Genomic Characterization and Antimicrobial Resistance Profile of Streptococcus uberis Strains Isolated from Cows with Mastitis from Northwestern Spain
Abstract
1. Introduction
2. Results
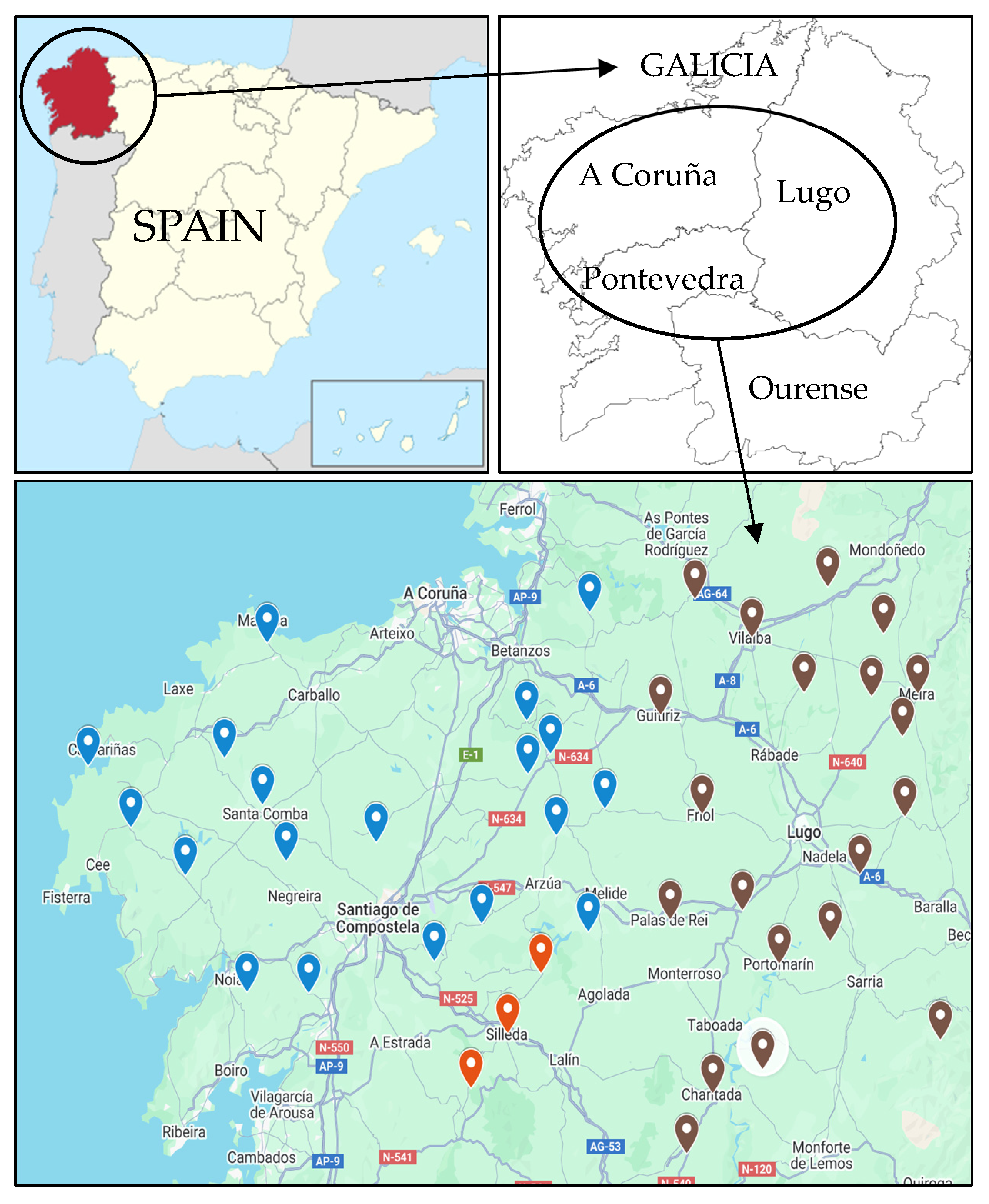
2.1. Descriptive Data
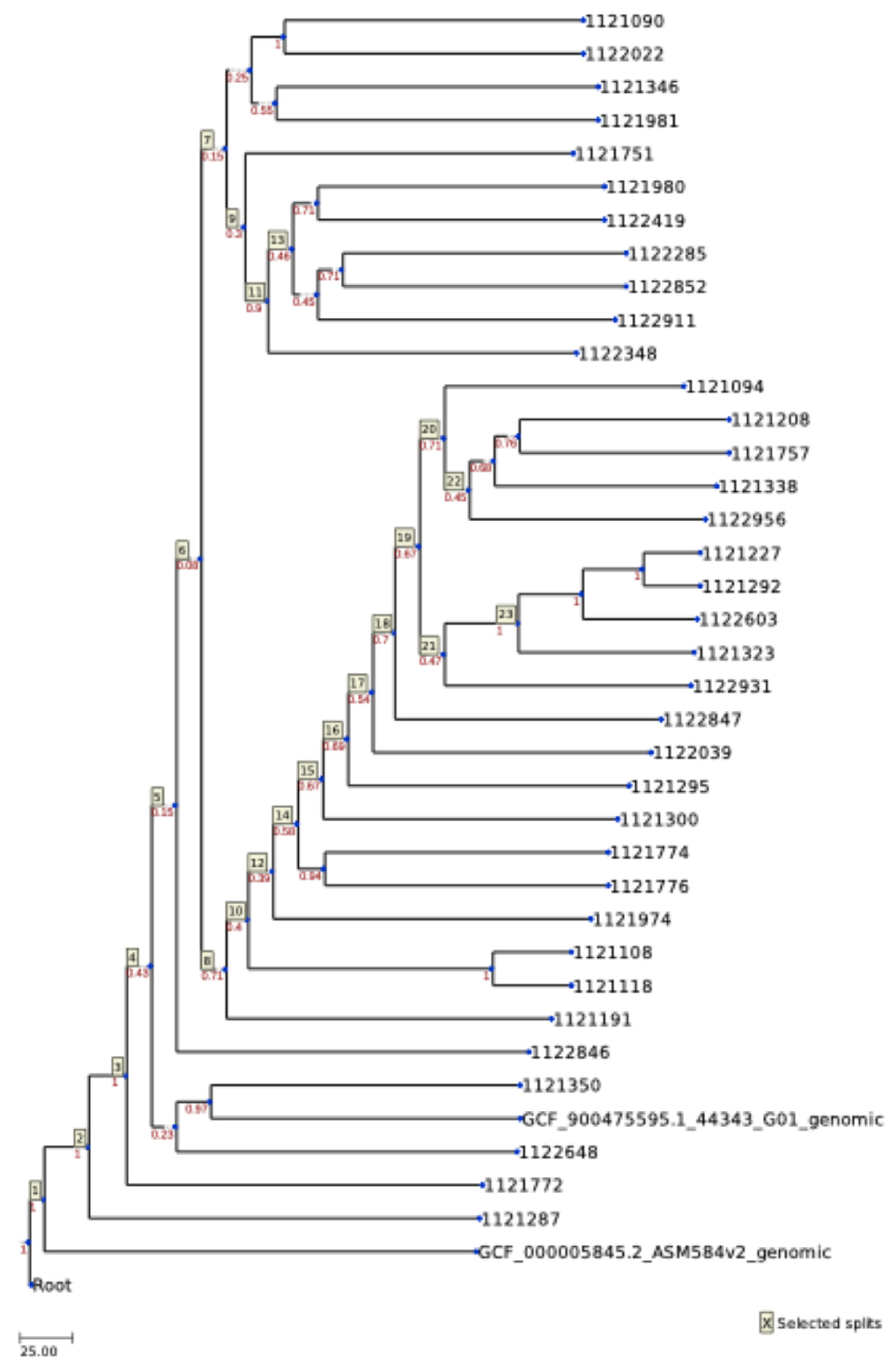
2.2. MLST
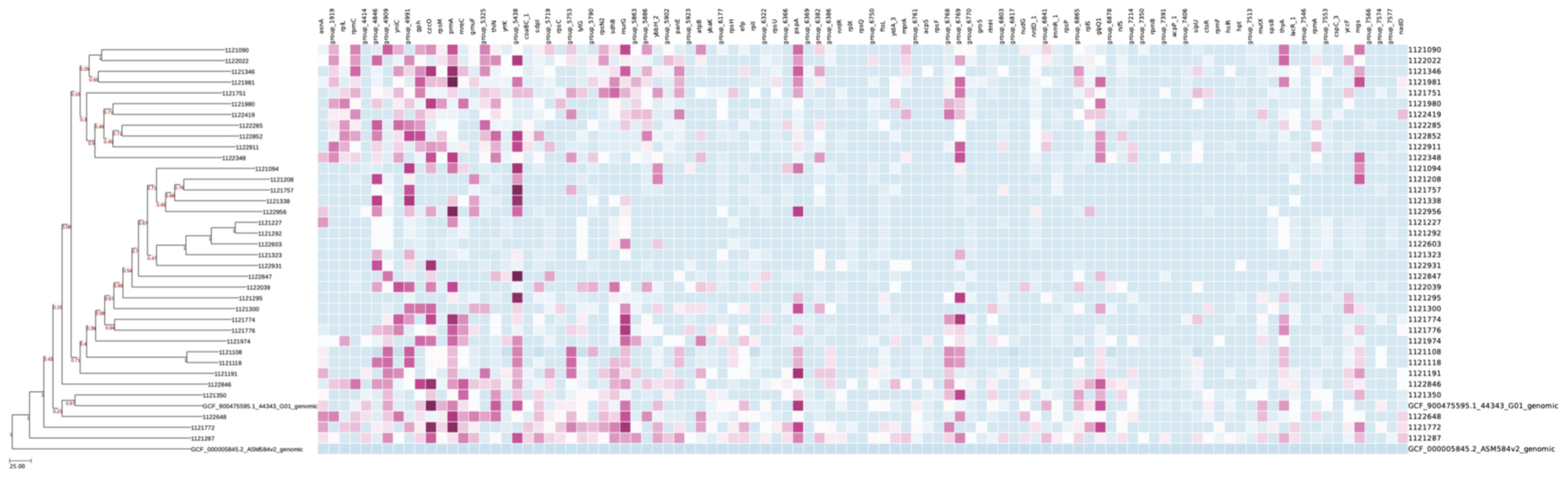
2.3. Antimicrobial Profile
2.4. Plasmids and Virulence
3. Discussion
4. Materials and Methods
4.1. Isolates and Strain Identification
4.2. Susceptibility to Antibiotics
4.3. DNA Isolation
4.4. Whole-Genome Sequencing
4.5. Bioinformatic Analysis
4.5.1. Assembly
4.5.2. Polishing and Annotation
4.5.3. Typing, Antimicrobial Resistance and Virulence
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gibbs, E.P.J. The Evolution of One Health: A Decade of Progress and Challenges for the Future. Vet. Rec. 2014, 174, 85–91. [Google Scholar] [CrossRef]
- FAO; OIE; WHO; UNICEF; UNSIC; World Bank. Contributing to One World, One Health—A Strategic Framework for Reducing Risks of Infectious Diseases at the Animal–Human–Ecosystems Interface 2008, pp. 1–68. Available online: https://www.fao.org/4/aj137e/aj137e00.htm (accessed on 1 March 2024).
- FAO. Dairy Market Review: Emerging Trends and Outlook 2022; FAO: Rome, Italy, 2022. [Google Scholar]
- Fundación Juana de Vega. Informe Do Sector Lácteo 2025. Available online: https://www.campogalego.gal/wp-content/uploads/2025/04/InformedoSectorLacteo2024_FundacionJuanadeVega.pdf (accessed on 1 March 2024).
- OECD/FAO Dairy and Dairy Products. In OECD-FAO Agricultural Outlook 2022–2031; OECE, FAO, Eds.; OECD: Paris, France; FAO: Rome, Italy, 2022; pp. 212–223. ISBN 978-92-5-136313-3. [Google Scholar]
- Ward, P.N.; Holden, M.T.G.; Leigh, J.A.; Lennard, N.; Bignell, A.; Barron, A.; Clark, L.; Quail, M.A.; Woodward, J.; Barrell, B.G.; et al. Evidence for Niche Adaptation in the Genome of the Bovine Pathogen Streptococcus uberis. BMC Genom. 2009, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, E.G.; Toma, L.; Prignano, G.; Pelagalli, L.; Police, A.; Cavallotti, C.; Torelli, R.; Sanguinetti, M.; Ensoli, F. Misidentification of Streptococcus uberis as a Human Pathogen: A Case Report and Literature Review. Int. J. Infect. Dis. 2015, 33, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Kabelitz, T.; Aubry, E.; van Vorst, K.; Amon, T.; Fulde, M. The Role of Streptococcus spp. in Bovine Mastitis. Microorganisms 2021, 9, 1497. [Google Scholar] [CrossRef]
- Bisharat, N.; Crook, D.W.; Leigh, J.; Harding, R.M.; Ward, P.N.; Coffey, T.J.; Maiden, M.C.; Peto, T.; Jones, N. Hyperinvasive Neonatal Group B Streptococcus Has Arisen from a Bovine Ancestor. J. Clin. Microbiol. 2004, 42, 2161–2167. [Google Scholar] [CrossRef]
- Gottschalk, M.; Segura, M.; Xu, J. Streptococcus Suis Infections in Humans: The Chinese Experience and the Situation in North America. Anim. Health Res. Rev. 2007, 8, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Valentiny, C.; Dirschmid, H.; Lhotta, K. Streptococcus uberis and Staphylococcus aureus Forefoot and Blood Stream Co-Infection in a Hemodialysis Patient: A Case Report. BMC Nephrol. 2015, 16, 73. [Google Scholar] [CrossRef]
- Murdoch, D.R.; Seaward, L.M.; MacFarlane, M.R. Human Wound Infection with Streptococcus uberis. Clin. Microbiol. Newsl. 1997, 19, 174–175. [Google Scholar] [CrossRef]
- Tamilselvam, B.; Almeida, R.A.; Dunlap, J.R.; Oliver, S.P. Streptococcus Uberis Internalizes and Persists in Bovine Mammary Epithelial Cells. Microb. Pathog. 2006, 40, 279–285. [Google Scholar] [CrossRef]
- De Jong, A.; El Garch, F.; Simjee, S.; Moyaert, H.; Rose, M.; Youala, M.; Siegwart, E. Monitoring of Antimicrobial Susceptibility of Udder Pathogens Recovered from Cases of Clinical Mastitis in Dairy Cows across Europe: VetPath Results. Vet. Microbiol. 2018, 213, 73–81. [Google Scholar] [CrossRef]
- Käppeli, N.; Morach, M.; Zurfluh, K.; Corti, S.; Nüesch-Inderbinen, M.; Stephan, R. Sequence Types and Antimicrobial Resistance Profiles of Streptococcus uberis Isolated from Bovine Mastitis. Front. Vet. Sci. 2019, 6, 234. [Google Scholar] [CrossRef]
- Reyes, J.; Rodriguez-Lecompte, J.C.; Blanchard, A.; McClure, J.T.; Sánchez, J. Molecular Variability of Streptococcus uberis Isolates from Intramammary Infections in Canadian Dairy Farms from the Maritime Region. Can. J. Vet. Res. 2019, 83, 168–176. [Google Scholar]
- Saini, V.; McClure, J.T.; Léger, D.; Dufour, S.; Sheldon, A.G.; Scholl, D.T.; Barkema, H.W. Antimicrobial Use on Canadian Dairy Farms. J. Dairy Sci. 2012, 95, 1209–1221. [Google Scholar] [CrossRef]
- Tomazi, T.; Freu, G.; Alves, B.G.; de Souza Filho, A.F.; Heinemann, M.B.; dos Santos, M.V. Genotyping and Antimicrobial Resistance of Streptococcus uberis Isolated from Bovine Clinical Mastitis. PLoS ONE 2019, 14, e0223719. [Google Scholar] [CrossRef]
- Pol, M.; Ruegg, P.L. Treatment Practices and Quantification of Antimicrobial Drug Usage in Conventional and Organic Dairy Farms in Wisconsin. J. Dairy Sci. 2007, 90, 249–261. [Google Scholar] [CrossRef]
- Fessia, A.S.; Odierno, L.M. Potential Factors Involved in the Early Pathogenesis of Streptococcus uberis Mastitis: A Review. Folia Microbiol. 2021, 66, 509–523. [Google Scholar] [CrossRef]
- Zadoks, R.N.; Tikofsky, L.L.; Boor, K.J. Ribotyping of Streptococcus Uberis from a Dairy’s Environment, Bovine Feces and Milk. Vet. Microbiol. 2005, 109, 257–265. [Google Scholar] [CrossRef]
- Lopez-Benavides, M.G.; Williamson, J.H.; Pullinger, G.D.; Lacy-Hulbert, S.J.; Cursons, R.T.; Leigh, J.A. Field Observations on the Variation of Streptococcus uberis Populations in a Pasture-Based Dairy Farm. J. Dairy Sci. 2007, 90, 5558–5566. [Google Scholar] [CrossRef] [PubMed]
- Ericsson Unnerstad, H.; Lindberg, A.; Persson Waller, K.; Ekman, T.; Artursson, K.; Nilsson-Öst, M.; Bengtsson, B. Microbial Etiology of Acute Clinical Mastitis and Agent-Specific Risk Factors. Vet. Microbiol. 2009, 137, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Monistero, V.; Barberio, A.; Cremonesi, P.; Castiglioni, B.; Morandi, S.; Lassen, D.C.K.; Astrup, L.B.; Locatelli, C.; Piccinini, R.; Filippa Addis, M.; et al. Genotyping and Antimicrobial Susceptibility Profiling of Streptococcus uberis Isolated from a Clinical Bovine Mastitis Outbreak in a Dairy Farm. Antibiotics 2021, 10, 644. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, R.N.; Gillespie, B.E.; Barkema, H.W.; Sampimon, O.C.; Oliver, S.P.; Schukken, Y.H. Clinical, Epidemiological and Molecular Characteristics of Streptococcus uberis Infections in Dairy Herds. Epidemiol. Infect. 2003, 130, 335–349. [Google Scholar] [CrossRef]
- Davies, P.L.; Leigh, J.A.; Bradley, A.J.; Archer, S.C.; Emes, R.D.; Green, M.J. Molecular Epidemiology of Streptococcus uberis Clinical Mastitis in Dairy Herds: Strain Heterogeneity and Transmission. J. Clin. Microbiol. 2016, 54, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Leelahapongsathon, K.; Schukken, Y.H.; Pinyopummintr, T.; Suriyasathaporn, W. Comparison of Transmission Dynamics between Streptococcus uberis and Streptococcus agalactiae Intramammary Infections. J. Dairy Sci. 2016, 99, 1418–1426. [Google Scholar] [CrossRef] [PubMed]
- Wente, N.; Klocke, D.; Paduch, J.H.; Zhang, Y.; Seeth, M.T.; Zoche-Golob, V.; Reinecke, F.; Mohr, E.; Krömker, V. Associations between Streptococcus uberis Strains from the Animal Environment and Clinical Bovine Mastitis Cases. J. Dairy Sci. 2019, 102, 9360–9369. [Google Scholar] [CrossRef]
- Wald, R.; Baumgartner, M.; Gutschireiter, J.; Bazzanella, B.; Lichtmannsperger, K.; Wagner, M.; Wittek, T.; Stessl, B. Comparison of the Population Structure of Streptococcus uberis Mastitis Isolates from Austrian Small-Scale Dairy Farms and a Slovakian Large-Scale Farm. J. Dairy Sci. 2020, 103, 1820–1830. [Google Scholar] [CrossRef]
- Díaz-Cao, J.M.; Barreal, M.L.; Pombo, B.; Prieto, A.; Alonso, J.M.; Iglesias, A.; Lorenzana, R.; López-Novo, C.; Díez-Baños, P.; Fernández, G. Evaluation and Cluster Analysis of Inflammatory Reactions of Dairy Cattle Mastitis Pathogens in Milk Samples Submitted for Microbiological Examination. Span. J. Agric. Res. 2019, 17, e0505. [Google Scholar] [CrossRef]
- Vélez, J.R.; Cameron, M.; Rodríguez-Lecompte, J.C.; Xia, F.; Heider, L.C.; Saab, M.; Trenton McClure, J.; Sánchez, J. Whole-Genome Sequence Analysis of Antimicrobial Resistance Genes in Streptococcus uberis and Streptococcus dysgalactiae Isolates from Canadian Dairy Herds. Front. Vet. Sci. 2017, 4, 63. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.C.C.; Yang, Y.; Rodrigues, M.X.; Tomazi, T.; Bicalho, R.C. Whole-Genome Sequencing Reveals High Genetic Diversity of Streptococcus uberis Isolated from Cows with Mastitis. BMC Vet. Res. 2021, 17, 321. [Google Scholar] [CrossRef]
- Galician Institute of Statistics Registro de Ganado Bovino. 2025. Available online: https://www.ige.gal/igebdt/igeapi/datos/2917/0:2022,1:4,2:1,3:11 (accessed on 1 June 2025).
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely Randomized Trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Huang, J.; Li, T.; Zhu, Y.; Li, Q.; Kuo, C.; Guo, X.; Wei, B.; Ni, P.; Dong, K. Molecular Characterization and Potential Host-switching of Swine Farm associated Clostridioides difficile ST11. Vet. Microbiol. 2024, 294, 110129. [Google Scholar] [CrossRef]
- Fenske, L.; Noll, I.; Blom, J.; Ewers, C.; Semmler, T.; Fawzy, A.; Eisenberg, T. A Dominant Clonal Lineage of Streptococcus uberis in Cattle in Germany. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2022, 115, 857–870. [Google Scholar] [CrossRef]
- Hossain, M.; Egan, S.A.; Coffey, T.; Ward, P.N.; Wilson, R.; Leigh, J.A.; Emes, R.D. Virulence Related Sequences; Insights Provided by Comparative Genomics of Streptococcus uberis of Differing Virulence. BMC Genom. 2015, 16, 334. [Google Scholar] [CrossRef]
- Vezina, B.; Al-harbi, H.; Ramay, H.R.; Soust, M.; Moore, R.J.; Olchowy, T.W.J.; Alawneh, J.I. Sequence Characterization and Novel Insights into Bovine Mastitis-Associated Streptococcus uberis in Dairy Herds. Sci. Rep. 2021, 11, 3046. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Cabezas, P.; Castro-Nallar, E.; Crandall, K.A. Pathogen Typing in the Genomics Era: MLST and the Future of Molecular Epidemiology. Infect. Genet. Evol. 2013, 16, 38–53. [Google Scholar] [CrossRef] [PubMed]
- Maiden, M.C.J.; Bygraves, J.A.; Feil, E.; Morelli, G.; Russell, J.E.; Urwin, R.; Zhang, Q.; Zhou, J.; Zurth, K.; Caugant, D.A.; et al. Multilocus Sequence Typing: A Portable Approach to the Identification of Clones within Populations of Pathogenic Microorganisms. Proc. Natl. Acad. Sci. USA 1998, 95, 3140–3145. [Google Scholar] [CrossRef] [PubMed]
- Didelot, X.; Maiden, M.C.J. Impact of Recombination on Bacterial Evolution. Trends Microbiol. 2010, 18, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Joshi, C.J.; Ke, W.; Drangowska-Way, A.; O’Rourke, E.J.; Lewis, N.E. What Are Housekeeping Genes? PLoS Comput. Biol. 2022, 18, e1010295. [Google Scholar] [CrossRef]
- Tomita, T.; Meehan, B.; Wongkattiya, N.; Malmo, J.; Pullinger, G.; Leigh, J.; Deighton, M. Identification of Streptococcus uberis Multilocus Sequence Types Highly Associated with Mastitis. Appl. Environ. Microbiol. 2008, 74, 114–124. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome Analysis of Multiple Pathogenic Isolates of Streptococcus agalactiae: Implications for the Microbial “Pan-Genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef]
- Maione, D.; Margarit, I.; Rinaudo, C.D.; Masignani, V.; Mora, M.; Scarselli, M.; Tettelin, H.; Brettoni, C.; Iacobini, E.T.; Rosini, R.; et al. Identification of a Universal Group B Streptococcus Vaccine by Multiple Genome Screen. Science 2005, 309, 148–150. [Google Scholar] [CrossRef]
- Lauer, P.; Rinaudo, C.D.; Soriani, M.; Margarit, I.; Maione, D.; Rosini, R.; Taddei, A.R.; Mora, M.; Rappuoli, R.; Grandi, G.; et al. Genome Analysis Reveals Pili in Group B Streptococcus. Science 2005, 309, 105. [Google Scholar] [CrossRef]
- Morneau, D. Pan-Genomes: Moving beyond the Reference. Nature Milestones, 10 February 2021; p. S19. [Google Scholar]
- Lang, P.; Lefébure, T.; Wang, W.; Zadoks, R.N.; Schukken, Y.; Stanhope, M.J. Gene Content Differences across Strains of Streptococcus uberis Identified Using Oligonucleotide Microarray Comparative Genomic Hybridization. Infect. Genet. Evol. 2009, 9, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Liu, X.; Simeneh, Z.M.; Yang, M.; Li, R. Benchmarking of Nanopore R10.4 and R9.4.1 Flow Cells in Single-Cell Whole-Genome Amplification and Whole-Genome Shotgun Sequencing. Comput. Struct. Biotechnol. J. 2023, 21, 2352–2364. [Google Scholar] [CrossRef] [PubMed]
- van Tonder, A.J.; Mistry, S.; Bray, J.E.; Hill, D.M.C.; Cody, A.J.; Farmer, C.L.; Klugman, K.P.; von Gottberg, A.; Bentley, S.D.; Parkhill, J.; et al. Defining the Estimated Core Genome of Bacterial Populations Using a Bayesian Decision Model. PLoS Comput. Biol. 2014, 10, e1003788. [Google Scholar] [CrossRef]
- Preska Steinberg, A.; Lin, M.; Kussell, E. Core Genes Can Have Higher Recombination Rates than Accessory Genes within Global Microbial Populations. Elife 2022, 11, e78533. [Google Scholar] [CrossRef]
- Panchal, J.; Patel, A.; Patel, S.; Goswami, D. Understanding Mastitis: Microbiome, Control Strategies, and Prevalence—A Comprehensive Review. Microb. Pathog. 2024, 187, 106533. [Google Scholar] [CrossRef]
- Kluytmans-van den Bergh, M.F.Q.; Rossen, J.W.A.; Bruijnin-Verhagen, P.C.J.; Bonten, M.J.M.; Friedrich, A.W.; Vandenbroucke-Grauls, C.M.J.E.; Willems, R.J.L.; Kluytmans, J.A.J.W. Whole-Genome Multilocus Sequence Typing of Extended-Spectrum-Beta-Lactamase-Producing Enterobacteriaceae. J. Clin. Microbiol. 2016, 12, 2919–2927. [Google Scholar] [CrossRef] [PubMed]
- Vulić, M.; Lenski, R.E.; Radman, M. Mutation, Recombination, and Incipient Speciation of Bacteria in the Laboratory. Proc. Natl. Acad. Sci. USA 1999, 96, 7348–7351. [Google Scholar] [CrossRef]
- Horton, J.S.; Taylor, T.B. Mutation Bias and Adaptation in Bacteria. Microbiology 2023, 169, 1404. [Google Scholar] [CrossRef]
- Ramiro, R.S.; Durão, P.; Bank, C.; Gordo, I. Low Mutational Load and High Mutation Rate Variation in Gut Commensal Bacteria. PLoS Biol. 2020, 18, e3000617. [Google Scholar] [CrossRef]
- Derakhshani, H.; Plaizier, J.C.; De Buck, J.; Barkema, H.W.; Khafipour, E. Composition and Co-Occurrence Patterns of the Microbiota of Different Niches of the Bovine Mammary Gland: Potential Associations with Mastitis Susceptibility, Udder Inflammation, and Teat-End Hyperkeratosis. Anim. Microbiome 2020, 2, 11. [Google Scholar] [CrossRef]
- Barlow, J. Mastitis Therapy and Antimicrobial Susceptibility: A Multispecies Review with a Focus on Antibiotic Treatment of Mastitis in Dairy Cattle. J. Mammary Gland. Biol. Neoplasia 2011, 16, 383–407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Tao, L.; Boonyayatra, S.; Niu, G. Antimicrobial Resistance of Streptococcus uberis Isolated from Bovine Mastitis: A Review. Indian J. Anim. Res. 2021, 56, 1435–1441. [Google Scholar] [CrossRef]
- Overesch, G.; Stephan, R.; Perreten, V. Antimicrobial Susceptibility of Gram-Positive Udder Pathogens from Bovine Mastitis Milk in Switzerland. Schweiz. Arch. Tierheilkd. 2013, 155, 339–350. [Google Scholar] [CrossRef]
- Molineri, A.I.; Camussone, C.; Zbrun, M.V.; Suárez Archilla, G.; Cristiani, M.; Neder, V.; Calvinho, L.; Signorini, M. Antimicrobial Resistance of Staphylococcus aureus Isolated from Bovine Mastitis: Systematic Review and Meta-Analysis. Prev. Vet. Med. 2021, 188, 105261. [Google Scholar] [CrossRef]
- Rosa, N.M.; Agnoletti, F.; Lollai, S.; Tola, S. Comparison of PCR-RFLP, API® 20 Strep and MALDI-TOF MS for Identification of Streptococcus Spp. Collected from Sheep and Goat Milk Samples. Small Rumin. Res. 2019, 180, 35–40. [Google Scholar] [CrossRef]
- Preine, F.; Herrera, D.; Scherpenzeel, C.; Kalmus, P.; McCoy, F.; Smulski, S.; Rajala-Schultz, P.; Schmenger, A.; Moroni, P.; Krömker, V. Different European Perspectives on the Treatment of Clinical Mastitis in Lactation. Antibiotics 2022, 11, 1107. [Google Scholar] [CrossRef] [PubMed]
- Boireau, C.; Cazeau, G.; Jarrige, N.; Calavas, D.; Madec, J.-Y.; Leblond, A.; Haenni, M.; Gay, É. Antimicrobial Resistance in Bacteria Isolated from Mastitis in Dairy Cattle in France, 2006–2016. J. Dairy Sci. 2018, 101, 9451–9462. [Google Scholar] [CrossRef] [PubMed]
- Rosa, N.M.; Duprè, I.; Azara, E.; Longheu, C.M.; Tola, S. Molecular Typing and Antimicrobial Susceptibility Profiles of Streptococcus uberis Isolated from Sheep Milk. Pathogens 2021, 10, 1489. [Google Scholar] [CrossRef]
- Gulen, D.; Kaya, A.D.; Aydin, M.; Tanriverdi, Y. Urinary Tract Infections Caused by Streptococcus uberis: A Pathogen of Bovine Mastitis-Report of Seven Cases. Afr. J. Microbiol. Res. 2013, 7, 3908–3912. [Google Scholar]
- Sánchez, J.; Moreno, J.J.; Roldán, A.; Ildefonso, J.A.; Florensa, J. Hepatic Abscess Caused by Streptococcus uberis. Enferm. Infecc. Microbiol. Clin. 1991, 9, 189–190. [Google Scholar]
- Sarkar, T.K.; Murarka, R.S.; Gilardi, G.L. Primary Streptococcus viridans Pneumonia. Chest 1989, 96, 831–834. [Google Scholar] [CrossRef]
- Bouskraoui, M.; Benbachir, M.; Abid, A. Endocardite a Streptococcus uberis Chez Un Nourrisson Atteint d’un Canal Atrioventriculaire. Arch. Pediatr. 1999, 6, 481. [Google Scholar] [CrossRef]
- Łazińska, B.; Ciszek, M.; Rokosz, A.; Sawicka-Grzelak, A.; Pa̧czek, L.; Łuczak, M. Bacteriological Urinalysis in Patients after Renal Transplantation. Pol. J. Microbiol. 2005, 54, 317–321. [Google Scholar]
- Huang, W.T.; Chang, L.Y.; Hsueh, P.R.; Lu, C.Y.; Shao, P.L.; Huang, F.Y.; Lee, P.I.; Chen, C.M.; Lee, C.Y.; Huang, L.M. Clinical Features and Complications of Viridans Streptococci Bloodstream Infection in Pediatric Hemato-Oncology Patients. J. Microbiol. Immunol. Infect. 2007, 40, 349–354. [Google Scholar]
- Kessel, S.; Wittenberg, C.E. Joint Infection in a Young Patient Caused by Streptococcus uberis, a Pathogen of Bovine Mastitis: A Case Report. Z. Orthop. Unfallchirurgie 2008, 146, 507–509. [Google Scholar] [CrossRef]
- Velez-Montoya, R.; Rascón-Vargas, D.; Mieler, W.F.; Fromow-Guerra, J.; Morales-Cantón, V. Intravitreal Ampicillin Sodium for Antibiotic-Resistant Endophthalmitis: Streptococcus uberis First Human Intraocular Infection Report. J. Ophthalmol. 2010, 2010, 169739. [Google Scholar] [CrossRef]
- Zadoks, R.N.; Middleton, J.R.; McDougall, S.; Katholm, J.; Schukken, Y.H. Molecular Epidemiology of Mastitis Pathogens of Dairy Cattle and Comparative Relevance to Humans. J. Mammary Gland Biol. Neoplasia 2011, 16, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Abueg, L.A.L.; Afgan, E.; Allart, O.; Awan, A.H.; Bacon, W.A.; Baker, D.; Bassetti, M.; Batut, B.; Bernt, M.; Blankenberg, D.; et al. The Galaxy Platform for Accessible, Reproducible, and Collaborative Data Analyses: 2024 Update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef] [PubMed]
- Blankenberg, D.; Gordon, A.; Von Kuster, G.; Coraor, N.; Taylor, J.; Nekrutenko, A.; Team, G. Manipulation of FASTQ Data with Galaxy. Bioinform. Appl. Note 2010, 26, 1783–1785. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- De Coster, W.; Rademakers, R. NanoPack2: Population-Scale Evaluation of Long-Read Sequencing Data. Bioinformatics 2023, 39, btad311. [Google Scholar] [CrossRef]
- Vaser, R.; Šikić, M. Time- and Memory-Efficient Genome Assembly with Raven. Nat. Comput. Sci. 2021, 1, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Waterman, M.S. Genomic Mapping by Fingerprinting Random Clones: A Mathematical Analysis. Genomics 1988, 2, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Arratia, R.; Lander, E.S.; Tavaré, S.; Waterman, M.S. Genomic Mapping by Anchoring Random Clones: A Mathematical Analysis. Genomics 1991, 11, 806–827. [Google Scholar] [CrossRef]
- Port, E.; Sun, F.; Martin, D.; Waterman, M.S. Genomic Mapping by End-Characterized Random Clones: A Mathematical Analysis. Genomics 1995, 26, 84–100. [Google Scholar] [CrossRef]
- Oxford Nanopore Technologies Ltd. Medaka. GitHub. 2018. Available online: https://github.com/nanoporetech/medaka (accessed on 1 March 2024).
- Cuccuru, G.; Orsini, M.; Pinna, A.; Sbardellati, A.; Soranzo, N.; Travaglione, A.; Uva, P.; Zanetti, G.; Fotia, G. Orione, a Web-Based Framework for NGS Analysis in Microbiology. Bioinformatics 2014, 30, 1928–1929. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate. GitHub. 2016. Available online: https://github.com/tseemann/abricate (accessed on 1 March 2024).
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Lin, J.W.; Chen, C.C. Cano-WgMLST_BacCompare: A Bacterial Genome Analysis Platform for Epidemiological Investigation and Comparative Genomic Analysis. Front. Microbiol. 2019, 10, 1687. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A General Classification Scheme for Bacterial Virulence Factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef] [PubMed]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder—Distinguishing Friend from Foe Using Bacterial Whole Genome Sequence Data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef]
| Strains Code | Phenotypic Antimicrobial Profile | Resistance Genes | Number of VF |
|---|---|---|---|
| 1121090 | TET, ERY, CLI | ant(6)-Ia_2; lnu(B)_2; lsa(E)_1 | 8 |
| 1121094 | - | erm(B)_18 | 11 |
| 1121108 | - | lnu(C)_1 | 10 |
| 1121118 | - | lnu(C)_1 | 11 |
| 1121191 | lnu(C)_1 | 13 | |
| 1121208 | mph(B)_1 | 11 | |
| 1121227 | tet(L)_2; tet(M)_5; ant(6)-Ia_2; lnu(B)_2; lsa(E)_1 | 14 | |
| 1121287 | - | erm(B)_18 | - |
| 1121292 | TET, CLI | lsa(E)_1; lnu(B)_2; ant(6)-Ia_2; tet(L)_2; tet(M)_5 | 13 |
| 1121295 | - | erm(B)_18; lnu(C)_1 | 10 |
| 1121300 | - | - | 12 |
| 1121323 | TET, CLI | lsa(E)_1; lnu(B)_2; ant(6)-Ia_2; tet(M)_5; tet(L)_2 | 11 |
| 1121338 | - | - | 10 |
| 1121346 | erm(B)_18;aph(3′)-III_1;ant(6)-Ia_1;tet(O)_3 | - | |
| 1121350 | lnu(C)_1;erm(B)_18;aph(3′)-III_1;ant(6)-Ia_1;tet(O)_3 | 11 | |
| 1121751 | CLI | lsa(E)_1; lnu(B)_2; ant(6)-Ia_2 | 6 |
| 1121757 | TET, CLI | lsa(E)_1; lnu(B)_2; ant(6)-Ia_2; mph(B)_1; tet(M)_5; tet(L)_2 | 13 |
| 1121772 | tet(L)_2; tet(M)_5; ant(6)-Ia_2; lnu(B)_2; lsa(E)_1; lnu(C)_1 | 10 | |
| 1121774 | lsa(E)_1;lnu(B)_2;ant(6)-Ia_2 | - | |
| 1121776 | lnu(B)_2; lsa(E)_1; ant(6)-Ia_2; erm(B)_18 | 11 | |
| 1121974 | tet(O)_3;erm(B)_18;ant(6)-Ia_3 | 13 | |
| 1121980 | TET, ERY, CLI | tet(O)_3;ant(6)-Ia_1;aph(3′)-III_1;erm(B)_18 | 13 |
| 1121981 | - | 10 | |
| 1122022 | erm(B)_18; aph(3′)-III_1; ant(6)-Ia_1; lnu(B)_2; lsa(E)_1 | 9 | |
| 1122039 | ant(6)-Ia_2; lnu(B)_2; lsa(E)_1; tet(S)_3 | 11 | |
| 1122285 | TET, ERY, CLI | lsa(E)_1; lnu(B)_2; ant(6)-Ia_2; erm(B)_18; aph(3′)-III_1; ant(6)-Ia_1; tet(O)_3 | 9 |
| 1122348 | TET, ERY, CLI | ant(6)-Ia_3;erm(B)_18;tet(O)_3;tet(L)_2;tet(M)_5; | 10 |
| 1122419 | ant(6)-Ia_2; lnu(B)_2; lsa(E)_1 | 10 | |
| 1122603 | TET, CLI | tet(L)_2; tet(M)_5; ant(6)-Ia_2; lnu(B)_2; lsa(E)_1 | 14 |
| 1122648 | ERY, CLI | erm(B)_18 | 8 |
| 1122846 | - | - | 9 |
| 1122847 | CLI | lsa(E)_1; lnu(B)_2; ant(6)-Ia_2 | 11 |
| 1122852 | TET, CLI, CRO | lsa(E)_1; lnu(B)_2; ant(6)-Ia_2; tet(L)_2; tet(M)_5; lnu(C)_1 | 9 |
| 1122911 | tet(L)_2;tet(M)_5; ant(6)-Ia_2; lnu(B)_2;lsa(E)_1;lnu(D)_1;mph(B)_1 | 10 | |
| 1122931 | tet(M)_5; lsa(E)_1; lnu(B)_2; ant(6)-Ia_2 | 10 | |
| 1122956 | erm(B)_18;lnu(D)_1 | 10 |
| Strains | lnu | ant(6)-Ia | lsa(E)_1 | tet | erm(B)_18 | aph(3′)-III_1 | mph(B)_1 | Strains | I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | XIII | XIV | XV | XVI | XVII | XVIII | XIX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1121090 | B | 2 | + | 1121090 | |||||||||||||||||||||||
| 1121094 | + | 1121094 | |||||||||||||||||||||||||
| 1121108 | C | 1121108 | |||||||||||||||||||||||||
| 1121118 | C | 1121118 | |||||||||||||||||||||||||
| 1121191 | C | 1121191 | |||||||||||||||||||||||||
| 1121208 | + | 1121208 | |||||||||||||||||||||||||
| 1121227 | B | 2 | + | M, L | 1121227 | ||||||||||||||||||||||
| 1121287 | + | 1121287 | |||||||||||||||||||||||||
| 1121292 | B | 2 | + | M, L | 1121292 | ||||||||||||||||||||||
| 1121295 | C | + | 1121295 | ||||||||||||||||||||||||
| 1121300 | 1121300 | ||||||||||||||||||||||||||
| 1121323 | B | 2 | + | M, L | 1121323 | ||||||||||||||||||||||
| 1121338 | 1121338 | ||||||||||||||||||||||||||
| 1121346 | 1 | O | + | + | 1121346 | ||||||||||||||||||||||
| 1121350 | C | 1 | O | + | + | 1121350 | |||||||||||||||||||||
| 1121751 | B | 2 | + | 1121751 | |||||||||||||||||||||||
| 1121757 | B | 2 | + | M, L | + | 1121757 | |||||||||||||||||||||
| 1121772 | B, C | 2 | + | M, L | 1121772 | ||||||||||||||||||||||
| 1121774 | B | 2 | + | 1121774 | |||||||||||||||||||||||
| 1121776 | B | 2 | + | + | 1121776 | ||||||||||||||||||||||
| 1121974 | 3 | O | + | 1121974 | |||||||||||||||||||||||
| 1121980 | 1 | O | + | + | 1121980 | ||||||||||||||||||||||
| 1121981 | 1121981 | ||||||||||||||||||||||||||
| 1122022 | B | 1 | + | + | + | 1122022 | |||||||||||||||||||||
| 1122039 | B | 2 | + | S | 1122039 | ||||||||||||||||||||||
| 1122285 | B | 2, 1 | + | O | + | + | 1122285 | ||||||||||||||||||||
| 1122348 | 3 | M, L, O | + | 1122348 | |||||||||||||||||||||||
| 1122419 | B | 2 | + | 1122419 | |||||||||||||||||||||||
| 1122603 | B | 2 | + | M, L | 1122603 | ||||||||||||||||||||||
| 1122648 | + | 1122648 | |||||||||||||||||||||||||
| 1122846 | 1122846 | ||||||||||||||||||||||||||
| 1122847 | B | 2 | + | 1122847 | |||||||||||||||||||||||
| 1122852 | B, C | 2 | + | M, L | 1122852 | ||||||||||||||||||||||
| 1122911 | B, D | 2 | + | M, L | + | 1122911 | |||||||||||||||||||||
| 1122931 | B | 2 | + | M | 1122931 | ||||||||||||||||||||||
| 1122956 | D | + | 1122956 |
| VF Class | VF | Related Genes | Strains (%) 1 |
|---|---|---|---|
| Adherence | Agglutinin receptor | Undetermined | 5 (15.15) |
| Fibronectin-binding proteins | fbp54 | 33 (100) | |
| Laminin-binding proteins | lmb | 30 (90.91) | |
| Streptococcal lipoprotein rotamase A | -/slrA | 13 (39.39) | |
| Streptococcal plasmid receptor/GAPDH | plr/gapA | 21 (63.64) | |
| rlrA islet | srtC | 1 (3.03) | |
| Enzyme | Hyaluronidase | hyIB | 12 (36.36) |
| Streptococcal enolase | eno | 31 (93.94) | |
| Immune evasion | Capsule | Undetermined | 33 (100) |
| Polysaccharide capsule (Bacillus) | /galE | 11 (33.33) | |
| Exopolysaccharide (Haemophilus) | galE | 2 (6.06) | |
| Immunoreactive antigen | Rib | rib | 4 (12.12) |
| Manganese uptake | Pneumococcal surface antigen A/Metal binding protein SloC | psaA | 33 (100) |
| Protease | C3-degrading protease | cppA | 33 (100) |
| C5a peptidase | scpA/scpB | 24 (72.73) | |
| Serine protease | htrA/degP | 33 (100) | |
| Trigger factor | tig/ropA | 21 (63.64) | |
| Cell surface components | Trehalose-recycling ABC transporter (Mycobacterium) | sugC | 10 (30.30) |
| Serum resistance and immune evasion | LPS (Francisella) | wbtP | 2 (6.06) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quinto, E.J.; Redondo del Río, P.; de Mateo Silleras, B.; Prieto, A.; López-Lorenzo, G.; Franco, C.M.; Vázquez, B.I. Genomic Characterization and Antimicrobial Resistance Profile of Streptococcus uberis Strains Isolated from Cows with Mastitis from Northwestern Spain. Antibiotics 2025, 14, 1059. https://doi.org/10.3390/antibiotics14111059
Quinto EJ, Redondo del Río P, de Mateo Silleras B, Prieto A, López-Lorenzo G, Franco CM, Vázquez BI. Genomic Characterization and Antimicrobial Resistance Profile of Streptococcus uberis Strains Isolated from Cows with Mastitis from Northwestern Spain. Antibiotics. 2025; 14(11):1059. https://doi.org/10.3390/antibiotics14111059
Chicago/Turabian StyleQuinto, Emiliano J., Paz Redondo del Río, Beatriz de Mateo Silleras, Alberto Prieto, Gonzalo López-Lorenzo, Carlos M. Franco, and Beatriz I. Vázquez. 2025. "Genomic Characterization and Antimicrobial Resistance Profile of Streptococcus uberis Strains Isolated from Cows with Mastitis from Northwestern Spain" Antibiotics 14, no. 11: 1059. https://doi.org/10.3390/antibiotics14111059
APA StyleQuinto, E. J., Redondo del Río, P., de Mateo Silleras, B., Prieto, A., López-Lorenzo, G., Franco, C. M., & Vázquez, B. I. (2025). Genomic Characterization and Antimicrobial Resistance Profile of Streptococcus uberis Strains Isolated from Cows with Mastitis from Northwestern Spain. Antibiotics, 14(11), 1059. https://doi.org/10.3390/antibiotics14111059








