Epidemiological Evidence Supports the Role of Microbial Interactions in Polymicrobial UTI Infections Revealed by In Vitro Research
Abstract
1. Introduction
2. Results
2.1. Patients and Urinary Samples
2.2. Microbial Genera and Species According to Types of Positive Cultures and Groups of Patients
2.3. Urinary Microbial Positivity, Frequency of Species Present and Number of Different Paired Organisms
2.4. Positive Pairwise Microbial Associations
2.5. Associations Between Individual Microorganisms and Groups of Patients
2.6. Associations Between Individual Microorganisms and Types of Positive Cultures
2.7. Associations of Individual Microorganisms According to Groups of Patients and Types of Positive Urinary Samples
3. Discussion
4. Methods
4.1. Database
4.2. Samples Collection and Microbial Cultures
4.3. Patients and Urinary Samples and Definitions
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, H.; Wang, Y.; Jiao, J.; Li, Z.; Cao, J.; Song, B.; Jin, J.; Liu, Y.; Wen, X.; et al. Prevalence, incidence, and risk factors of urinary tract infection among immobile inpatients in China: A prospective, multi-centre study. J. Hosp. Inf. 2019, 104, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. Urinary tract infection syndromes: Occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect. Dis. Clin. North Am. 2014, 28, 1–13. [Google Scholar] [CrossRef]
- Tariq, R.; Pardi, D.S.; Tosh, P.K.; Walker, R.C.; Razonable, R.R.; Khanna, S. Fecal microbiota transplantation for recurrent Clostridium difficile infection reduces recurrent urinary tract infection frequency. Clin. Infect. Dis. 2017, 65, 1745–1747. [Google Scholar] [CrossRef]
- Kline, K.A.; Lewis, A.L. Gram-positive uropathogens, polymicrobial urinary tract infection, and the emerging microbiota of the urinary tract. Microbiol. Spectrum. 2012, 4. [Google Scholar] [CrossRef]
- Riley, L.W. Distinguishing Pathovars from Nonpathovars: Escherichia coli. Microbiol. Spectrum. 2020, 8, 10-1128. [Google Scholar] [CrossRef]
- Marrs, C.F.; Zhang, L.; Foxman, B. Escherichia coli urinary tract infections: Are there distinct uropathogenic E. coli (UPEC) pathotypes? FEMS Microbiol. Lett. 2005, 252, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Kaur, M.; Datta, P.; Chander, J. Chronic urinary carrier state due to Salmonella Typhi causing urinary tract infection in an immunocompetent healthy woman. Trop. Doct. 2018, 48, 236–238. [Google Scholar] [CrossRef]
- Stamatiou, K.; Polyzois, K.; Dahanis, S.; Lambou, T.; Skolarikos, A. Brucella melitensis: A rarely suspected cause of infections of genitalia and the lower urinary tract. Braz. J. Infect. Dis. 2009, 13, 86–89. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, A.; Andini, N.; Yang, S.A. ‘culture’ shift: Application of molecular techniques for diagnosing polymicrobial infections. Biotechnol. Adv. 2019, 37, 476–490. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, Y.; Kang, Y.; Wu, G.; He, J.; Wang, Z.; Yang, J.; Wang, Y.; Yang, X.; Jia, W. A retrospective study of the detection of sepsis pathogens comparing blood culture and culture independent digital PCR. Heliyon. 2024, 10, e27523. [Google Scholar] [CrossRef]
- Duan, J.; Ding, J.; Wei, Y.; Zhang, Y.; You, Z.; Li, D.; Chen, C. Metagenomic analysis identifying a polymicrobial pulmonary infection in a non-HIV immunocompromised patient: A case report. BMC Pulmonary Medicine. 2025, 25, 12. [Google Scholar] [CrossRef]
- Hunt, B.C.; Brix, V.; Vath, J.; Guterman, B.L.; Taddei, S.M.; Learman, B.S.; Brauer, A.L.; Shen, S.; Qu, J.; Armbruster, C.E. Metabolic interplay between Proteus mirabilis and Enterococcus faecalis facilitates polymicrobial biofilm formation and invasive disease. mBio. 2024, 15, e0216424. [Google Scholar] [CrossRef]
- Cowan, S.E.; Gilbert, E.; Liepmann, D.; Keasling, J.D. Commensal interactions in a dual-species biofilm exposed to mixed organic compounds. Appl. Environ. Microbiol. 2000, 66, 4481–4485. [Google Scholar] [CrossRef]
- Ferreira, R.B.R.; Antunes, L.C.M.; Sal-Man, N. Pathogen-pathogen interactions during co-infections. ISME J. 2025, 19, wraf104. [Google Scholar] [CrossRef]
- Kumar, H.; Kawai, T.; Akira, S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009, 388, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Lewin, G.R.; Evans, E.R.; Whiteley, M. Microbial interactions impact stress tolerance in a model oral community. Microbiol. Spectr. 2024, 12, e0100524. [Google Scholar] [CrossRef] [PubMed]
- Kline, K.A.; Schwartz, D.J.; Gilbert, N.M.; Hultgren, S.J.; Lewis, A.L. Immune modulation by group B Streptococcus influences host susceptibility to urinary tract infection. Infect. Immun. 2012, 80, 4186–4194. [Google Scholar] [CrossRef]
- Na, K.; Oh, B.C.; Jung, Y. Multifaceted role of CD14 in innate immunity and tissue homeostasis. Cytokine Growth Factor Rev. 2023, 74, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Tay, W.H.; Chong, K.K.; Kline, K.A. Polymicrobial-host interactions during infection. J. Mol. Biol. 2016, 428, 3355–3371. [Google Scholar] [CrossRef] [PubMed]
- Nye, T.M.; Zou, Z.; Obernuefemann, C.L.; Pinkner, J.S.; Lowry, E.; Kleinschmidt, K.; Bergeron, K.; Klim, A.; Dodson, K.W.; Flores-Mireles, A.L.; et al. Microbial co-occurrences on catheters from long-term catheterized patients. Nat. Commun. 2024, 15, 61. [Google Scholar] [CrossRef] [PubMed]
- Laudisio, A.; Marinosci, F.; Fontana, D.; Gemma, A.; Zizzo, A.; Coppola, A.; Rodano, L.; Antonelli Incalzi, R. The burden of comorbidity is associated with symptomatic polymicrobial urinary tract infection among institutionalized elderly. Aging Clin. Exp. Res. 2015, 27, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, C.E.; Smith, S.N.; Yep, A.; Mobley, H.L. Increased incidence of urolithiasis and bacteremia during Proteus mirabilis and Providencia stuartii coinfection due to synergistic induction of urease activity. J. Infect. Dis. 2014, 209, 1524–1532. [Google Scholar] [CrossRef]
- Tsuchimori, N.; Hayashi, R.; Shino, A.; Yamazaki, T.; Okonogi, K. Enterococcus faecalis aggravates pyelonephritis caused by Pseudomonas aeruginosa in experimental ascending mixed urinary tract infection in mice. Infect. Immun. 1994, 62, 4534–4541. [Google Scholar] [CrossRef]
- Sloan, T.J.; Turton, J.C.; Tyson, J.; Musgrove, A.; Fleming, V.M.; Lister, M.M.; Loose, M.W.; Sochett, R.E.; Digle, M.; Game, F.L.; et al. Examining diabetic heel ulcers through an ecological lens: Microbial community dynamics associated with healing and infection. J. Med. Microbiol. 2019, 68, 230–240. [Google Scholar] [CrossRef]
- Chang, Z.; Deng, J.; Zhang, J.; Wu, H.; Wu, Y.; Bin, L.; Sun, B. Rapid and accurate diagnosis of urinary tract infections using targeted next-generation sequencing: A multicenter comparative study with metagenomic sequencing and traditional culture methods. J. Infect. 2025, 90, 106459. [Google Scholar] [CrossRef]
- Radera, S.; Srivastava, S.; Agarwal, J. Virulence genotyping and multidrug resistance pattern of Escherichia coli isolated from community-acquired and hospital-acquired urinary tract infections. Cureus 2022, 14, e29404. [Google Scholar] [CrossRef]
- Pettigrew, M.M.; Johnson, J.K.; Harris, A.D. The human microbiota: Novel targets for hospital-acquired infections and antibiotic resistance. Ann. Epidemiol. 2016, 26, 342–347. [Google Scholar] [CrossRef]
- Drew, S.J.; Poutanen, S.M.; Mazzulli, T.; McGeer, A.M.; Sarabia, A.; Pong-Porter, S.; Rzayev, Y.; Willey, B.; Green, K.; Low, D.E. Decreased prevalence of virulence factors among ciprofloxacin-resistant uropathogenic Escherichia coli isolates. J. Clin. Microbiol. 2005, 43, 4218–4220. [Google Scholar] [CrossRef]
- Armbruster, C.E.; Forsyth-DeOrnellas, V.; Johnson, A.O.; Smith, S.N.; Zhao, L.; Wu, W.; Mobley, H.L.T. Genome-wide transposon mutagenesis of Proteus mirabilis: Essential genes, fitness factors for catheter-associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements. PLoS Pathog. 2017, 13, e1006434. [Google Scholar] [CrossRef] [PubMed]
- Lewin, G.R.; Stacy, A.; Michie, K.L.; Lamont, R.J.; Whiteley, M. Large-scale identification of pathogen essential genes during coinfection with sympatric and allopatric microbes. Proc. Natl. Acad. Sci. USA 2019, 116, 19685–19694. [Google Scholar] [CrossRef]
- Stacy, A.; McNally, L.; Darch, S.E.; Brown, S.P.; Whiteley, M. The biogeography of polymicrobial infection. Nat. Rev. Microbiol. 2016, 14, 93–105. [Google Scholar] [CrossRef]
- Hanson, C.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Martiny, J.B. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat. Rev. Microbiol. 2012, 10, 497–506. [Google Scholar] [CrossRef]
- Walch, P.D.K.; Broz, P. Molecular mechanisms of co-infections. EMBO reports 2025, 26, 3714–3729. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, B.G.; Anderson, M.; Ferguson, J.K. A predictive model of days from infection to discharge in patients with healthcare-associated urinary tract infections: A structural equation modelling approach. J. Hosp. Infect. 2017, 97, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Jeepipalli, S.; Gurusamy, P.; Luz Martins, A.R.; Colella, E.; Nadakuditi, S.R.; Desaraju, T.; Yada, A.; Onime, J.; William, J.; Bhattacharyya, I.; et al. Altered microRNA Expression Correlates with Reduced TLR2/4-Dependent Periodontal Inflammation and Bone Resorption Induced by Polymicrobial Infection. Microbiol. Spectr. 2025, 14, e0016025. [Google Scholar] [CrossRef] [PubMed]
- Thapa, B.; Pak, S.; Chung, D.; Shin, H.K.; Lee, S.H.; Lee, K. Cell-penetrating TLR inhibitor peptide alleviates ulcerative colitis by the functional modulation of macrophages. Front. Immunol. 2023, 14, 1165667. [Google Scholar] [CrossRef]
- Petruk, G.; Puthia, M.; Samsudin, F.; Petrlova, J.; Olm, F.; Mittendorfer, M.; Hyllén, S.; Edström, D.; Strömdahl, A.C.; Diehl, C.; et al. Targeting Toll-like receptor-driven systemic inflammation by engineering an innate structural fold into drugs Thrombin-derived C-terminal peptides (TCPs) are endogenous anti-infective immunomodulators interfering with CD14-mediated TLR-dependent immune responses. Nat. Commun. 2023, 14, 6097. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Q.; Li, M.; Lao, J.; Tang, H.; Ming, S.; Wu, M.; Gong, S.; Li, L.; Liu, L.; et al. SLAMF7 regulates the inflammatory response in macrophages during polymicrobial sepsis. J. Clin. Invest. 2023, 133, e150224. [Google Scholar] [CrossRef]
- Schilling, J.D.; Martin, S.M.; Hunstad, D.A.; Patel, K.P.; Mulvey, M.A.; Justice, S.S.; Lorenz, S.G.; Hultgren, S.J. CD14-and Toll-like receptor-dependent activation of bladder epithelial cells by lipopolysaccharide and Type 1 piliated Escherichia coli. Infect. Immun. 2003, 71, 1470–1480. [Google Scholar] [CrossRef]
- Hunstad, D.A.; Justice, S.S.; Hung, C.S.; Lauer, S.R.; Hultgren, S.J. Suppression of bladder epithelial cytokine responses by uropathogenic Escherichia coli. Infect. Immun. 2005, 73, 3999–4006. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.M.; Carfrae, L.A.; Rachwalski, K.; French, S.; Catacutan, D.; Gordzevich, R.; MacNair, C.R.; Speagle, M.E.; Werah, F.; Stokes, J.M.; et al. Exploiting the fitness cost of metallo-β-lactamase expression can overcome antibiotic resistance in bacterial pathogens. Nat. Microbiol. 2025, 10, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Fleiss, J.L.; Levin, B.; Paik, M.C. Statistical Methods for Rates and Proportion, 3rd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2003. [Google Scholar]
- StataCorp LP. Stata Statistical Software: Release 14; StataCorp LP: College Station, TX, USA, 2015. [Google Scholar]
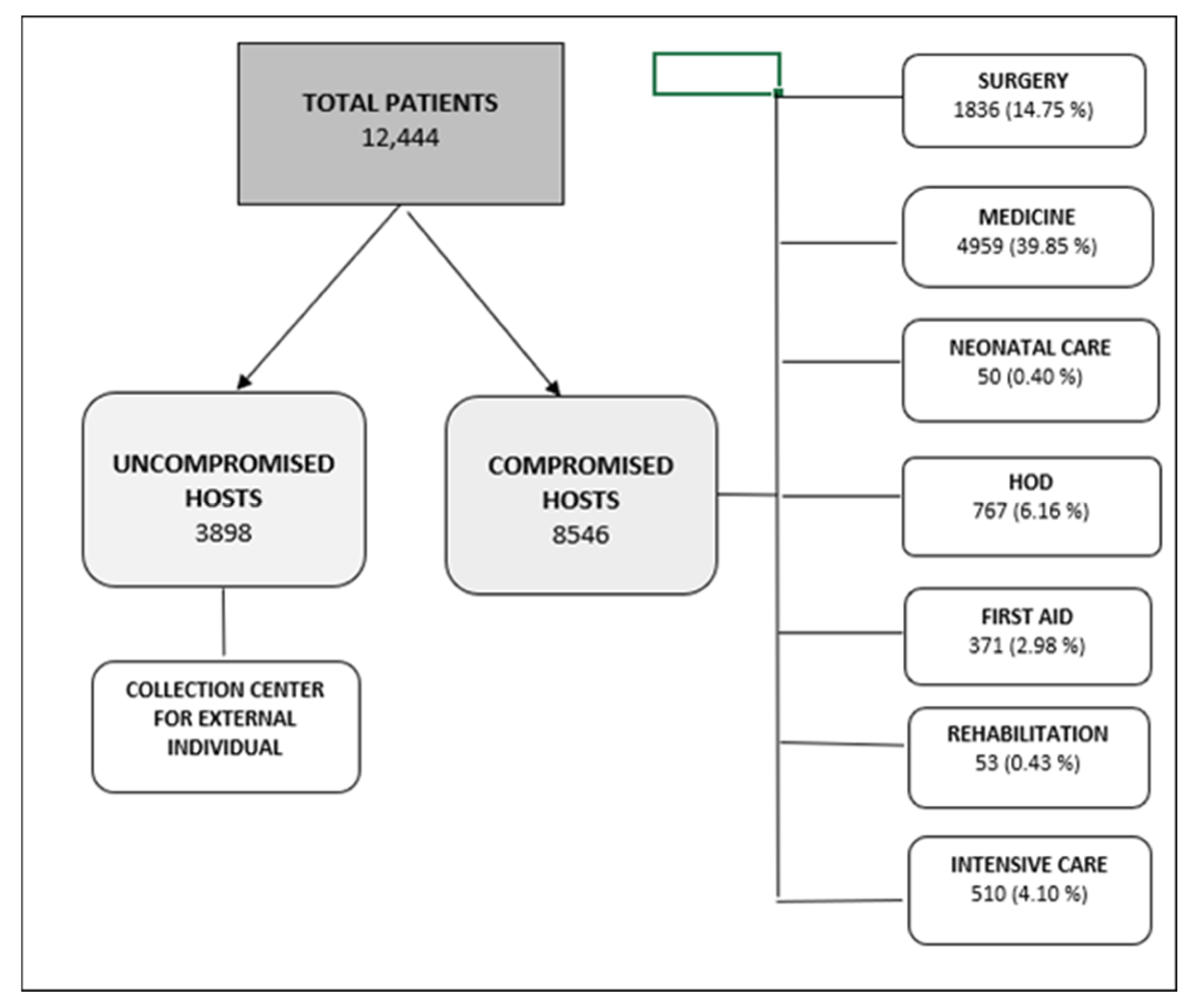
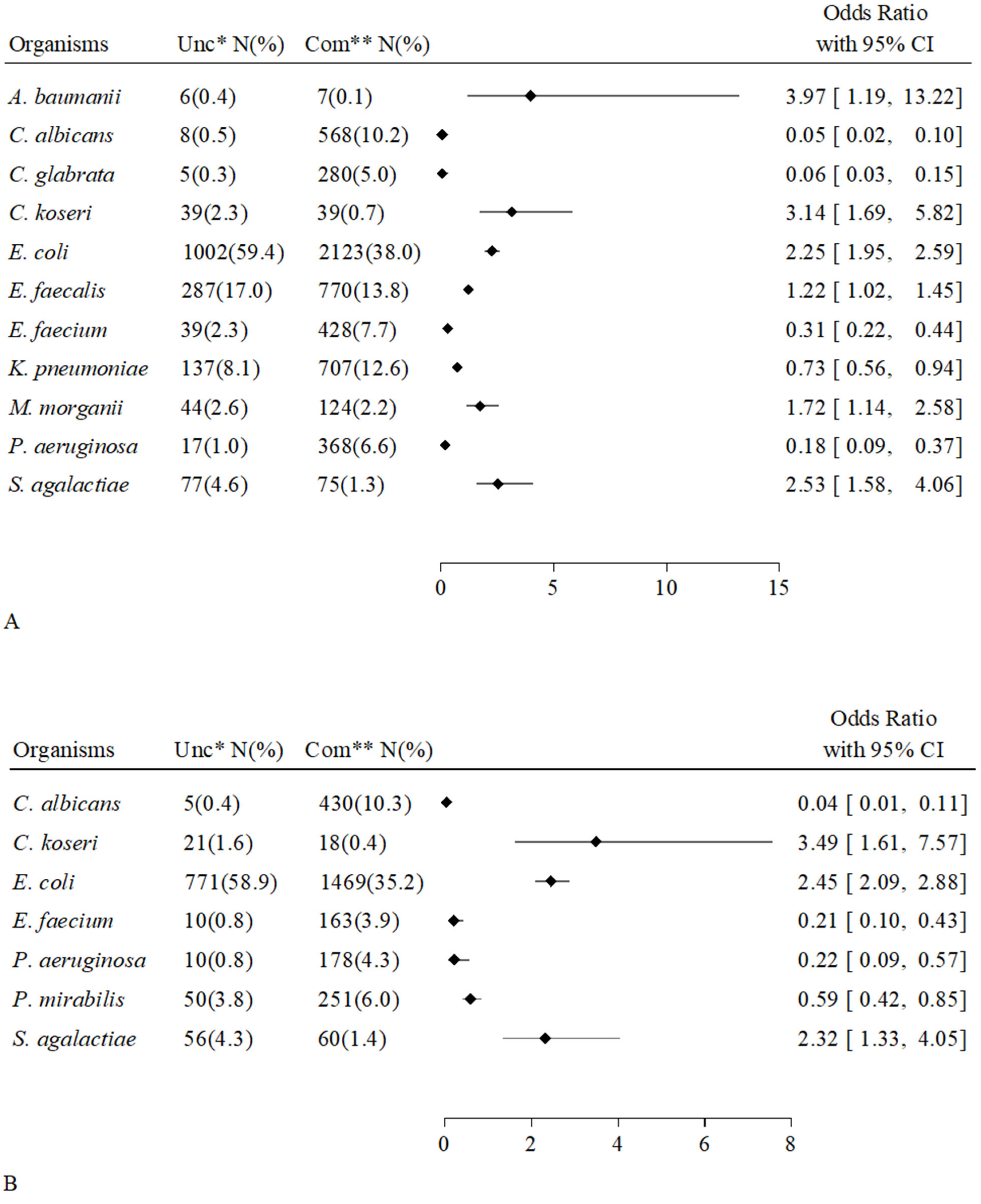
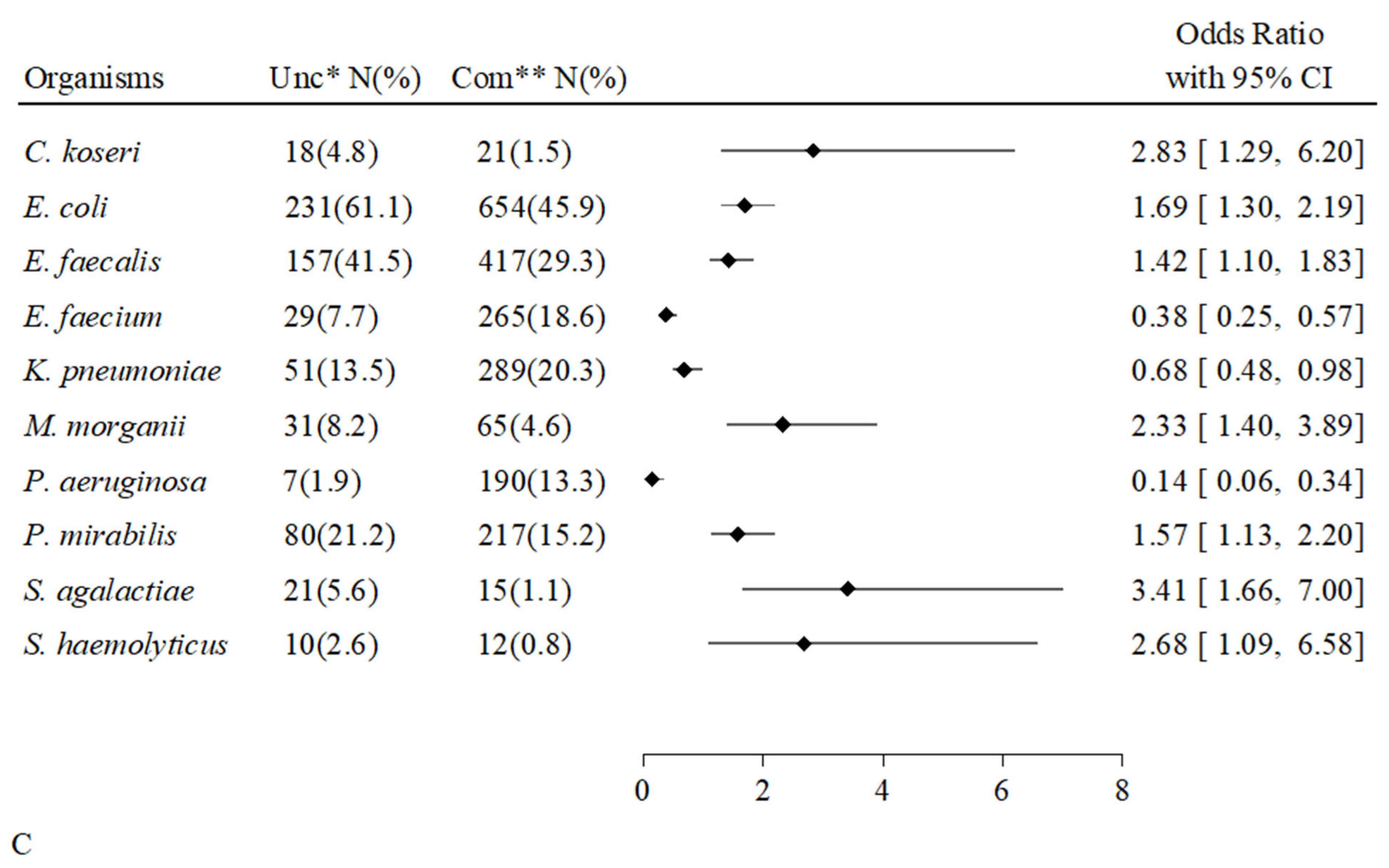
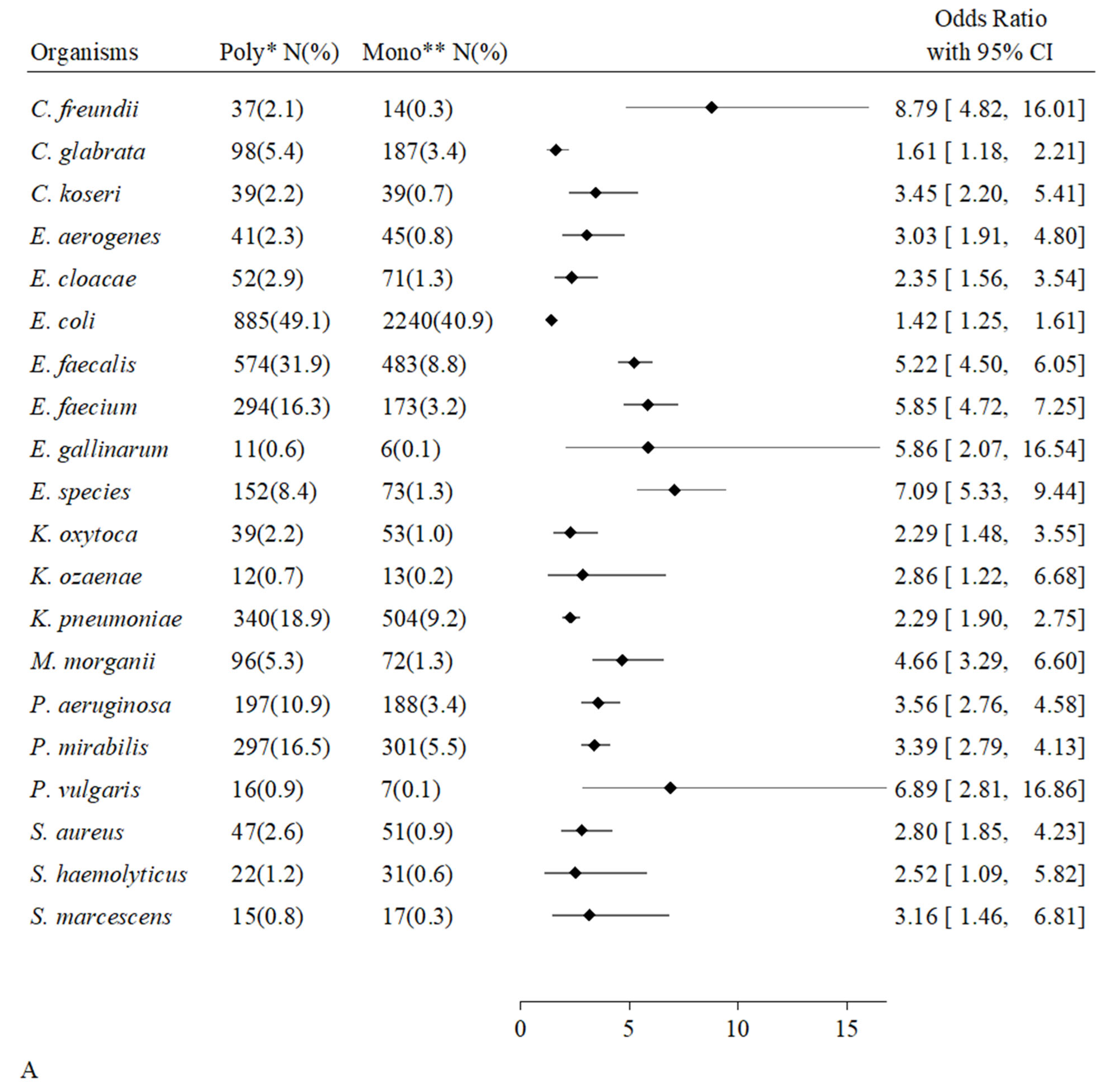
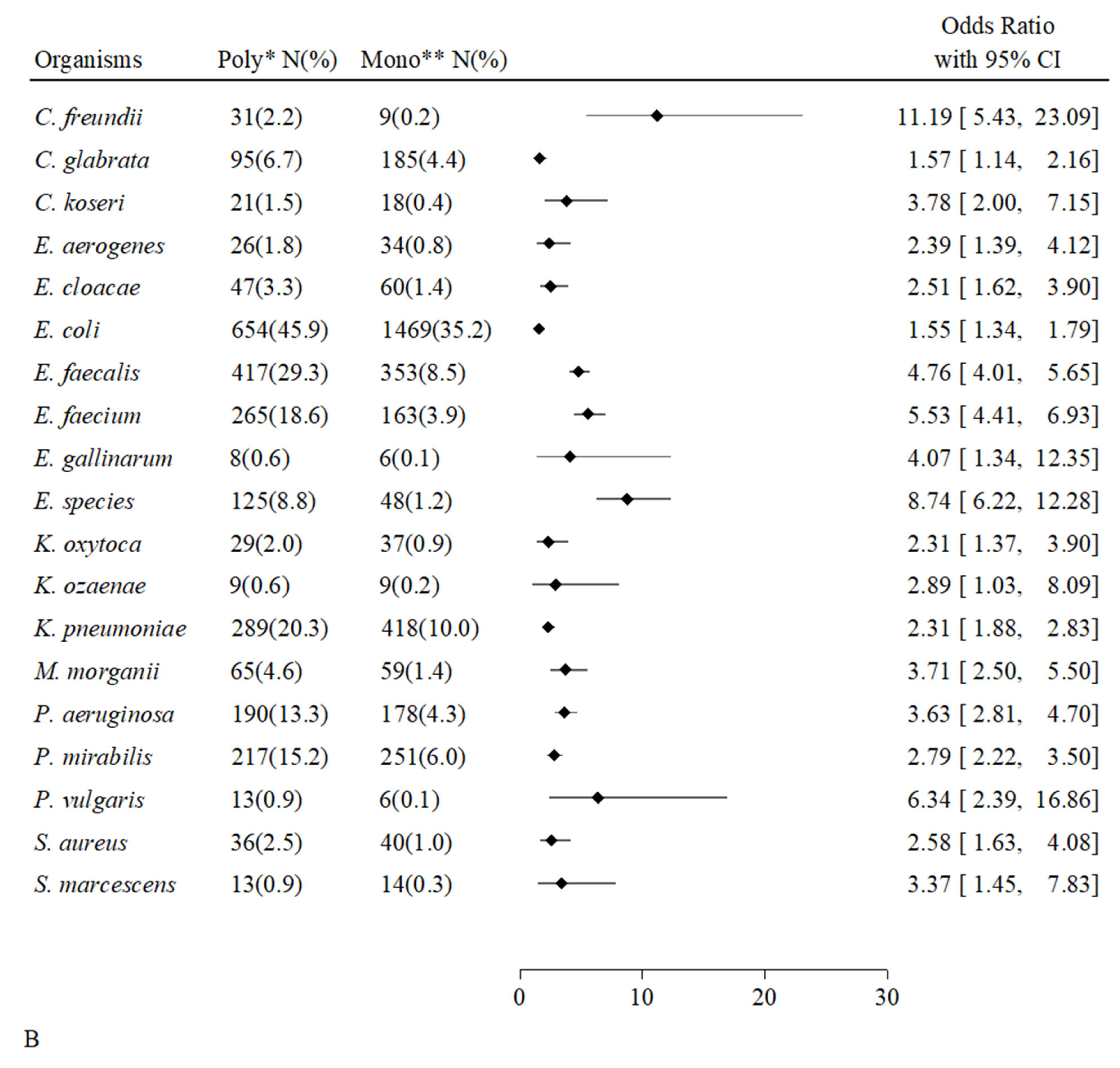
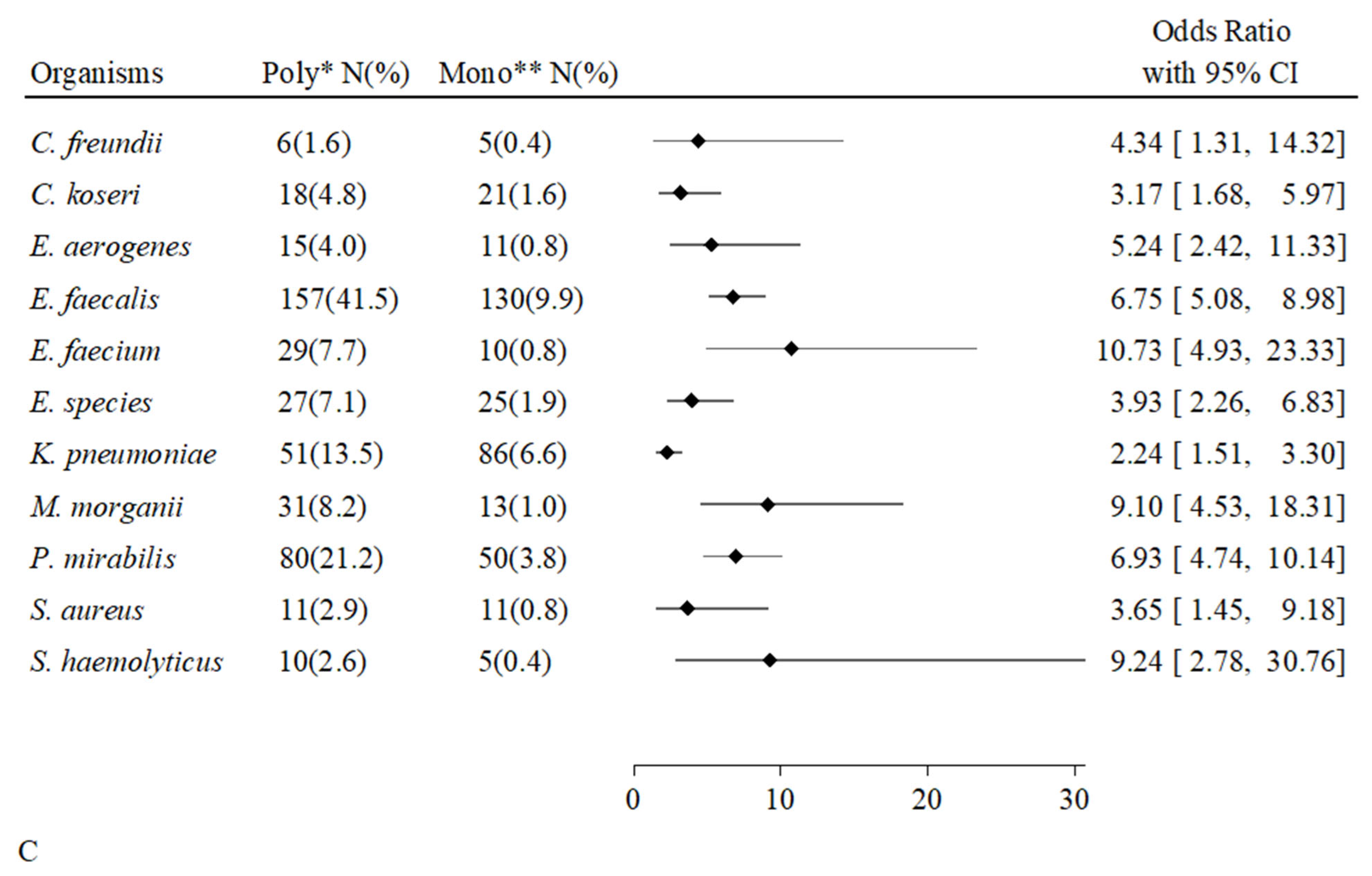
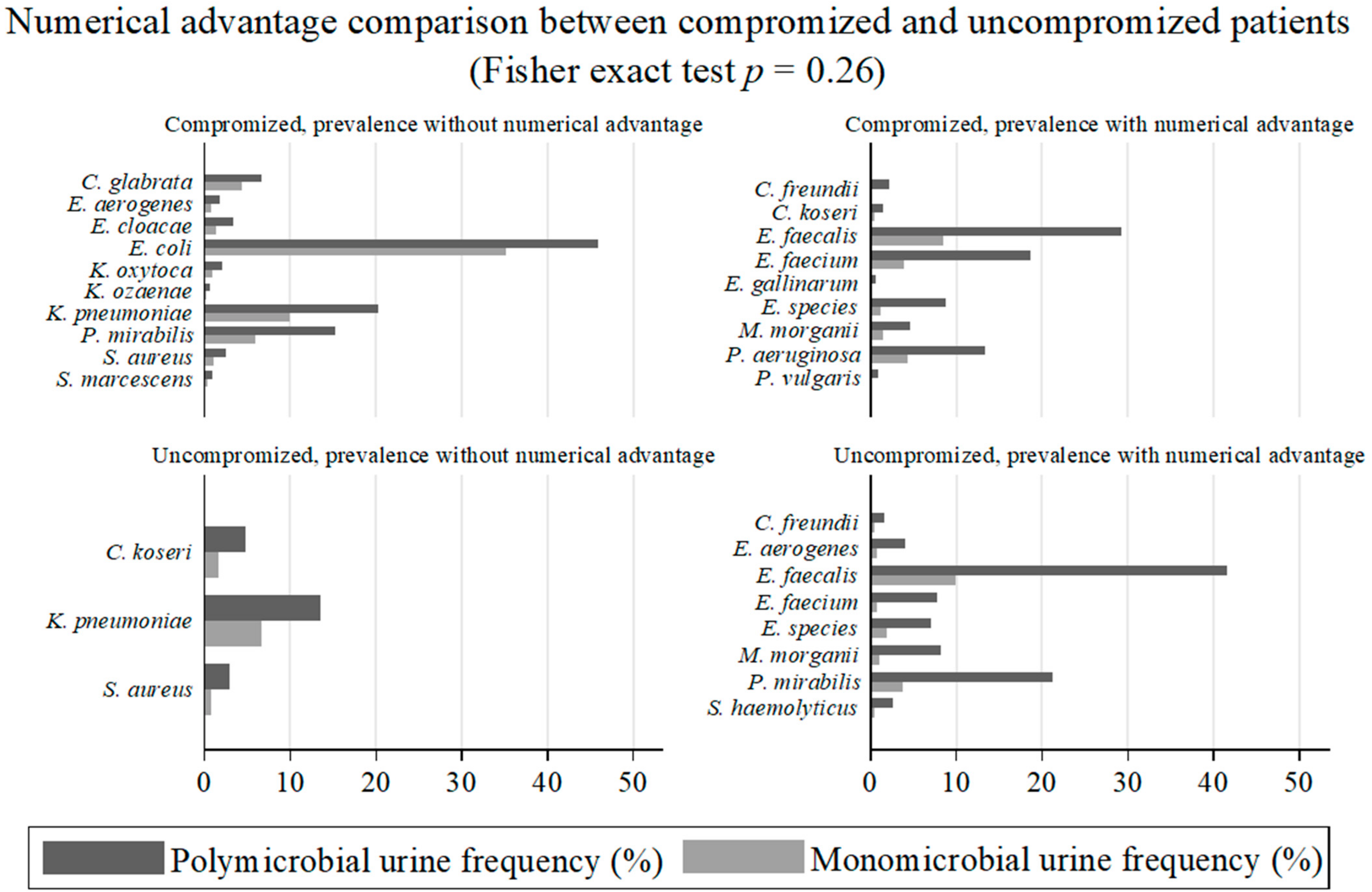
| Overall Hosts | Uncompromised Hosts | Compromised Hosts | p-Value | ||||
|---|---|---|---|---|---|---|---|
| A | |||||||
| N | % | N | % | N | % | ||
| Total hosts | 12,444 | 3898 | 8546 | ||||
| females | 6417 | (52) | 2366 | (61) | 4051 | (47) | <0.001 |
| males | 6027 | (48) | 1532 | (39) | 4495 | (53) | |
| ≥65 years | 5301 | (43) | 1892 | (49) | 3409 | (40) | <0.001 |
| <65 years | 7143 | (57) | 2006 | (51) | 5137 | (60) | |
| B | |||||||
| Total cultures | 24,067 | 5658 | 18,409 | ||||
| negative cultures | 16,787 | 3970 | 12,817 | ||||
| from | |||||||
| total hosts | 10,004 | (69) | 3107 | (71) * | 6897 | (68) * | 0.001 * |
| females | 4730 | (47) | 1734 | (56) | 2996 | (43) | <0.001 |
| males | 5274 | (53) | 1373 | (44) | 3901 | (57) | |
| ≥65 years | 4583 | (46) | 1577 | (51) | 3006 | (44) | <0.001 |
| <65 years | 5421 | (54) | 1530 | (49) | 3891 | (56) | |
| positive cultures | 7280 | 1688 | 5592 | ||||
| from | |||||||
| total hosts | 4431 | (31) * | 1250 | (29) * | 3181 | (32) * | |
| females | 2831 | (64) | 968 | (77) | 1863 | (59) | <0.001 |
| males | 1600 | (36) | 282 | (23) | 1318 | (41) | |
| ≥65 years | 1431 | (32) | 496 | (40) | 935 | (29) | <0.001 |
| <65 years | 3000 | (68) | 754 | (60) | 2246 | (71) | |
| monomicrobial cultures | 5478 | 1310 | 4168 | ||||
| from | |||||||
| total hosts | 3609 | (72) | 1010 | (75) ** | 2599 | (72) ** | |
| females | 2279 | (63) | 781 | (77) | 1498 | (58) | <0.001 |
| males | 1330 | (37) | 229 | (23) | 1101 | (42) | |
| ≥65 years | 1184 | (33) | 403 | (40) | 781 | (30) | <0.001 |
| <65 years | 2425 | (67) | 607 | (60) | 1818 | (70) | |
| polymicrobial cultures | 1802 | 378 | 1424 | ||||
| from | |||||||
| total hosts | 1377 | (28) ** | 344 | (25) ** | 1033 | (28) ** | 0.036 ** |
| females | 893 | (65) | 268 | (78) | 625 | (61) | <0.001 |
| males | 484 | (35) | 76 | (22) | 408 | (39) | |
| ≥65 years | 400 | (29) | 128 | (37) | 272 | (26) | <0.001 |
| <65 years | 977 | (71) | 216 | (63) | 761 | (74) |
| Positive Urine Cultures | |||
|---|---|---|---|
| polymicrobial N (%) | monomicrobial N (%) | p-value 1 | |
| Microbial genera | |||
| from Hosts | |||
| Uncompromised | 17 (4.5) | 20 (1.5) | <0.001 |
| Compromised | 21 (1.5) | 28 (0.7) | 0.005 |
| p-value 2 | <0.001 | 0.004 | |
| Microbial species | |||
| from Hosts | |||
| Uncompromised | 41 (10.8) | 42 (3.2) | <0.001 |
| Compromised | 61 (4.3) | 69 (1.7) | <0.001 |
| p-value 2 | <0.001 | <0.001 | |
| Microbial Species | Isolated from Samples N | % | Coupled with 2nd Species N | Microbial Species | Isolated from Samples N | % | Coupled with 2nd Species N |
|---|---|---|---|---|---|---|---|
| E. coli | 3125 | 42.93 | 45 | K. oxytoca | 92 | 1.26 | 10 |
| E. faecalis | 1057 | 14.52 | 44 | S. maltophilia | 20 | 0.27 | 9 |
| E. faecium | 467 | 6.41 | 31 | C. parapsilosis | 94 | 1.29 | 9 |
| K. pneumoniae | 844 | 11.59 | 31 | S. haemolyticus | 53 | 0.73 | 9 |
| P. mirabilis | 598 | 8.21 | 29 | S. marcescens | 32 | 0.44 | 9 |
| P. aeruginosa | 385 | 5.29 | 27 | P. putida | 10 | 0.14 | 8 |
| C. glabrata | 285 | 3.91 | 19 | P. stuartii | 18 | 0.25 | 7 |
| E. species | 225 | 3.09 | 19 | A. baumanii | 13 | 0.18 | 7 |
| M. morganii | 168 | 2.31 | 19 | P. vulgaris | 23 | 0.32 | 6 |
| C. freundii | 51 | 0.70 | 19 | S. fonticola | 8 | 0.11 | 5 |
| C. albicans | 576 | 7.91 | 17 | K. ozaenae | 25 | 0.34 | 5 |
| E. cloacae | 123 | 1.69 | 14 | S. hominis | 13 | 0.18 | 5 |
| S. agalactiae | 152 | 2.09 | 12 | S. epidermidis | 45 | 0.62 | 5 |
| E. aerogenes | 86 | 1.18 | 11 | C. tropicalis | 44 | 0.60 | 5 |
| C. koseri | 78 | 1.07 | 11 | P. rettgeri | 11 | 0.15 | 5 |
| S. aureus | 98 | 1.35 | 11 | E. avium | 6 | 0.08 | 5 |
| First Organism from Hosts | Second Organism | First AND Second Organisms | First Organism Alone | Odds Ratio (95% CI) |
|---|---|---|---|---|
| All | ||||
| C. albicans | C. glabrata | 30 (21.3) | 68 (4.1) | 4.29 (2.48–7.40) |
| C. albicans | C. tropicalis | 7 (5.0) | 8 (0.5) | 7.93 (2.92–21.5) |
| C. albicans | E. faecium | 61 (43.3) | 233 (14.0) | 4.09 (2.59–6.46) |
| C. glabrata | E. faecium | 34 (34.7) | 260 (15.3) | 2.62 (1.48–4.64) |
| Compromised | ||||
| C. albicans | C. glabrata | 30 (21.7) | 65 (5.1) | 4.24 (2.45–7.33) |
| C. albicans | C. tropicalis | 7 (5.1) | 6 (0.5) | 8.08 (2.86–22.82) |
| C. albicans | E. faecium | 61 (44.2) | 204 (15.9) | 4.19 (2.62–6.68) |
| C. glabrata | E. faecium | 34 (35.8) | 231 (17.4) | 2.67 (1.48–4.79) |
| Uncompromised | ||||
| S. haemolyticus | E. faecalis | 8 (80.0) | 149 (40.5) | 5.25 (1.09–25.16) |
| Monomicrobial Samples | Polymicrobial Samples | |||
|---|---|---|---|---|
| Compromised Hosts | Uncompromised Hosts | Compromised Hosts | Uncompromised Hosts | |
| ref | OR (95%CI) | OR (95%CI) | OR (95%CI) | |
| E. coli | 1 | 2.44 (2.08–2.86) | 1.56 (1.35–1.81) | 2.67 (2.11–3.38) |
| E. species | 1 | 1.48 (0.89–2.44) | 8.43 (5.99–11.86) | 5.9 (3.59–9.7) |
| M. morganii | 1 | 0.99 (0.51–1.91) | 3.63 (2.44–5.39) | 9.03 (5.5–14.82) |
| P. mirabilis | 1 | 0.62 (0.44–0.88) | 2.87 (2.29–3.6) | 4.23 (3.05–5.86) |
| S. haemolyticus | 1 | 0.89 (0.31–2.51) | 1.55 (0.57–4.23) | 6.99 (3.21–15.21) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piatti, G.; Mannini, A.; Vitale, A.; Bruzzone, M.; Schito, A.M.; Ceppi, M. Epidemiological Evidence Supports the Role of Microbial Interactions in Polymicrobial UTI Infections Revealed by In Vitro Research. Antibiotics 2025, 14, 1028. https://doi.org/10.3390/antibiotics14101028
Piatti G, Mannini A, Vitale A, Bruzzone M, Schito AM, Ceppi M. Epidemiological Evidence Supports the Role of Microbial Interactions in Polymicrobial UTI Infections Revealed by In Vitro Research. Antibiotics. 2025; 14(10):1028. https://doi.org/10.3390/antibiotics14101028
Chicago/Turabian StylePiatti, Gabriella, Alessandro Mannini, Alberto Vitale, Marco Bruzzone, Anna Maria Schito, and Marcello Ceppi. 2025. "Epidemiological Evidence Supports the Role of Microbial Interactions in Polymicrobial UTI Infections Revealed by In Vitro Research" Antibiotics 14, no. 10: 1028. https://doi.org/10.3390/antibiotics14101028
APA StylePiatti, G., Mannini, A., Vitale, A., Bruzzone, M., Schito, A. M., & Ceppi, M. (2025). Epidemiological Evidence Supports the Role of Microbial Interactions in Polymicrobial UTI Infections Revealed by In Vitro Research. Antibiotics, 14(10), 1028. https://doi.org/10.3390/antibiotics14101028






