virulence traits in canine and feline enterococci in Tunisia. Acta Vet. Hung. 2017, 65, 173–184. [Google Scholar] [CrossRef]
- Almeida, L.M.; Lebreton, F.; Gaca, A.; Bispo, P.M.; Saavedra, J.T.; Calumby, R.N.; Grillo, L.M.; Nascimento, T.G.; Filsner, P.H.; Moreno, A.M.; et al. Transferable Resistance Gene optrA in Enterococcus faecalis from Swine in Brazil. Antimicrob. Agents Chemother. 2020, 64, 10–1128. [Google Scholar] [CrossRef]
- Barlow, R.S.; McMillan, K.E.; Duffy, L.L.; Fegan, N.; Jordan, D.; Mellor, G.E. Antimicrobial resistance status of Enterococcus from Australian cattle populations at slaughter. PLoS ONE 2017, 12, e0177728. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, C.A.; Tanomaru-Filho, M.; Faria-Junior, N.B.; Faria, G.; Guerreiro-Tanomaru, J.M. Antimicrobial activity of root canal irrigants associated with cetrimide against biofilm and planktonic Enterococcus faecalis. J. Contemp. Dent. Pract. 2014, 15, 603–607. [Google Scholar] [CrossRef]
- Osman, M.; Yassine, I.; Hassan, J.; Ericson, J.E.; Schiff, S.J.; Ghorbani Tajani, A.; Bieder, B.; Hamza, M.; Kassem, I.I. Critical genomic insights into vancomycin-resistant Enterococcus faecium in Lebanon. Microbiol. Spectr. 2025, 13, e0017125. [Google Scholar] [CrossRef]
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655.
- Kim, H.J.; Youn, H.Y.; Kang, H.J.; Moon, J.S.; Jang, Y.S.; Song, K.Y.; Seo, K.H. Prevalence and Virulence Characteristics of Enterococcus faecalis and Enterococcus faecium in Bovine Mastitis Milk Compared to Bovine Normal Raw Milk in South Korea. Animals 2022, 12, 1407. [Google Scholar] [CrossRef]
- Stępień-Pyśniak, D.; Bertelloni, F.; Dec, M.; Cagnoli, G.; Pietras-Ożga, D.; Urban-Chmiel, R.; Ebani, V.V. Characterization and Comparison of Enterococcus spp. Isolates from Feces of Healthy Dogs and Urine of Dogs with UTIs. Animals 2021, 11, 2845. [Google Scholar] [CrossRef] [PubMed]
- Molale, L.G.; Bezuidenhout, C.C. Virulence determinants and production of extracellular enzymes in Enterococcus spp. from surface water sources. Water Sci. Technol. 2016, 73, 1817–1824. [Google Scholar] [CrossRef]
- Ogura, A.; Iwasaki, M.; Ogihara, H.; Teramura, H. Evaluation of the Novel Dry Sheet Culture Method for the Enumeration of Enterobacteriaceae. Biocontrol. Sci. 2018, 23, 235–240. [Google Scholar] [CrossRef]
- Boulianne, M.; Arsenault, J.; Daignault, D.; Archambault, M.; Letellier, A.; Dutil, L. Drug use and antimicrobial resistance among Escherichia coli and Enterococcus spp. isolates from chicken and turkey flocks slaughtered in Quebec, Canada. Can. J. Vet. Res. 2016, 80, 49–59. [Google Scholar]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of Changes to the Clinical and Laboratory Standards In-stitute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 2021, 59, e0021321. [Google Scholar] [CrossRef] [PubMed]
- Stępień-Pyśniak, D.; Hauschild, T.; Dec, M.; Marek, A.; Urban-Chmiel, R.; Kosikowska, U. Phenotypic and genotypic characterization of Enterococcus spp. from yolk sac infections in broiler chicks with a focus on virulence factors. Poult. Sci. 2021, 100, 100985. [Google Scholar] [CrossRef] [PubMed]
- Margos, G.; Binder, K.; Dzaferovic, E.; Hizo-Teufel, C.; Sing, A.; Wildner, M.; Fingerle, V.; Jolley, K. PubMLST.org—The new home for the Borrelia MLSA database. Ticks Tick Borne Dis. 2015, 6, 869–871. [Google Scholar] [CrossRef]
- Zhong, Z.; Zhang, W.; Song, Y.; Liu, W.; Xu, H.; Xi, X.; Menghe, B.; Zhang, H.; Sun, Z. Comparative genomic analysis of the genus Enterococcus. Microbiol. Res. 2017, 196, 95–105. [Google Scholar] [CrossRef]
- Dong, N.; Lin, D.; Zhang, R.; Chan, E.W.; Chen, S. Carriage of blaKPC-2 by a virulence plasmid in hypervirulent Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 3317–3321. [Google Scholar] [CrossRef]
- Tyson, G.H.; Sabo, J.L.; Rice-Trujillo, C.; Hernandez, J.; McDermott, P.F. Whole-genome sequencing based characterization of antimicrobial resistance in Enterococcus. Pathog. Dis. 2018, 76, fty018. [Google Scholar] [CrossRef]
- Agius, J.E.; Phalen, D.N.; Rose, K.; Eden, J.S. Genomic Insights into the Pathogenicity of a Novel Biofilm-Forming Enterococcus sp. Bact. (Enterococcus lacertideformus) Identified Reptiles. Front. Microbiol. 2021, 12, 635208. [Google Scholar]
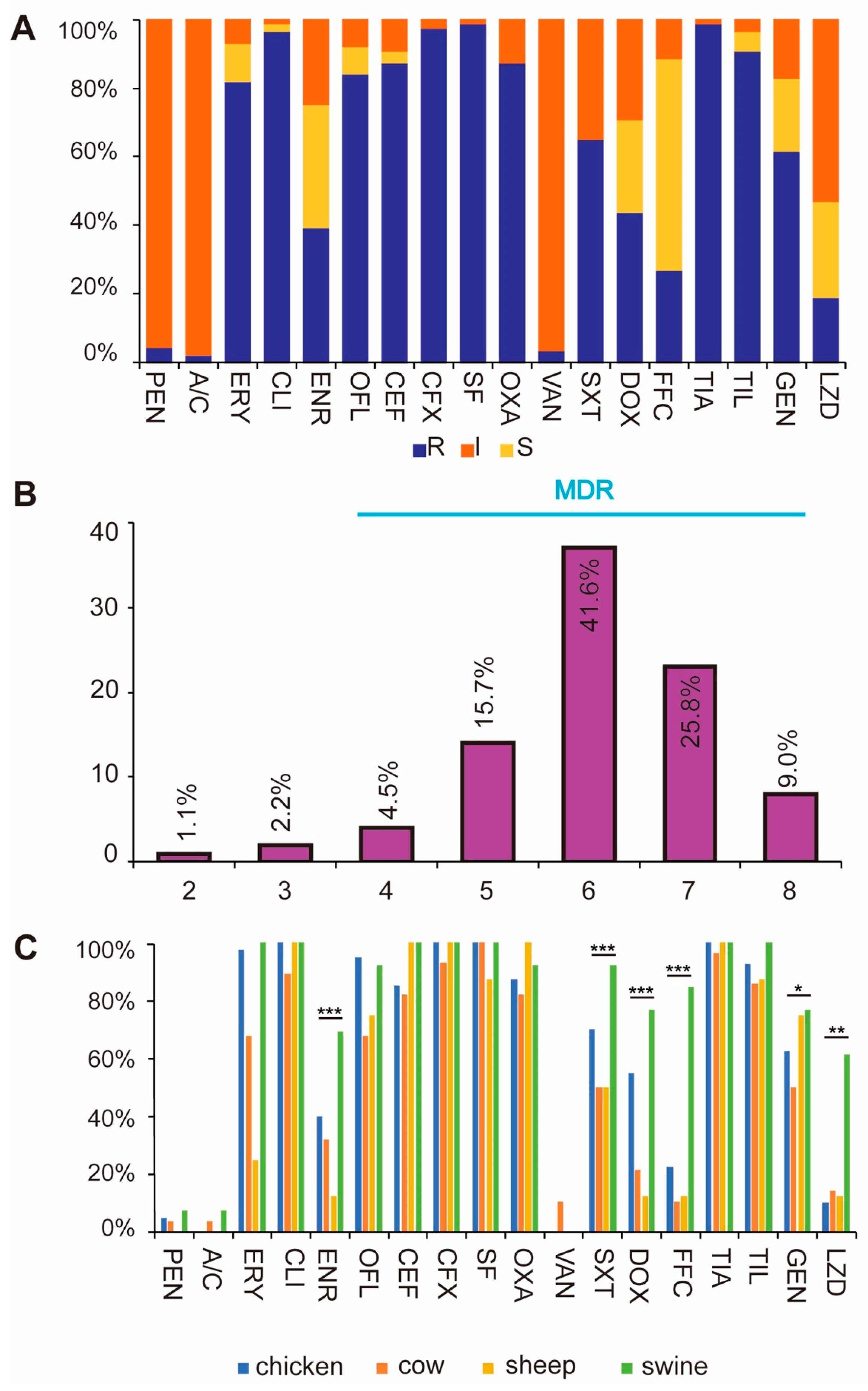
| Antibiotic Type Drug | Name | Overall Sensitivity | ||
|---|---|---|---|---|
| R | I | S | ||
| Penicillin | Benzylpenicillin | 87.6% | 0 | 12.4% |
| Penicillin | 4.5% | 0 | 95.5% | |
| Augmentin | 2.2% | 0 | 97.8% | |
| Clindamycin | Clindamycin | 96.6% | 2.2% | 1.1% |
| Quinolones | Enrofloxacin | 39.3% | 36.0% | 24.7% |
| Ofloxacin | 84.3% | 7.9% | 7.9% | |
| beta-lactams | Ceftiofur | 87.6% | 3.4% | 9.0% |
| Cefoxitin | 97.8% | 0 | 2.2% | |
| Sulfonamides | Sulfamethoxazole | 98.9% | 0 | 1.1% |
| Glycopeptides | Vancomycin | 3.4% | 0 | 96.6% |
| Sulfonamides | Cotrimoxazole | 65.2% | 0 | 36.0% |
| Tetracycline | Doxycycline | 43.8% | 27.0% | 29.2% |
| Florfenicol | 27.0% | 61.8% | 11.2% | |
| Macrolides | Erythromycin | 82.0% | 11.2% | 6.7% |
| Taimiocin | 98.9% | 0 | 1.1% | |
| Temicosin | 91.0% | 5.6% | 3.4% | |
| Aminoglycosides | Gentamycin | 61.8% | 21.3% | 16.9% |
| Oxazolidinones | Linezolid | 19.1% | 28.1% | 52.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Zhang, W.; Tang, T.; Zhang, L.; Cui, S.; Chen, S.; Hao, H.; Deng, Y.; Zhang, W.; Yang, Q.; et al. Genomic Characterization of Multiple Antibiotic-Resistant Enterococcus in Farm Animals in Ningxia Province, China. Antibiotics 2025, 14, 1137. https://doi.org/10.3390/antibiotics14111137
Zhao H, Zhang W, Tang T, Zhang L, Cui S, Chen S, Hao H, Deng Y, Zhang W, Yang Q, et al. Genomic Characterization of Multiple Antibiotic-Resistant Enterococcus in Farm Animals in Ningxia Province, China. Antibiotics. 2025; 14(11):1137. https://doi.org/10.3390/antibiotics14111137
Chicago/Turabian StyleZhao, Haoyu, Wen Zhang, Tianran Tang, Likun Zhang, Shengling Cui, Shengli Chen, Huafang Hao, Yating Deng, Weimin Zhang, Qi Yang, and et al. 2025. "Genomic Characterization of Multiple Antibiotic-Resistant Enterococcus in Farm Animals in Ningxia Province, China" Antibiotics 14, no. 11: 1137. https://doi.org/10.3390/antibiotics14111137
APA StyleZhao, H., Zhang, W., Tang, T., Zhang, L., Cui, S., Chen, S., Hao, H., Deng, Y., Zhang, W., Yang, Q., Yang, Z., Shao, Q., & Wang, J. (2025). Genomic Characterization of Multiple Antibiotic-Resistant Enterococcus in Farm Animals in Ningxia Province, China. Antibiotics, 14(11), 1137. https://doi.org/10.3390/antibiotics14111137





