Phage Therapy as a Novel Alternative to Antibiotics Through Adaptive Evolution and Fitness Trade-Offs
Abstract
1. Introduction
2. Adaptive Evolution of Phages to Overcome Bacterial Resistance Mechanisms
3. Adaptive Evolution and Phage/Host Interactions
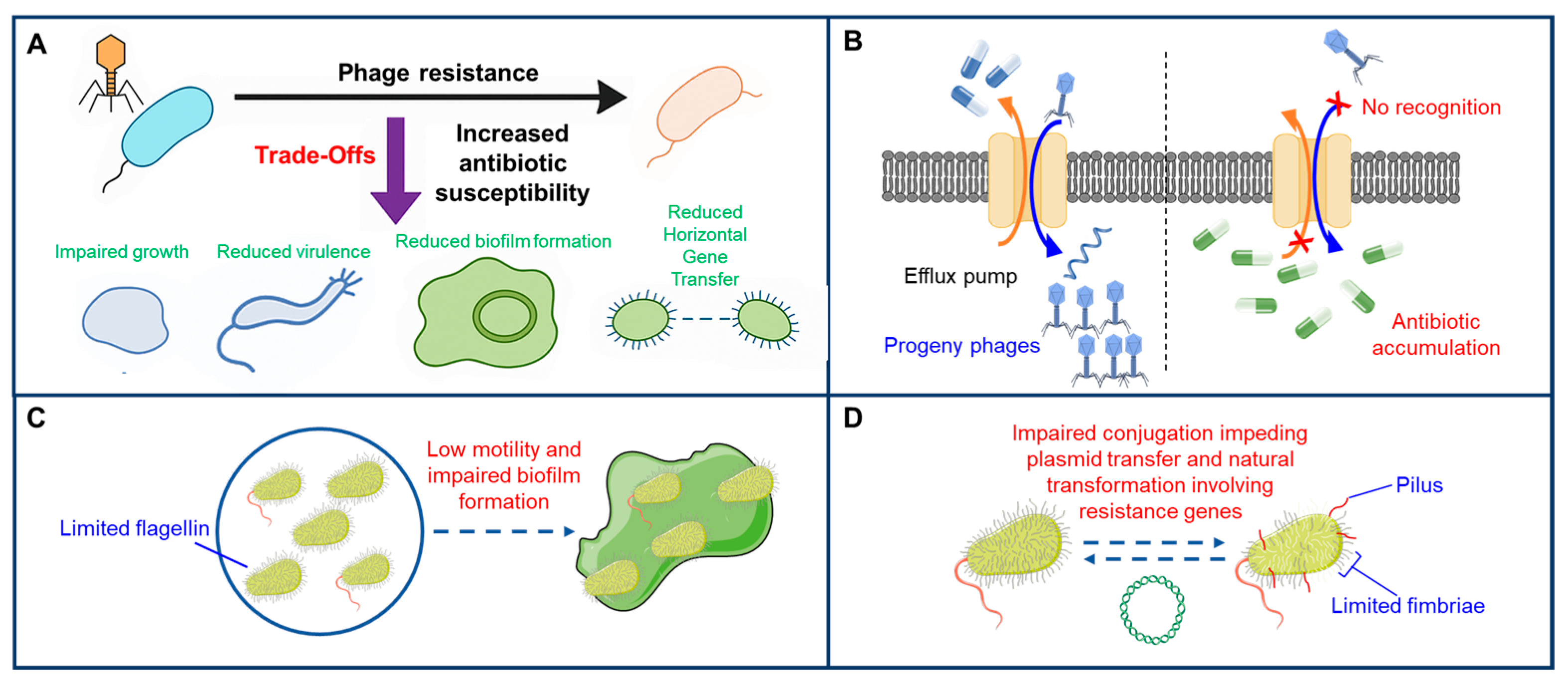
4. Antagonistic Pleiotropy as an Evolutionary Double-Edged Sword
5. Innovative Phage Therapy Through Adaptive Evolution and Fitness Trade-Offs
6. Challenges and Paradigm Shift in Antimicrobial Strategy
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Akram, F.; Imtiaz, M.; Haq, I.U. Emergent crisis of antibiotic resistance: A silent pandemic threat to 21st century. Microb. Pathog. 2023, 174, 105923. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- de Kraker, M.E.A.; Stewardson, A.J.; Harbarth, S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016, 13, e1002184. [Google Scholar] [CrossRef]
- Roope, L.S.J.; Smith, R.D.; Pouwels, K.B.; Buchanan, J.; Abel, L.; Eibich, P.; Butler, C.C.; Tan, P.S.; Walker, A.S.; Robotham, J.V.; et al. The challenge of antimicrobial resistance: What economics can contribute. Science 2019, 364, eaau4679. [Google Scholar] [CrossRef]
- Zhen, X.; Stålsby Lundborg, C.; Sun, X.; Zhu, N.; Gu, S.; Dong, H. Economic burden of antibiotic resistance in China: A national level estimate for inpatients. Antimicrob. Resist. Infect. Control 2021, 10, 5. [Google Scholar] [CrossRef]
- Hao, L.; Yang, X.; Chen, H.; Wei, S.; Xu, B.; Zhao, Z. Distribution and drug resistance of bacterial infection in hospitalized patients at the respiratory department before and after the COVID-19 pandemic in Guangzhou, China. Microorganisms 2023, 11, 2542. [Google Scholar] [CrossRef]
- Li, Z.; Cai, H.; Xu, B.; Dong, Q.; Jia, K.; Lin, Z.; Wang, X.; Liu, Y.; Qin, X. Prevalence, antibiotic resistance, resistance and virulence determinants of Campylobacter jejuni in China: A systematic review and meta-analysis. One Health 2025, 20, 100990. [Google Scholar] [CrossRef]
- Kumar, N.R.; Balraj, T.A.; Kempegowda, S.N.; Prashant, A. Multidrug-resistant sepsis: A critical healthcare challenge. Antibiotics 2024, 13, 46. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial resistance: A growing serious threat for global public health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Bobate, S.; Mahalle, S.; Dafale, N.A.; Bajaj, A. Emergence of environmental antibiotic resistance: Mechanism, monitoring and management. Environ. Adv. 2023, 13, 100409. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.M.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Anastassopoulou, C.; Ferous, S.; Petsimeri, A.; Gioula, G.; Tsakris, A. Phage-based therapy in combination with antibiotics: A promising alternative against multidrug-resistant Gram-negative pathogens. Pathogens 2024, 13, 896. [Google Scholar] [CrossRef]
- Romero-Calle, D.; Guimarães Benevides, R.; Góes-Neto, A.; Billington, C. Bacteriophages as alternatives to antibiotics in clinical care. Antibiotics 2019, 8, 138. [Google Scholar] [CrossRef]
- Hibstu, Z.; Belew, H.; Akelew, Y.; Mengist, H.M. Phage therapy: A different approach to fight bacterial infections. Biologics 2022, 16, 173–186. [Google Scholar]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Therapeut. 2017, 8, 162–173. [Google Scholar] [CrossRef]
- Olawade, D.B.; Fapohunda, O.; Egbon, E.; Ebiesuwa, O.A.; Usman, S.O.; Faronbi, A.O.; Fidelis, S.C. Phage therapy: A targeted approach to overcoming antibiotic resistance. Microb. Pathog. 2024, 197, 107088. [Google Scholar] [CrossRef]
- Oechslin, F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef]
- Borin, J.M.; Avrani, S.; Barrick, J.E.; Petrie, K.L.; Meyer, J.R. Coevolutionary phage training leads to greater bacterial suppression and delays the evolution of phage resistance. Proc. Natl. Acad. Sci. USA 2021, 118, e2104592118. [Google Scholar]
- Vu, T.N.; Clark, J.R.; Jang, E.; D’Souza, R.; Nguyen, L.P.; Pinto, N.A.; Yoo, S.; Abadie, R.; Maresso, A.W.; Yong, D. Appelmans protocol—A directed in vitro evolution enables induction and recombination of prophages with expanded host range. Virus Res. 2024, 339, 199272. [Google Scholar]
- Kering, K.K.; Zhang, X.; Nyaruaba, R.; Yu, J.; Wei, H. Application of adaptive evolution to improve the stability of bacteriophages during storage. Viruses 2020, 12, 423. [Google Scholar] [CrossRef]
- Friman, V.P.; Soanes-Brown, D.; Sierocinski, P.; Molin, S.; Johansen, H.K.; Merabishvili, M.; Pirnay, J.P.; De Vos, D.; Buckling, A. Pre-adapting parasitic phages to a pathogen leads to increased pathogen clearance and lowered resistance evolution with Pseudomonas aeruginosa cystic fibrosis bacterial isolates. J. Evol. Biol. 2016, 29, 188–198. [Google Scholar] [CrossRef]
- Hasan, M.; Ahn, J. Evolutionary dynamics between phages and bacteria as a possible approach for designing effective phage therapies against antibiotic-resistant bacteria. Antibiotics 2022, 11, 915. [Google Scholar] [CrossRef]
- Dupuis, M.-È.; Villion, M.; Magadán, A.H.; Moineau, S. CRISPR-Cas and restriction–modification systems are compatible and increase phage resistance. Nat. Commun. 2013, 4, 2087. [Google Scholar] [CrossRef]
- Yirmiya, E.; Leavitt, A.; Lu, A.; Ragucci, A.E.; Avraham, C.; Osterman, I.; Garb, J.; Antine, S.P.; Mooney, S.E.; Hobbs, S.J.; et al. Phages overcome bacterial immunity via diverse anti-defence proteins. Nature 2024, 625, 352–359. [Google Scholar] [CrossRef]
- Camara-Wilpert, S.; Mayo-Munoz, D.; Russel, J.; Fagerlund, R.D.; Madsen, J.S.; Fineran, P.C.; Sorensen, S.J.; Pinilla-Redondo, R. Bacteriophages suppress CRISPR-Cas immunity using RNA-based anti-CRISPRs. Nature 2023, 623, 601–607. [Google Scholar] [CrossRef]
- Maestri, A.; Pons, B.J.; Pursey, E.; Chong, C.E.; Gandon, S.; Custodio, R.; Olina, A.; Agapov, A.; Chisnall, M.A.W.; Grasso, A.; et al. The bacterial defense system MADS interacts with CRISPR-Cas to limit phage infection and escape. Cell Host Microbe 2024, 32, 1412–1426.e11. [Google Scholar] [CrossRef]
- Enikeeva, F.N.; Severinov, K.V.; Gelfand, M.S. Restriction-modification systems and bacteriophage invasion: Who wins? J. Theor. Biol. 2010, 266, 550–559. [Google Scholar] [CrossRef]
- Birkholz, N.; Jackson, S.A.; Fagerlund, R.D.; Fineran, P.C. A mobile restriction-modification system provides phage defence and resolves an epigenetic conflict with an antagonistic endonuclease. Nucleic Acids Res. 2022, 50, 3348–3361. [Google Scholar] [CrossRef]
- Zhong, Y.; Lauschke, V.M. The phage anti-restriction induced system: New insights into bacterial immunity and bacteriophage escape strategies. Signal Transduct. Target. Ther. 2024, 9, 269. [Google Scholar] [CrossRef]
- Islam, M.M.; Mahbub, N.U.; Shin, W.S.; Oh, M.H. Phage-encoded depolymerases as a strategy for combating multidrug-resistant Acinetobacter baumannii. Front. Cell. Infect. Microb. 2024, 14, 1462620. [Google Scholar] [CrossRef]
- Huang, T.; Zhang, Z.; Tao, X.; Shi, X.; Lin, P.; Liao, D.; Ma, C.; Cai, X.; Lin, W.; Jiang, X.; et al. Structural and functional basis of bacteriophage K64-ORF41 depolymerase for capsular polysaccharide degradation of Klebsiella pneumoniae K64. Int. J. Biol. Macromol. 2024, 265, 130917. [Google Scholar] [CrossRef]
- Fujiki, J.; Yokoyama, D.; Yamamoto, H.; Kimura, N.; Shimizu, M.; Kobayashi, H.; Nakamura, K.; Iwano, H. Biocontrol of phage resistance in Pseudomonas infections: Insights into directed breaking of spontaneous evolutionary selection in phage therapy. Viruses 2025, 17, 1080. [Google Scholar] [CrossRef]
- Li, X.; He, Y.; Wang, Z.; Wei, J.; Hu, T.; Si, J.; Tao, G.; Zhang, L.; Xie, L.; Abdalla, A.E.; et al. A combination therapy of phages and antibiotics: Two is better than one. Int. J. Biol. Sci. 2021, 17, 3573–3582. [Google Scholar] [CrossRef]
- Peng, H.; Chen, I.A.; Qimron, U. Engineering phages to fight multidrug-resistant bacteria. Chem. Rev. 2025, 125, 933–971. [Google Scholar] [CrossRef]
- Beata, O.; Manal, M. The War between Bacteria and Bacteriophages. In Growing and Handling of Bacterial Cultures; Madhusmita, M., Ed.; IntechOpen: Rijeka, Croatia, 2019; Chapter 6. [Google Scholar] [CrossRef]
- Latka, A.; Lemire, S.; Grimon, D.; Dams, D.; Maciejewska, B.; Lu, T.; Drulis-Kawa, Z.; Briers, Y. Engineering the modular receptor-binding proteins of Klebsiella phages switches their capsule serotype specificity. mBio 2021, 12, e00455-21. [Google Scholar] [CrossRef]
- Abdelsattar, A.S.; Dawooud, A.; Rezk, N.; Makky, S.; Safwat, A.; Richards, P.J.; El-Shibiny, A. How to train your phage: The recent efforts in phage training. Biologics 2021, 1, 70–88. [Google Scholar] [CrossRef]
- Burrowes, B.H.; Molineux, I.J.; Fralick, J.A. Directed in vitro evolution of therapeutic bacteriophages: The Appelmans protocol. Viruses 2019, 11, 241. [Google Scholar] [CrossRef] [PubMed]
- Dicks, L.M.T.; Vermeulen, W. Bacteriophage-host interactions and the therapeutic potential of bacteriophages. Viruses 2024, 16, 478. [Google Scholar] [CrossRef]
- Palma, M.; Qi, B. Advancing phage therapy: A comprehensive review of the safety, efficacy, and future prospects for the targeted treatment of bacterial infections. Infect. Dis. Rep. 2024, 16, 1127–1181. [Google Scholar] [CrossRef] [PubMed]
- Taslem Mourosi, J.; Awe, A.; Guo, W.; Batra, H.; Ganesh, H.; Wu, X.; Zhu, J. Understanding bacteriophage tail fiber interaction with host surface receptor: The key “blueprint” for reprogramming phage host range. Int. J. Mol. Sci. 2022, 23, 12146. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Ahn, J. Adaptive trade-offs between bacteriophage and antibiotic resistance in Salmonella Typhimurium. Microb. Pathog. 2025, 207, 107886. [Google Scholar] [CrossRef]
- Chen, L.; Zhao, X.; Wongso, S.; Lin, Z.; Wang, S. Trade-offs between receptor modification and fitness drive host-bacteriophage co-evolution leading to phage extinction or co-existence. ISME J. 2024, 18, wrae214. [Google Scholar] [CrossRef]
- van den Berg, B.; Silale, A.; Baslé, A.; Brandner, A.F.; Mader, S.L.; Khalid, S. Structural basis for host recognition and superinfection exclusion by bacteriophage T5. Proc. Natl. Acad. Sci. USA 2022, 119, e2211672119. [Google Scholar] [CrossRef]
- Dunne, M.; Prokhorov, N.S.; Loessner, M.J.; Leiman, P.G. Reprogramming bacteriophage host range: Design principles and strategies for engineering receptor binding proteins. Curr. Opin. Biotechnol. 2021, 68, 272–281. [Google Scholar] [CrossRef]
- Abedon, S.T. Bacteriophage adsorption: Likelihood of virion encounter with bacteria and other factors affecting rates. Antibiotics 2023, 12, 723. [Google Scholar] [CrossRef] [PubMed]
- Naureen, Z.; Dautaj, A.; Anpilogov, K.; Camilleri, G.; Dhuli, K.; Tanzi, B.; Maltese, P.E.; Cristofoli, F.; De Antoni, L.; Beccari, T.; et al. Bacteriophages presence in nature and their role in the natural selection of bacterial populations. Acta Biomed. 2020, 91, e2020024. [Google Scholar]
- Cao, L.; Mi, J.; He, Y.; Xuan, G.; Wang, J.; Li, M.; Tong, Y. Quorum sensing inhibits phage infection by regulating biofilm formation of P. aeruginosa PAO1. J. Virol. 2025, 99, e01872-24. [Google Scholar] [PubMed]
- Bürkle, M.; Korf, I.H.E.; Lippegaus, A.; Krautwurst, S.; Rohde, C.; Weissfuss, C.; Nouailles, G.; Tene, X.M.; Gaborieau, B.; Ghigo, J.M.; et al. Phage-phage competition and biofilms affect interactions between two virulent bacteriophages and Pseudomonas aeruginosa. ISME J. 2025, 19, wraf065. [Google Scholar] [CrossRef]
- Bołoz, A.; Lannoy, V.; Olszak, T.; Drulis-Kawa, Z.; Augustyniak, D. The interplay between bacterial extracellular vesicles and phages: Receptors, mechanisms, and implications. Viruses 2025, 17, 1180. [Google Scholar] [CrossRef]
- Jdeed, G.; Kravchuk, B.; Tikunova, N.V. Factors affecting phage-bacteria coevolution dynamics. Viruses 2025, 17, 235. [Google Scholar] [CrossRef]
- Gonzales, M.E.M.; Ureta, J.C.; Shrestha, A.M.S. PHIStruct: Improving phage-host interaction prediction at low sequence similarity settings using structure-aware protein embeddings. Bioinformatics 2025, 41, btaf016. [Google Scholar] [CrossRef]
- Cucić, S.; Putzeys, L.; Boon, M.; Lepp, D.; Lavigne, R.; Khursigara, C.M.; Anany, H. Multi-omics characterization of a lytic phage targeting Listeria monocytogenes. mSystems 2025, 10, e0058725. [Google Scholar] [CrossRef]
- Doud, M.B.; Robertson, J.M.; Strathdee, S.A. Optimizing phage therapy with artificial intelligence: A perspective. Front. Cell. Infect. Microb. 2025, 15, 1611857. [Google Scholar] [CrossRef]
- Zou, H.; Huang, X.; Xiao, W.; He, H.; Liu, S.; Zeng, H. Recent advancements in bacterial anti-phage strategies and the underlying mechanisms altering susceptibility to antibiotics. Microbiol. Res. 2025, 295, 128107. [Google Scholar] [CrossRef]
- Fujiki, J.; Nakamura, K.; Nakamura, T.; Iwano, H. Fitness trade-offs between phage and antibiotic sensitivity in phage-resistant variants: Molecular action and insights into clinical applications for phage therapy. Int. J. Mol. Sci. 2023, 24, 15628. [Google Scholar] [CrossRef]
- Laure, N.N.; Ahn, J. Phage resistance-mediated trade-offs with antibiotic resistance in Salmonella Typhimurium. Microb. Pathog. 2022, 171, 105732. [Google Scholar] [CrossRef] [PubMed]
- Perry, E.B.; Barrick, J.E.; Bohannan, B.J.M. The molecular and genetic basis of repeatable coevolution between Escherichia coli and bacteriophage T3 in a laboratory microcosm. PLoS ONE 2015, 10, e0130639. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, A.R.; Fortier, A.; Roush, C.; Lessing, A.J.; Bender, R.G.; Barahman, R.; Grant, R.; Chan, B.K.; Turner, P.E. Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 11207–11216. [Google Scholar] [CrossRef]
- Gao, D.; Ji, H.; Wang, L.; Li, X.; Hu, D.; Zhao, J.; Wang, S.; Tao, P.; Li, X.; Qian, P. Fitness trade-offs in phage cocktail-resistant Salmonella enterica serovar Enteritidis results in increased antibiotic susceptibility and reduced virulence. Microbiol. Spectr. 2022, 10, e0291422. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef]
- García-Cruz, J.C.; Rebollar-Juarez, X.; Limones-Martinez, A.; Santos-Lopez, C.S.; Toya, S.; Maeda, T.; Ceapă, C.D.; Blasco, L.; Tomás, M.; Díaz-Velásquez, C.E.; et al. Resistance against two lytic phage variants attenuates virulence and antibiotic resistance in Pseudomonas aeruginosa. Front. Cell. Infect. Microb. 2023, 13, 1280265. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Feng, Y.; McNally, A.; Zong, Z. Characterization of phage resistance and phages capable of intestinal decolonization of carbapenem-resistant Klebsiella pneumoniae in mice. Commun. Biol. 2022, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zeng, Y.; Wang, M.; Bao, R.; Chen, Y.; Li, X.; Pan, J.; Zhu, T.; Hu, B.; Tan, D. Characterization of phage resistance and their impacts on bacterial fitness in Pseudomonas aeruginosa. Microbiol. Spectr. 2022, 10, e0207222. [Google Scholar] [CrossRef]
- Kortright, K.E.; Done, R.E.; Chan, B.K.; Souza, V.; Turner, P.E.; Vives, M. Selection for phage resistance reduces virulence of Shigella flexneri. Appl. Environ. Microbiol. 2022, 88, e01514-21. [Google Scholar] [CrossRef]
- Majkowska-Skrobek, G.; Markwitz, P.; Sosnowska, E.; Lood, C.; Lavigne, R.; Drulis-Kawa, Z. The evolutionary trade-offs in phage-resistant Klebsiella pneumoniae entail cross-phage sensitization and loss of multidrug resistance. Environ. Microbiol. 2021, 23, 7723–7740. [Google Scholar] [CrossRef]
- Zhu, S.; Yang, B.; Wang, Z.; Liu, Y. Augmented dissemination of antibiotic resistance elicited by non-antibiotic factors. Ecotoxicol. Environ. Saf. 2023, 262, 115124. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, M.; Ju, L.; Li, M.; Zhao, M.; Deng, H.; Rensing, C.; Yang, Q.E.; Zhou, S. Phage-mediated virulence loss and antimicrobial susceptibility in carbapenem-resistant Klebsiella pneumoniae. mBio 2025, 16, e02957-24. [Google Scholar] [CrossRef]
- Hasan, M.; Dawan, J.; Ahn, J. Assessment of the potential of phage-antibiotic synergy to induce collateral sensitivity in Salmonella Typhimurium. Microb. Pathog. 2023, 180, 106134. [Google Scholar] [CrossRef]
- Fu, Y.; Yin, M.; Cao, L.; Lu, Y.; Li, Y.; Zhang, L. Capsule mutations serve as a key strategy of phage resistance evolution of K54 hypervirulent Klebsiella pneumoniae. Commun. Biol. 2025, 8, 257. [Google Scholar] [CrossRef] [PubMed]
- Oromí-Bosch, A.; Antani, J.D.; Turner, P.E. Developing phage therapy that overcomes the evolution of bacterial resistance. Annu. Rev. Virol. 2023, 10, 503–524. [Google Scholar] [CrossRef] [PubMed]
- Fatima, R.; Hynes, A.P. Temperate phage-antibiotic synergy is widespread-extending to Pseudomonas- but varies by phage, host strain, and antibiotic pairing. mBio 2025, 16, e02559-24. [Google Scholar] [CrossRef] [PubMed]
- Loganathan, A.; Bozdogan, B.; Manohar, P.; Nachimuthu, R. Phage-antibiotic combinations in various treatment modalities to manage MRSA infections. Front. Pharmacol. 2024, 15, 1356179. [Google Scholar] [CrossRef]
- Hall, A.R.; De Vos, D.; Friman, V.P.; Pirnay, J.P.; Buckling, A. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl. Environ. Microbiol. 2012, 78, 5646–5652. [Google Scholar] [CrossRef]
- Gou, Z.; Yao, P.; Xiong, L.; Wang, X.; Yuan, Q.; Sun, F.; Cheng, Y.; Xia, P. Potential of a phage cocktail in the treatment of multidrug-resistant Klebsiella pneumoniae pulmonary infection in mice. BMC Microbiol. 2025, 25, 151. [Google Scholar] [CrossRef]
- Lopes, A.; Pereira, C.; Almeida, A. Sequential combined effect of phages and antibiotics on the inactivation of Escherichia coli. Microorganisms 2018, 6, 125. [Google Scholar] [CrossRef]
- Kim, J.; Jo, A.; Chukeatirote, E.; Ahn, J. Assessment of antibiotic resistance in Klebsiella pneumoniae exposed to sequential in vitro antibiotic treatments. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 60. [Google Scholar] [CrossRef]
- Akturk, E.; Pinto, G.; Ostyn, L.; Crabbé, A.; Melo, L.D.R.; Azeredo, J.; Coenye, T. Combination of phages and antibiotics with enhanced killing efficacy against dual-species bacterial communities in a three-dimensional lung epithelial model. Biofilm 2025, 9, 100245. [Google Scholar] [CrossRef]
- Kang, Y.S.; Park, W. Trade-off between antibiotic resistance and biological fitness in Acinetobacter sp. strain DR1. Environ. Microbiol. 2010, 12, 1304–1318. [Google Scholar] [CrossRef]
- Stepanyan, K.; Wenseleers, T.; Duéñez-Guzmán, E.A.; Muratori, F.; Van den Bergh, B.; Verstraeten, N.; De Meester, L.; Verstrepen, K.J.; Fauvart, M.; Michiels, J. Fitness trade-offs explain low levels of persister cells in the opportunistic pathogen Pseudomonas aeruginosa. Mol. Ecol. 2015, 24, 1572–1583. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, D.; Kim, M. The application of adaptively evolved thermostable bacteriophage ΦYMFM0293 to control Salmonella spp. in poultry skin. Food Res. Int. 2023, 167, 112665. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.B.; Bhaskar, R.; Han, S.S. Bacteriophages: Natural antimicrobial bioadditives for food preservation in active packaging. Int. J. Biol. Macromol. 2024, 276, 133945. [Google Scholar] [CrossRef] [PubMed]
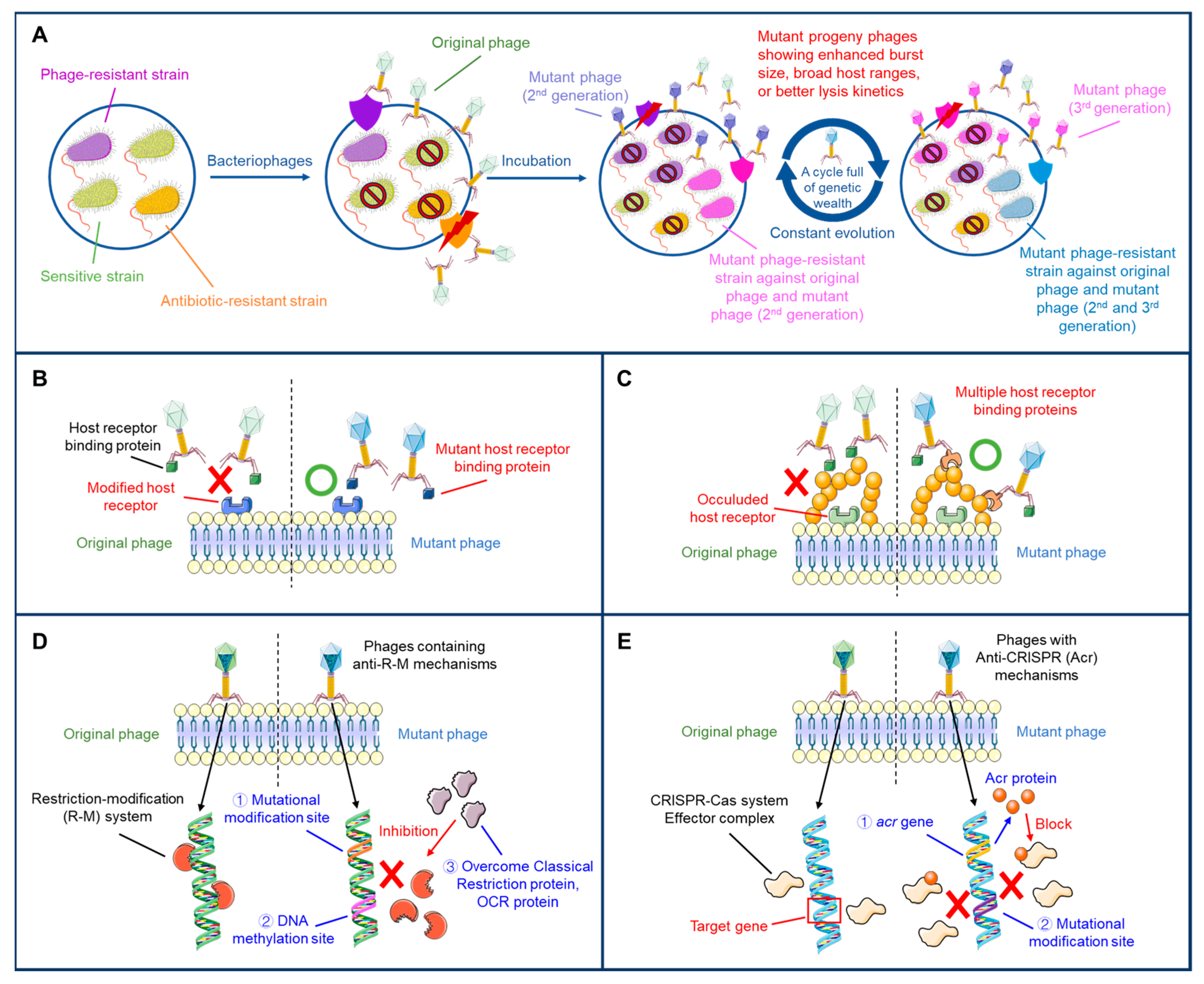
| Bacterial Resistance Mechanism | Description | Phage Adaptation | Reference |
|---|---|---|---|
| Surface receptor modification | Alteration or loss of phage-binding receptors (LPS, outer membrane proteins, pili, flagella) | Mutation of tail fibers, baseplate proteins, or spikes to recognize modified or alternative receptors | [22] |
| CRISPR-Cas immune systems | Sequence-specific degradation of phage genomes using bacterial CRISPR arrays | Evolution of anti-CRISPR proteins or genome sequence modification to evade CRISPR targeting | [25,26] |
| Restriction-modification systems | Cleavage of foreign DNA at specific recognition sites | Phage DNA methylation or mutation of restriction sites to escape cleavage | [27,28,29] |
| Biofilm formation | Extracellular polymeric substances shielding cells and receptors | Production of depolymerases or enzymes to degrade biofilm matrix | [30,31] |
| Efflux pump-related resistance | Enhanced drug efflux or metabolic adaptations that indirectly limit phage entry | No direct phage adaptation; selection of phages exploiting alternative receptors or enhanced adsorption | [32,33] |
| Stress-response-mediated resistance | Altered receptor expression under nutrient limitation, quorum sensing, or host-mimicking stress | Selection for phages capable of infecting low-receptor-expressing or metabolically dormant cells | [34] |
| Receptor masking via capsules or extracellular polysaccharides | Physical shielding of receptor sites | Evolution of phages with enzymatic activity to degrade capsules or polysaccharides | [35,36] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Ahn, J. Phage Therapy as a Novel Alternative to Antibiotics Through Adaptive Evolution and Fitness Trade-Offs. Antibiotics 2025, 14, 1040. https://doi.org/10.3390/antibiotics14101040
Zhang S, Ahn J. Phage Therapy as a Novel Alternative to Antibiotics Through Adaptive Evolution and Fitness Trade-Offs. Antibiotics. 2025; 14(10):1040. https://doi.org/10.3390/antibiotics14101040
Chicago/Turabian StyleZhang, Song, and Juhee Ahn. 2025. "Phage Therapy as a Novel Alternative to Antibiotics Through Adaptive Evolution and Fitness Trade-Offs" Antibiotics 14, no. 10: 1040. https://doi.org/10.3390/antibiotics14101040
APA StyleZhang, S., & Ahn, J. (2025). Phage Therapy as a Novel Alternative to Antibiotics Through Adaptive Evolution and Fitness Trade-Offs. Antibiotics, 14(10), 1040. https://doi.org/10.3390/antibiotics14101040








