Influence of Florfenicol Treatments on Marine-Sediment Microbiomes: A Metagenomic Study of Bacterial Communities in Proximity to Salmon Aquaculture in Southern Chile
Abstract
1. Introduction
2. Results
2.1. 16S rRNA Sequencing of Marine Sediment Samples
2.2. Shannon Index (H′) Analysis
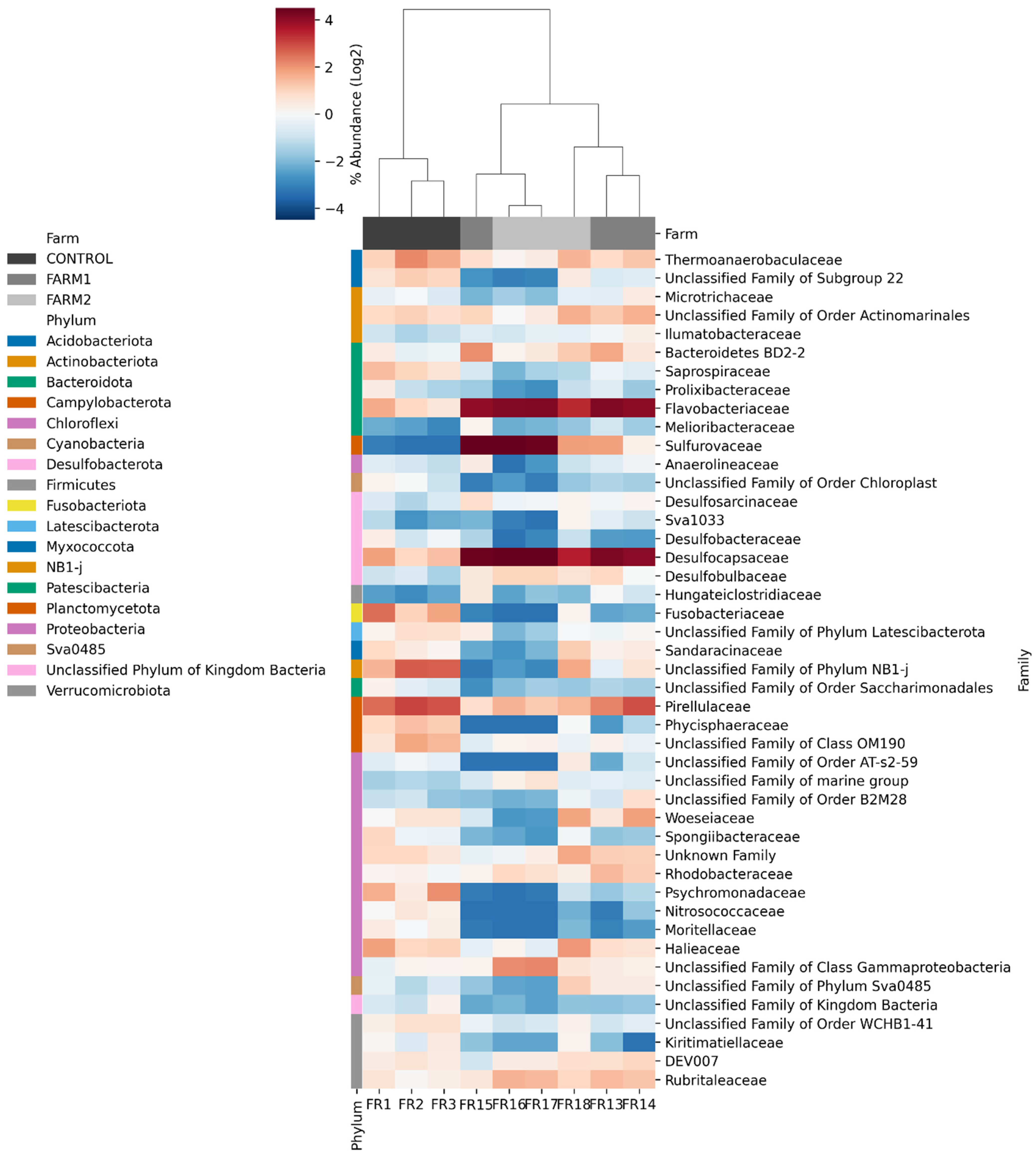
2.3. Relative Abundance Analysis
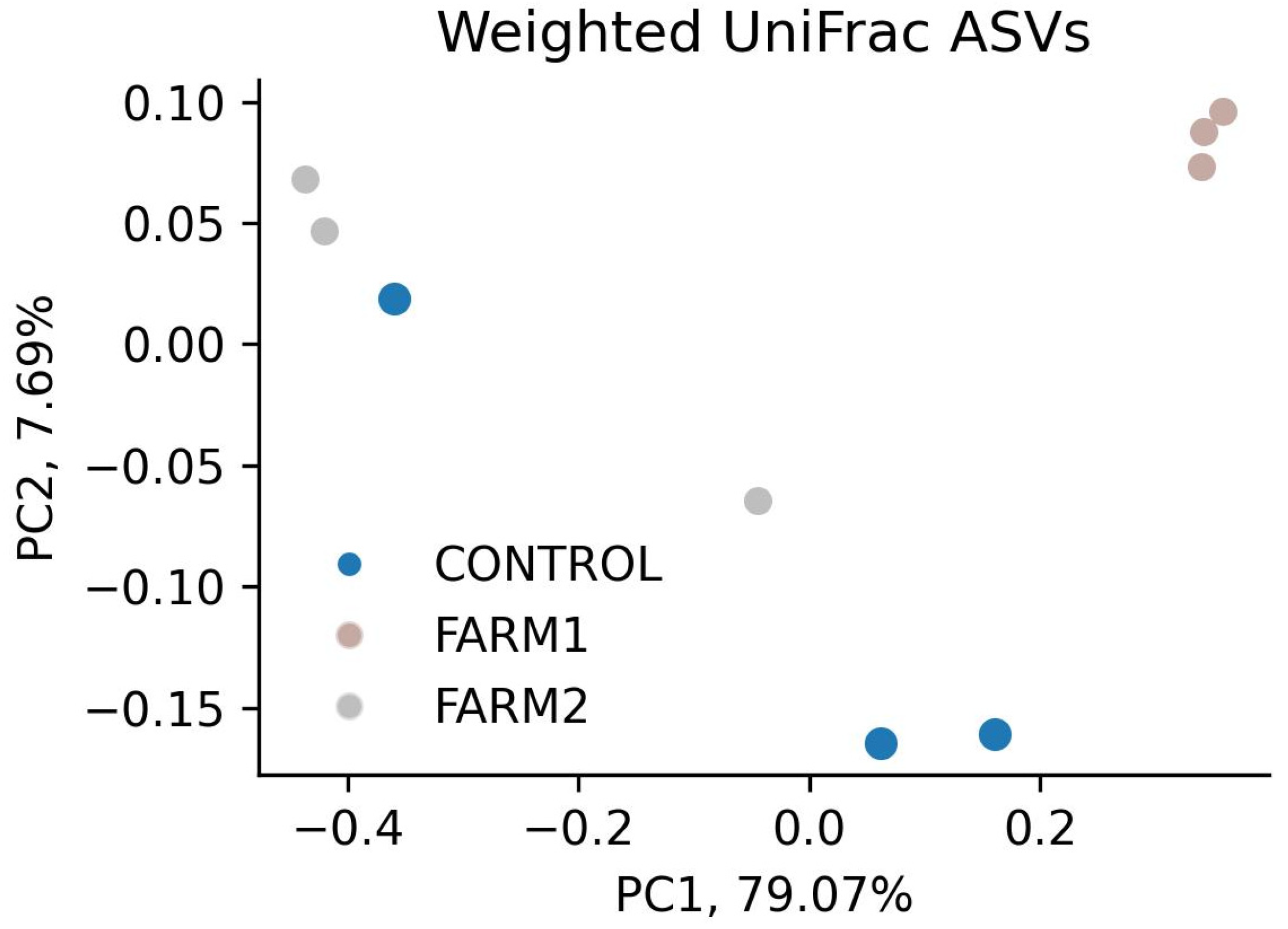
2.4. Weighted UniFrac Analysis
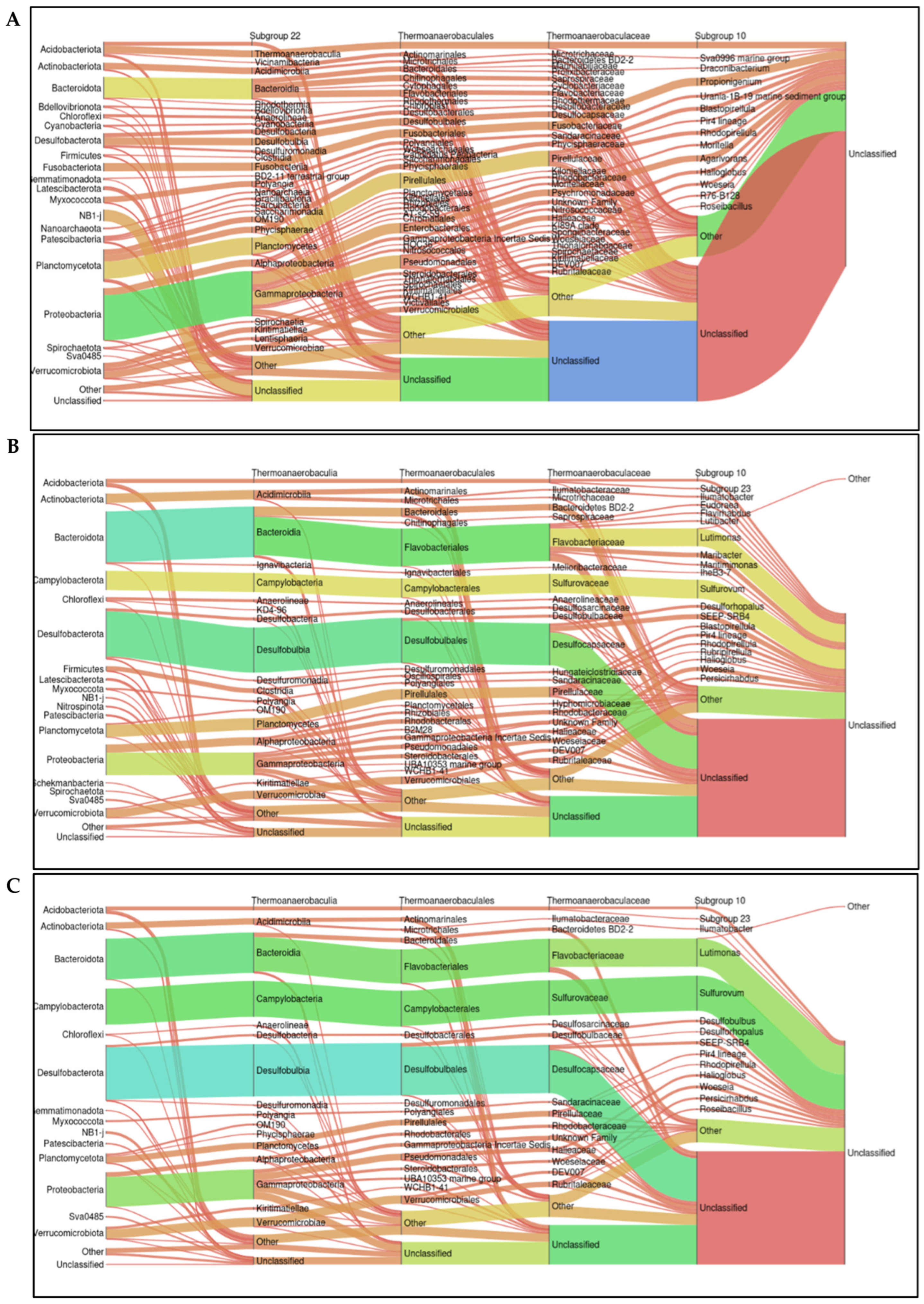
2.5. Relative Abundance at the Taxonomic Level
2.6. Predicted Functional Pathways of Bacterial Communities
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Collection of Sediment Samples from Salmon Farms
4.3. Total DNA Extraction
4.4. Preparation of 16S rDNA Sequencing Libraries and Bioinformatic Analysis
4.5. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vincent, S.G.T.; Jennerjahn, T.C.; Ramasamy, K. Microbial Communities in Coastal Sediments: Structure and Functions, 1st ed.; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Nowicki, M.; DeVries, T.; Siegel, D.A. Quantifying the carbon export and sequestration pathways of the ocean’s biological carbon pump. Glob. Biogeochem. Cycles 2022, 36, e2021GB007083. [Google Scholar] [CrossRef]
- Schultz, P.; Urban, N.R. Effects of bacterial dynamics on organic matter decomposition and nutrient release from sediments: A modeling study. Ecol. Model. 2008, 210, 1–14. [Google Scholar] [CrossRef]
- Grossart, H.P.; Massana, R.; McMahon, K.D.; Walsh, D.A. Linking metagenomics to aquatic microbial ecology and biogeochemical cycles. Limnol. Oceanogr. 2020, 65, S2–S20. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture 2024—Blue Transformation in Action; FAO: Rome, Italy, 2024. [Google Scholar]
- Bondad-Reantaso, M.G.; MacKinnon, B.; Karunasagar, I.; Fridman, S.; Alday-Sanz, V.; Brun, E.; Le Groumellec, M.; Li, A.; Surachetpong, W.; Karunasagar, I.; et al. Review of alternatives to antibiotic use in aquaculture. Rev. Aquacult. 2023, 15, 1421–1451. [Google Scholar] [CrossRef]
- Chen, H.; Ma, K.; Huang, Y.; Yao, Z.; Chu, C. Stable soil microbial functional structure responding to biodiversity loss based on metagenomic evidences. Front. Microbiol. 2021, 12, 716764. [Google Scholar] [CrossRef]
- Iavicoli, I.; Fontana, L.; Agathokleous, E.; Santocono, C.; Russo, F.; Vetrani, I.; Fedele, M.; Calabrese, E.J. Hormetic dose responses induced by antibiotics in bacteria: A phantom menace to be thoroughly evaluated to address the environmental risk and tackle the antibiotic resistance phenomenon. Sci. Total Environ. 2021, 798, 149255. [Google Scholar] [CrossRef]
- Zhang, T.; Ding, Y.; Peng, J.; Dai, Y.; Luo, S.; Liu, W.; Ma, Y. Effects of broad-spectrum antibiotic (florfenicol) on resistance genes and bacterial community structure of water and sediments in an aquatic microcosm model. Antibiotics 2022, 11, 1299. [Google Scholar] [CrossRef]
- Guo, X.; Chen, H.; Tong, Y.; Wu, X.; Tang, C.; Qin, X.; Guo, J.; Li, P.; Wang, Z.; Liu, W.; et al. A review on the antibiotic florfenicol: Occurrence, environmental fate, effects, and health risks. Environ. Res. 2024, 244, 117934. [Google Scholar] [CrossRef]
- Baquero, F.; Coque, T.M.; Martínez, J.L.; Aracil-Gisbert, S.; Lanza, V.F. Gene transmission in the one health microbiosphere and the channels of antimicrobial resistance. Front. Microbiol. 2019, 10, 2892. [Google Scholar] [CrossRef]
- Marmen, S.; Fadeev, E.; Al Ashhab, A.; Benet-Perelberg, A.; Naor, A.; Patil, H.J.; Cytryn, E.; Viner-Mozzini, Y.; Sukenik, A.; Lalzar, M.; et al. Seasonal dynamics are the major driver of microbial diversity and composition in intensive freshwater aquaculture. Front. Microbiol. 2021, 12, 679743. [Google Scholar] [CrossRef]
- Zhang, M.; Pan, L.; Huang, F.; Gao, S.; Su, C.; Zhang, M.; He, Z. Metagenomic analysis of composition, function and cycling processes of microbial community in water, sediment and effluent of Litopenaeus vannamei farming environments under different culture modes. Aquaculture 2019, 506, 280–293. [Google Scholar] [CrossRef]
- Subsecretaría de Pesca y Acuicultura (SUBPESCA). Informe Sectorial de Pesca y Acuicultura; Departamento de análisis sectorial: Valparaíso, Chile, 2024; Available online: https://www.subpesca.cl/portal/616/articles-125035_documento.pdf (accessed on 20 August 2025).
- Servicio Nacional de Pesca y Acuicultura (SERNAPESCA). Informe Sobre Uso de Antimicrobianos y Antiparasitarios en la Salmonicultura Nacional; Subdirección de acuicultura; Departamento de salud Animal: Valparaíso, Chile, 2025; Available online: https://www.sernapesca.cl/app/uploads/2025/07/Informe-Uso-Antimicrobianos-y-Antiparasitarios-Ano-2024.pdf (accessed on 20 August 2025).
- Avendaño-Herrera, R.; Mancilla, M.; Miranda, C.D. Use of antimicrobials in Chilean salmon farming: Facts, myths and perspectives. Rev. Aquacult. 2023, 15, 89–111. [Google Scholar] [CrossRef]
- Nakayama, T.; Hoa, T.T.T.; Harada, K.; Warisaya, M.; Asayama, M.; Hinenoya, A.; Lee, J.W.; Phu, T.M.; Ueda, S.; Sumimura, Y.; et al. Water metagenomic analysis reveals low bacterial diversity and the presence of antimicrobial residues and resistance genes in a river containing wastewater from backyard aquacultures in the Mekong Delta, Vietnam. Environ. Pollut. 2017, 222, 294–306. [Google Scholar] [CrossRef]
- Domínguez, M.; Miranda, C.D.; Fuentes, O.; de la Fuente, M.; Godoy, F.A.; Bello-Toledo, H.; González-Rocha, G. Occurrence of transferable integrons and sul and dfr genes among sulfonamide-and/or trimethoprim-resistant bacteria isolated from Chilean salmonid farms. Front. Microbiol. 2019, 10, 748. [Google Scholar] [CrossRef]
- Ortiz-Severín, J.; Hodar, C.; Stuardo, C.; Aguado-Norese, C.; Maza, F.; González, M.; Cambiazo, V. Impact of salmon farming in the antibiotic resistance and structure of marine bacterial communities from surface seawater of a northern Patagonian area of Chile. Biol. Res. 2024, 57, 84. [Google Scholar] [CrossRef]
- Chen, J.; Yang, Y.; Jiang, X.; Ke, Y.; He, T.; Xie, S. Metagenomic insights into the profile of antibiotic resistomes in sediments of aquaculture wastewater treatment system. J. Environ. Sci. 2022, 113, 345–355. [Google Scholar] [CrossRef]
- Xiong, W.; Sun, Y.; Zhang, T.; Ding, X.; Li, Y.; Wang, M.; Zeng, Z. Antibiotics, antibiotic resistance genes, and bacterial community composition in fresh water aquaculture environment in China. Microb. Ecol. 2015, 70, 425–432. [Google Scholar] [CrossRef]
- Cheng, C.; Ying, Y.; Zhou, D.; Zhu, L.; Lu, J.; Li, A.; Bao, Q.; Zhu, M. RamA, a transcriptional regulator conferring florfenicol resistance in Leclercia adecarboxylata R25. Folia Microbiol. 2020, 65, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Thomassen, G.; Reiche, T.; Tennfjord, C.; Mehli, L. Antibiotic resistance properties among Pseudomonas spp. associated with salmon processing environments. Microorganisms 2022, 10, 1420. [Google Scholar] [CrossRef]
- Ojasanya, R.; Gardner, I.; Groman, D.; Saksida, S.; Saab, M.; Thakur, K. Antimicrobial susceptibility profiles of bacteria commonly isolated from farmed salmonids in Atlantic Canada (2000–2021). Vet. Sci. 2022, 9, 159. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Liao, C.; Terhune, J.; Wang, L. Impacts of florfenicol on the microbiota landscape and resistome as revealed by metagenomic analysis. Microbiome 2019, 7, 155. [Google Scholar] [CrossRef]
- Ye, M.; Chen, G.; Du, Z. Effects of antibiotics on the bacterial community, metabolic functions and antibiotic resistance genes in mariculture sediments during enrichment culturing. J. Mar. Sci. Eng. 2020, 8, 604. [Google Scholar] [CrossRef]
- Poole, K. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 2012, 67, 2069–2089. [Google Scholar] [CrossRef]
- Zhou, Q.; Xie, X.; Feng, F.; Huang, S.; Sun, Y. Impact of acyl-homoserine lactones on the response of nitrogen cycling in sediment to florfenicol stress. Sci. Total Environ. 2021, 785, 147294. [Google Scholar] [CrossRef]
- Fierer, N.; Lennon, J. The generation and maintenance of diversity in microbial communities. Am. J. Bot. 2011, 98, 439–448. [Google Scholar] [CrossRef]
- Cao, H.; Liu, M.; Tong, M.; Jiang, S.; Chow, K.; To, K.; Tse, C.; Ho, P. Comprehensive investigation of antibiotic resistance gene content in cfiA-harboring Bacteroides fragilis isolates of human and animal origins by whole genome sequencing. Int. J. Med. Microbiol. 2022, 312, 151559. [Google Scholar] [CrossRef] [PubMed]
- Heckman, T.; Yazdi, Z.; Pomaranski, E.; De Alexandre Sebastião, F.; Mukkatira, K.; Vuglar, B.; Cain, K.; Loch, T.; Soto, E. Atypical flavobacteria recovered from diseased fish in the Western United States. Front. Cell. Infect. Microbiol. 2023, 13, 1149032. [Google Scholar] [CrossRef]
- Ozuolmez, D.; Stams, A.; Plugge, C. Propionate converting anaerobic microbial communities enriched from distinct biogeochemical zones of Aarhus Bay, Denmark under sulfidogenic and methanogenic conditions. Microorganisms 2020, 8, 394. [Google Scholar] [CrossRef] [PubMed]
- Slobodkin, A.; Ratnikova, N.; Slobodkina, G.; Klyukina, A.; Chernyh, N.; Merkel, A. Composition and metabolic potential of Fe(III)-reducing enrichment cultures of methanotrophic ANME-2a Archaea and associated Bacteria. Microorganisms 2023, 11, 555. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Han, T.; Xu, S.; Huang, H.; Qi, Z.; Zhu, Q. Bacterial community responses to the redox profile changes of mariculture sediment. Mar. Pollut. Bull. 2021, 166, 112250. [Google Scholar] [CrossRef]
- Zhang, T.; Peng, J.; Dai, Y.; Xie, X.; Luo, S.; Ding, Y.; Ma, Y. Effect of florfenicol on nirS-type denitrifying communities structure of water in an aquatic microcosm model. Front. Vet. Sci. 2023, 10, 1205394. [Google Scholar] [CrossRef]
- Verner-Jeffreys, D.; Brazier, T.; Perez, R.; Ryder, D.; Card, R.; Welch, T.; Hoare, R.; Ngo, T.; Mclaren, N.; Ellis, R.; et al. Detection of the florfenicol resistance gene floR in Chryseobacterium isolates from rainbow trout. Exception to the general rule? FEMS Microbiol. Ecol. 2017, 93, fix015. [Google Scholar] [CrossRef]
- Zárate, A.; Molina, V.; Valdés, J.; Icaza, G.; Vega, S.E.; Castillo, A.; Ugalde, J.A.; Dorador, C. Spatial co-occurrence patterns of benthic microbial assemblage in response to trace metals in the Atacama Desert Coastline. Front. Microbiol. 2023, 13, 1020491. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Yan, K.; Kong, L.; Gai, Y.; Jin, T.; He, Y.; Wang, Y.; Chen, F.; Lin, L.; Lin, Z.; et al. Metabolic tuning of a stable microbial community in the surface oligotrophic Indian Ocean revealed by integrated meta-omics. Mar. Life Sci. Technol. 2022, 4, 277–290. [Google Scholar] [CrossRef]
- Wirtz, M.; Droux, M. Synthesis of the sulfur amino acids: Cysteine and methionine. Photosynth. Res. 2005, 86, 345–362. [Google Scholar] [CrossRef]
- Choi, A.; Lee, T.; Cho, H.; Lee, W.; Hyun, J. Shifts in benthic bacterial communities associated with farming stages and a microbiological proxy for assessing sulfidic sediment conditions at fish farms. Mar. Pollut. Bull. 2022, 178, 113603. [Google Scholar] [CrossRef] [PubMed]
- Brewster, J.; Pachl, P.; McKellar, J.; Selmer, M.; Squire, C.; Patrick, W. Structures and kinetics of Thermotoga maritima MetY reveal new insights into the predominant sulfurylation enzyme of bacterial methionine biosynthesis. J. Biol. Chem. 2021, 296, 100797. [Google Scholar] [CrossRef]
- Anton, B.P.; Roberts, R.J. Beyond restriction modification: Epigenomic roles of DNA methylation in prokaryotes. Ann. Rev. Microbiol. 2021, 75, 129–149. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Lu, P.; Wang, R.; Liu, X.; Yuan, T. Sulfur: A neglected driver of the increased abundance of antibiotic resistance genes in agricultural reclaimed subsidence land located in coal mines with high phreatic water levels. Heliyon 2023, 9, e14364. [Google Scholar] [CrossRef]
- Sekar, K.; Linker, S.; Nguyen, J.; Gruenhagen, A.; Stocker, R.; Sauer, U. Bacterial glycogen provides short-term benefits in changing environments. Appl. Environ. Microbiol. 2019, 86, e00049-20. [Google Scholar] [CrossRef]
- Machado, D.; Maistrenko, O.; Andrejev, S.; Kim, Y.; Bork, P.; Patil, K.; Patil, K. Polarization of microbial communities between competitive and cooperative metabolism. Nat. Ecol. Evol. 2020, 5, 195–203. [Google Scholar] [CrossRef]
- Won, N.I.; Kim, K.H.; Kang, J.H.; Park, S.R.; Lee, H.J. Exploring the impacts of anthropogenic disturbance on seawater and sediment microbial communities in Korean coastal waters using metagenomics analysis. Int. J. Environ. Res. Public Health 2017, 14, 130. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. NaïveBayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lynch, S.; Thomson, P.; Santibañez, R.; Avendaño-Herrera, R. Influence of Florfenicol Treatments on Marine-Sediment Microbiomes: A Metagenomic Study of Bacterial Communities in Proximity to Salmon Aquaculture in Southern Chile. Antibiotics 2025, 14, 1016. https://doi.org/10.3390/antibiotics14101016
Lynch S, Thomson P, Santibañez R, Avendaño-Herrera R. Influence of Florfenicol Treatments on Marine-Sediment Microbiomes: A Metagenomic Study of Bacterial Communities in Proximity to Salmon Aquaculture in Southern Chile. Antibiotics. 2025; 14(10):1016. https://doi.org/10.3390/antibiotics14101016
Chicago/Turabian StyleLynch, Sergio, Pamela Thomson, Rodrigo Santibañez, and Ruben Avendaño-Herrera. 2025. "Influence of Florfenicol Treatments on Marine-Sediment Microbiomes: A Metagenomic Study of Bacterial Communities in Proximity to Salmon Aquaculture in Southern Chile" Antibiotics 14, no. 10: 1016. https://doi.org/10.3390/antibiotics14101016
APA StyleLynch, S., Thomson, P., Santibañez, R., & Avendaño-Herrera, R. (2025). Influence of Florfenicol Treatments on Marine-Sediment Microbiomes: A Metagenomic Study of Bacterial Communities in Proximity to Salmon Aquaculture in Southern Chile. Antibiotics, 14(10), 1016. https://doi.org/10.3390/antibiotics14101016





