Abstract
Objective: In the present study, we used phenotypic and molecular methods to determine susceptibility to oxacillin in coagulase-negative staphylococci (CoNS) and estimate the prevalence of strains with low-level resistance to oxacillin, mecA-positive oxacillin-susceptible methicillin-resistant (OS-MRCoNS), and borderline oxacillin-resistant (BORCoNS). Methods: One hundred one CoNS strains were screened for oxacillin and cefoxitin susceptibility using phenotypic (disk diffusion, agar dilution, latex agglutination, and chromagar) and molecular (detection of mecA, mecB, and mecC) methods. Staphylococcal cassette chromosome mec (SCCmec) typing was performed. Results: Sixteen (15.8%) CoNS strains were mecA-positive, and 85 (84.2%) were mec-negative. Seven (6.9%) were classified as OS-MRCoNS, accounting for 43.8% of all mecA-positive strains. Twelve (11.9%) mec-negative strains were classified as borderline oxacillin resistant (BORCoNS). Compared with MRCoNS and BORCoNS, OS-MRCoNS strains demonstrated lower resistance to non-beta-lactams. SCCmec type I cassette was predominant. The disc-diffusion method with oxacillin accurately predicted OS-MRCoNS strains but did not provide reliable results for BORCoNS strains. Meanwhile, the latex agglutination test and CHROMagar culture accurately identified BORCoNS but not OS-MRCoNS. Conclusions: Finally, our findings imply that the recognition of methicillin resistance in CoNS requires a meticulous approach and that further research is needed to develop unified laboratory diagnostic algorithms to prevent the misreporting of borderline CoNS.
1. Introduction
Coagulase-negative staphylococci (CoNS) are a heterogeneous group comprising over 80 species. In contrast to coagulase-positive staphylococci, such as Staphylococcus aureus, CoNS have been considered less pathogenic or non-pathogenic for years. Most CoNS species colonize human skin and mucosal membranes and are rarely involved in clinically symptomatic infections [1,2]. However, given new groups of immunocompromised patients and the growing complexity of medical procedures, CoNS have emerged as a leading group of nosocomial pathogens nowadays [3]. CoNS often causes foreign body-associated infections and infections in preterm neonates [1,4,5]. While the infections are usually subacute and associated with only mild inflammation, they constitute a substantial clinical burden due to their widespread occurrence and problems with choosing an effective antibiotic therapy. Acquired resistance of CNoS to antimicrobial agents is a common problem and spreads quickly between species [5].
Among multiple mechanisms of acquired drug resistance displayed by CoNS, the resistance to beta-lactam antibiotics appears the most important from a clinical and epidemiological perspective [6]. The two principal mechanisms of staphylococcal resistance to beta-lactams include the production of enzymes, beta-lactamases, and mutated penicillin-binding proteins (PBP) [7]. Due to the generation of new PBP, with low affinity to beta-lactams, staphylococci become resistant to all beta-lactam antibiotics used in clinical practice nowadays, except for fifth-generation cephalosporins (ceftaroline and ceftobiprole) [8]. This type of resistance is referred to as methicillin or oxacillin resistance, as it was first documented in the case of methicillin, the first semisynthetic anti-staphylococcal penicillin [9]. CoNS displaying this type of resistance are called methicillin-resistant coagulase-negative staphylococci (MRCoNS). Methicillin resistance is determined by the presence of acquired mecA, mecB, or mecC genes (previously known as mecALGA251) that encode additional penicillin-binding proteins, PBP2a (PBP2’), PBP2b, or PBP2c, respectively [10,11]. mecA and mecC are chromosomal genes that are part of the staphylococcal cassette chromosome mec (SCCmec) [12].
Traditional phenotype-based methods have reduced sensitivity and specificity for the recognition of methicillin resistance in CoNS. In some cases, identifying the resistance can be challenging and inconclusive. For example, emerging oxacillin-susceptible methicillin-resistant S. aureus (OS-MRSA) strains were reported to carry mecA or mecC genes despite oxacillin MICs corresponding to sensitivity (up to 2 mg/L). Furthermore, some borderline oxacillin-resistant S. aureus (BORSA) strains may present with borderline oxacillin MICs yet not harbor the mec genes. Usually, the BORSA strains display low oxacillin (2–12 mg/L) and cefoxitin MICs (4–8 mg/L). On top of that, some authors use the term BORSA to describe both groups (OS-MRSA and BORSA) mentioned above, which makes their appropriate identification challenging and associated with a high risk of bias [13,14]. Meanwhile, accurate identification of methicillin (oxacillin) resistance is crucial for effective epidemiological intervention (eradication of MRSA) and implementation of a successful anti-staphylococcal therapy.
In principle, neither EUCAST nor CLSI guidelines recommend systematic screening of Staphylococcus spp. strains for borderline resistance to methicillin [15,16]. However, EUCAST recommends testing for the BORSA phenotype if methicillin resistance screening with cefoxitin disc (30 μg) yields a negative result, but oxacillin MIC exceeds 4 mg/L whenever clinical indications exist [15].
In the present study, we used phenotypic and molecular methods to determine susceptibility to oxacillin in various coagulase-negative staphylococci (CoNS) species and the occurrence of strains with low-level resistance to oxacillin, mecA-positive oxacillin-susceptible methicillin-resistant (OS-MRCoNS), and borderline oxacillin-resistant (BORCoNS) strains.
2. Results
Of the 101 coagulase-negative staphylococci, 16 (15.8%) strains were mecA-positive, and 85 (84.2%) did not carry any mec gene (mecA, mecB, or mecC) (Figure 1).
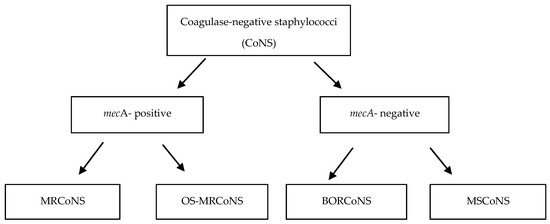
Figure 1.
Detection of methicillin resistance in coagulase-negative staphylococci (CoNS).
A comparative analysis of different methicillin resistance tests showed that oxacillin-based methods were highly sensitive in detecting mecA-positive phenotypes. Oxacillin disk diffusion and serial dilution methods showed 100% and 81.3% sensitivity, respectively, with PCR for mecA as a reference. The cefoxitin disk-diffusion test provided lower sensitivity (68.8%) but higher specificity (94.1%) than the oxacillin-based methods. Similarly, CHROMagar culture yielded lower sensitivity and higher specificity, 61.5% and 100%, respectively. The disc-diffusion method with oxacillin accurately predicted OS-MRCoNS strains but did not provide reliable results for BORCoNS strains. Meanwhile, the latex agglutination test and CHROMagar culture accurately identified BORCoNS but not OS-MRCoNS (Table 1).

Table 1.
Screening of methicillin resistance in coagulase-negative staphylococci (CoNS) strains.
2.1. Oxacillin-Susceptible Methicillin-Resistant Coagulase-Negative Staphylococci (OS-MRCoNS)
Seven (6.9%) mecA-positive strains demonstrated oxacillin MICs ≤ 0.5 mg/L or sensitivity to cefoxitin and/or oxacillin in the disk diffusion method. These strains, classified as oxacillin-susceptible strains (OS-MRCoNS), represented 43.8% of all mecA-positive strains. The OS-MRCoNS included S. saprophyticus (n = 3), S. epidermidis (n = 1), S. haemolyticus (n = 1), S. pasteurii (n = 1), and S. warneri (n = 1) species. All S. saprophyticus strains were resistant to oxacillin in the disk diffusion method but sensitive to cefoxitin. Other discrepancies in the results obtained with phenotypic methods are summarized in Table 2. Three OS-MRCoNS strains (S. epidermidis, S. pasteurii, and S. saprophyticus) carried type I SCCmec, one strain harbored type II SCCmec, and one strain presented with a unique type of SCCmec cassette. Three strains yielded false-negative results in the latex agglutination test, and four strains failed to grow on a CHROMagar (Table 2, Figure 1).

Table 2.
Phenotypic and genotypic characteristics of mecA-positive oxacillin-susceptible methicillin-resistant coagulase-negative staphylococci (OS-MRCoNS).
2.2. Methicillin-Resistant Coagulase-Negative Staphylococci (MRCoNS)
The remaining nine mecA-positive strains with oxacillin MICs of 1–24 µg/mL were classified as MRCoNS strains. All but one strain from this group produced PBP2a protein and demonstrated resistance to cefoxitin and oxacillin in the disc-diffusion method. S. haemolyticus (n = 5) was the most common species among the MRCoNS strains, followed by S. hominis, S. epidermidis, S. saprophyticus, and S. warneri (n = 1 each). The MRCoNS strains harbored predominantly type V (n = 3) and type I SCCmec (n = 1), with type II SCCmec carried by only one strain. All MRCoNS were correctly identified by latex agglutination test, and all but one were grown on a CHROMagar (Table 3, Figure 1).

Table 3.
Phenotypic and genotypic characteristics of mecA-positive methicillin-resistant coagulase-negative staphylococci (MRCoNS).
2.3. Borderline Oxacillin-Resistant Coagulase-Negative Staphylococci (BORCoNS)
Twelve (11.9%) mec-negative strains with oxacillin MICs between 0.5 µg/mL and 2 µg/mL were classified as borderline oxacillin-resistant (BORCoNS). These strains accounted for 14.1% of all mecA-negative strains. Although none of the strains carried the mecA, mecB or mecC genes or produced PBP2a protein, almost all of them (91.6%) were resistant to oxacillin, and 41.6% displayed resistance to cefoxitin in the disc-diffusion method. The BORCoNS strains included S. warneri (n = 4), S. equorum (n = 2), S. saprophyticus (n = 2), S. succinus (n = 2), S. cohnii (n = 1), and S. xylosus (n = 1) species. S. warneri was the most frequently detected species among the BORCoNS identified, with all strains having oxacillin MICs of 0.5 µg/mL. Other discrepancies in the results obtained using phenotypic methods are summarized in Table 3. Moreover, the strains neither yielded positive results in the latex agglutination test nor grew on a CHROMagar (Table 4, Figure 1).

Table 4.
Phenotypic and genotypic characteristics of borderline oxacillin-resistant coagulase-negative staphylococci strains (BORCoNS).
2.4. Antimicrobial Susceptibility Tests
All BORCoNS and MRCoNS strains produced penicillinase. Resistance to aminoglycosides and macrolides predominated in all groups of strains, displayed by approximately 50% of the isolates. Compared with MRCoNS and BORCoNS, OS-MRCoNS strains presented lower resistance to non-beta-lactam antibiotics, such as chloramphenicol, ciprofloxacin, clindamycin, and gentamycin. BORCoNS strains were more often resistant to tetracyclines (41.7%). All tested CoNS were susceptible to vancomycin, daptomycin, and linezolid (Table 5).

Table 5.
Comparison of antimicrobial-resistance of OS-MRCoNS MRCoNS and BORCoNS strains.
3. Discussion
Although CoNS have fewer virulence factors than S. aureus, they still play a significant role in opportunistic nosocomial infections, primarily due to the frequent occurrence of strains that constitute a reservoir of antimicrobial resistance [3]. Therefore, continuous attempts are undertaken at various levels of healthcare to accurately evaluate and monitor drug resistance, especially resistance to oxacillin, in CoNS [17]. CoNS strains with borderline resistance to oxacillin (OS-MRCoNS and BORCoNS) constitute a challenge for several reasons. First, EUCAST and CLSI guidelines defining laboratory standards for interpreting bacterial drug resistance are incomplete and inconclusive regarding CoNS [15,16]. Second, the strains with borderline resistance to oxacillin are difficult to detect and, as such, may be misreported as beta-lactam-sensitive or -resistant strains, mainly based on screening with phenotypic methods. This may result in prescribing antibiotics that are ineffective and eventually lead to treatment failure [18,19].
According to the recent EUCAST and CLSI guidelines, the disc-diffusion method with cefoxitin (30 µg) is a routine assay to identify resistance to oxacillin. This method is also considered a better predictor of mecA presence in most Staphylococcus spp. than the disc-diffusion method with oxacillin (1 μg) [15,16]. EUCAST recommends using the cefoxitin disc-diffusion method to identify nearly all CoNS species, with the differentiation between S. epidermidis and non-epidermidis strains based on species-specific zone diameters. The only exception pertains to S. pseudintermedius, in which EUCAST recommends using the oxacillin disc-diffusion test [15]. Similarly, CLSI advocates using the disc-diffusion method with cefoxitin for the identification of most species except for S. pseudintermedius and S. schleiferi, in the case of which oxacillin disc-diffusion test is recommended [16]. Regarding the borderline oxacillin MIC, EUCAST defined it at >2 mg/L for S. aureus and S. lugdunensis and at >0.25 for S. epidermidis. Unfortunately, the EUCAST guidelines do not specify borderline MICs for other CoNS species [15]. Meanwhile, CLSI defined borderline oxacillin MIC at ≥4 mg/L for S. aureus and S. lugdunensis and at ≥1 for the other CoNS, and this is the latter value that was used in our present study [16].
Due to the lack of a unified definition and diagnostic algorithm, microbiological laboratories face the problem of identifying and reporting CoNS strains with borderline resistance to oxacillin [20]. CLSI does not address the issue of borderline strains in their guidelines, whereas the EUCAST guidelines contain solely recommendations for borderline S. aureus (BORSA). Neither CLSI nor EUCAST guidelines provide recommendations for detecting borderline CoNS strains and further therapeutic approaches [15,16]. The lack of respective guidelines has been reflected by a substantial variety of interpretations and reporting of borderline resistance to oxacillin by various laboratories. Similarly, published research papers differ considerably in terms of used nomenclature, reported rates of borderline resistance, and approaches to treating infections caused by borderline-resistant pathogens.
mecA-positive oxacillin-sensitive S. aureus (OS-MRSA) strains have been reported in various geographical regions, including Europe, the United States, Brazil, Africa, Iran, India, and China [21,22,23,24,25,26]. The isolation rates of OS-MRSA from humans are typically estimated at a few to several percent (2–14.9%). The nearly 7% isolation rate documented in our present study fits within this range, but it needs to be emphasized that our current knowledge of the prevalence of OS-MRCoNS is considerably limited. In our present study, OS-MRCoNS strains belonged to various species, with the most common being S. saprophyticus, considerably less frequently found among classic MRCoNS, i.e., mecA-positive oxacillin-resistant strains. All OS-MR strains of S. saprophyticus were shown to be sensitive to cefoxitin on the disc-diffusion method and, hence, would be misreported as methicillin-sensitive (MSCoNS) on routine laboratory testing. Additionally, these strains yielded false negative results on CHROMagar, another factor contributing to their potential misidentification. Thus, despite being considered the most accurate predictor of mecA-positive strains, including S. saprophyticus strains, the disc-diffusion method with cefoxitin did not produce reliable results in our present study. Meanwhile, the disc-diffusion method with oxacillin accurately predicted the antibiotic resistance profile in most borderline strains. Misreporting OS-MRCoNS as methicillin-sensitive in routine laboratory practice may lead to failure in antibiotic therapy. Despite published reports about the effectiveness of beta-lactam antibiotics in treating experimentally induced infections with strains being borderline resistant to oxacillin, the application of this therapeutic option raises many concerns [27,28]. The resistance of OS-MRSA to oxacillin is considered inducible. It is postulated that after being exposed to this antibiotic, some OS-MRSA may transform into highly resistant clones insensitive to beta-lactams. The results of a recent study published by Gostev et al. suggest that the fast transformation of OS-MRSA to MRSA results from the preexistence of a small bacterial subpopulation with high MICs rather than from the selection of new mutants [29]. Variable levels of staphylococcal resistance to oxacillin may result from an instability in the DNA fragment that determines the resistance expression. The presence of a bacterial subpopulation with high MICs poses a high risk of transforming OS-MRSA into MRSA with a high level of resistance to beta-lactam antibiotics; this puts into question the usefulness of beta-lactams in the treatment of OS-MRSA infections. Thus, according to most authors, infections caused by OS-MRSA should be treated with antibiotics that are effective against MRSA, such as linezolid or vancomycin, rather than beta-lactams [27,29]. While, to the best of our knowledge, none of the published studies analyzed the problem in question with regards to OS-MRCoNS species, the same therapeutic approach seems applicable, especially given that compared to classic MRCoNS, the OS-MRCoNS isolated in our present study were more sensitive to non-beta-lactam antibiotics.
The second most frequently isolated group of strains with borderline resistance to oxacillin in the present study were BORCoNS (12%), i.e., mecA-negative strains showing resistance to oxacillin on phenotypic testing. According to the literature, the isolation rates of S. aureus with borderline resistance to oxacillin (BORSA) in a hospital setting vary from 1% to 12.5% or are even higher [19,30,31,32]. According to Khorvash et al., mec-negative oxacillin-resistant strains constituted up to 25.5% of all MRSA [33]. In another study, the isolation rate of BORSA from clinical material reached up to 50% [34]. The discrepancies in the isolation rates probably reflect the lack of unified diagnostic criteria for BORSA. The problem is even more evident in the case of BORCoNS strains that may be misidentified as methicillin-resistant and, thus, as non-eligible for beta-lactam antibiotic therapy. On the one hand, high-dose beta-lactams are considered an effective therapeutic option in non-complicated BORSA infections. On the other hand, Konstantinovski et al. demonstrated that such a therapy may be ineffective in severe infections, such as endocarditis, whereby beta-lactams should be substituted by vancomycin [35]. Similarly, Skinner et al. reported on a mecA-negative S. aureus strain with oxacillin MIC of 12 µg/mL, isolated from a patient with endocarditis, that did not respond to high-dose oxacillin therapy yet was successfully eradicated with vancomycin [36]. The BORCoNS isolated in our present study showed similar sensitivity to non-beta-lactam antibiotics, including vancomycin, as MRCoNS. Our findings imply that managing infections caused by mec-negative strains with borderline resistance to oxacillin requires a meticulous approach. The treatment protocol should be based on the results of antibiotic resistance testing, including oxacillin MIC.
Many SCCmec cassettes (I-VII) in coagulase-negative staphylococci were identified and reported. However, new combinations of the ccr and mec genes are detected; thus, unambiguous and appropriate identification is challenging. Additionally, it is indicated that those seven types described so far are not all types distributed worldwide. In our study, the SCCmec type I cassette was predominant. The occurrence of type I in many CoNS species has been described previously. The SCCmec type V was only found in S. haemolyticus. According to the literature, S. haemolyticus is a reservoir of SCCmec type V cassettes [37]. In our study, there were six strains with unknown SCCmec types, both OS-MRCoNS and MRCoNS. Many SCCmec cassettes found in CoNS cannot be classified as existing types, as they most likely contain undescribed allotypes or a mixture of existing ones [12].
According to EUCAST and CLSI guidelines, detection of the mec gene by PCR is considered a gold standard in evaluating methicillin resistance. It should be applied whenever the results of phenotypic testing are inconclusive, both for S. aureus and other Staphylococcus species. In routine screening, the disc-diffusion method with cefoxitin is considered the most accurate predictor of mec-positive staphylococci [15,16]. However, this method did not yield the desired predictive values in the present study, providing merely 68.8% sensitivity versus 100% sensitivity for the disc-diffusion method with oxacillin. Secchi previously reported the equally high sensitivity of the disc-diffusion method with oxacillin, whereas according to Swenson, this method produced a 94% sensitivity yet lower specificity (79%) [38,39]. Other authors also postulated using diagnostic criteria other than the disc-diffusion method with cefoxitin to minimize the risk of overlooking methicillin-resistant strains. According to Pinheiro et al., using multiple diagnostic criteria is remarkably advisable in the case of CoNS, which are more difficult to detect because of their heterogeneity and borderline resistance to oxacillin [17]. In our study, CHROMagar culture was the least sensitive method to detect mec-positive strains (61.5%). Other authors reported higher sensitivity to this test, but their studies involved S. aureus rather than CoNS and used CHROMagar from other manufacturers [40]. Notably, phenotypic methods, such as CHROMagar and latex agglutination method, accurately identified BORCoNS and MRCoNS as methicillin-sensitive and -resistant, respectively, providing 100% specificity. Identifying the resistance profile in OS-MRCoNS was more challenging, as the latex agglutination test and CHROMagar culture yielded false negative results. According to Nair, the sensitivity of the latex agglutination test was high (98.9%), with accurate positive results obtained for all OS-MRSA isolates [40]. The results of our present study and findings reported by other authors point to discrepancies between various tests and problems in the identification of borderline strains, implying that further research is needed before any harmonized recommendations could be published on this subject matter.
In summary, the present study demonstrated that CoNS strains with low levels of resistance to oxacillin may constitute a few to several percent of CoNS isolates and belong to various species. Our findings imply that the recognition of methicillin resistance in these strains requires a meticulous approach. The disc-diffusion method with oxacillin accurately predicted OS-MRCoNS strains but did not provide reliable results for BORCoNS strains. Meanwhile, the latex agglutination test and CHROMagar culture accurately identified BORCoNS but not OS-MRCoNS. Thus, the diagnostic protocol should be based not only on the results of phenotypic methods. The results presented herein warrant further research on borderline CoNS to develop unified laboratory diagnostic algorithms and to prevent the misidentification of these strains and antibiotic therapy failure.
4. Materials and Methods
4.1. Bacterial Strains
A total of 101 non-duplicate coagulase-negative staphylococci strains originating from the bacterial collection of the Department of Oral Microbiology, Medical University of Gdansk (MUG), were analyzed. The strains were isolated between 2016 and 2017, mainly from oral specimens during routine clinical laboratory procedures, not specifically for the present study. The collection included twelve CoNS species: Staphylococcus warneri (n = 43), S. haemolyticus (n = 12), S. saprophyticus (n = 9), S. epidermidis (n = 9), S. pasteurii (n = 8), S. hominis (n = 5), S. xylosus (n = 6), S. equorum (n = 3), S. kloosii (n = 2), S. succinus (n = 2), S. cohnii (n = 1), and S. simulans (n = 1). All strains were preliminarily identified using conventional methods. Identification of the strain at a species level was performed using the API system (bioMeriux, Marcy-l’Etoile, France) and confirmed by the matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) method (Bruker Daltonics, Bremen, Germany). The reference strains S. aureus ATCC 43300 and S. aureus ATCC 29213 were used as positive and negative controls, respectively. The isolates were stored at −80 °C in tryptic soy broth (Becton Dickinson, Franklin Lakes, NJ, USA) containing 20% glycerol.
4.2. Phenotypic Method of Methicillin Resistance Detection
4.2.1. Disk Diffusion Method (DDM)
Methicillin resistance was identified using cefoxitin (30 μg) and oxacillin (1 μg) by the disk diffusion method on Mueller–Hinton agar (Becton Dickinson, Franklin Lakes, NJ, USA) per CLSI recommendations [16]. MHA plates were incubated at 35 to 37 °C at 16 to 18 h for oxacillin and 24 h for cefoxitin. Both the oxacillin and cefoxitin zones of inhibition were read using reflected light.
4.2.2. Agar Dilution Method (ADM)
Oxacillin MICs were determined by agar dilution method according to CLSI recommendations [M07-A10]. All strains were plated on Mueller–Hinton agar supplemented with 2% sodium chloride. Accordingly, oxacillin was added to the media in concentrations in double dilution from 64 mg/L to 0.125 mg/L.
4.2.3. PBP2a Latex Agglutination Test
To detect PBP2a expression, OXOID PBP2’ Latex Agglutination Test Kit (Basingstoke, UK) was used according to the manufacturer’s instructions. For mecA-positive strains with latex agglutination negative, the assay was repeated as recommended by the manufacturer following overnight oxacillin induction.
4.2.4. CHROMagar MR
CHROMagar MR (GrasoBiotech, Starogard Gd., Poland) was used to screen methicillin-resistant strains. A 0.5 McFarland suspension of the bacterial colony prepared from a 24 h culture on 5% sheep blood agar was streaked on the CHROMagar MRSA. After 24 h of aerobic incubation at 35 °C, the grown blue colonies were indicated as MR strains.
4.3. Molecular Analysis of Methicillin Resistance
4.3.1. Genomic DNA Extraction
For genomic DNA extraction, strains were grown for 20 h at 37 °C on blood agar plates. A full inoculation loop of 10 μL of bacterial colonies was homogenized with a TissueLyser II (Qiagen, Germantown, MD, USA). The Qiagen DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD, USA) was used for genomic DNA extraction. The subsequent steps were performed according to the manufacturer’s instructions. Purified DNA was stored at −20 °C.
4.3.2. Detection of mecA, mecB, and mecC Genes
To detect the mecA gene, primers mecA-f1: TGGCCAATACAGGAACAGCA; mecA-r1: ACGTTGTAACCACCCCAAGA were designed for S. cohnii, S. epidermidis, S. equorum, S. haemolyticus, S. hominis, S. saprophyticus, S. succinus, S. warneri, and S. xylosus species (based on selected species with following GenBank accession numbers: NZ_CP073878.1; NZ_CP073863.1; CP045187.2; NZ_CP035541.1; NZ_CP065797.1; NZ_CP093539.1; CP014567.1). For mecA-negative strains, mecC and mecB genes were tested by simplex PCR according to Ito et al. and Becker et al., respectively [41,42].
4.3.3. Detection of SCCmec Cassettes
The SCCmec cassettes were typed using two independent methods described by Milheirico et al. [43] and Kondo et al. [44], with the USA300 3956/13 strain as a positive control. The PCR products were resolved by electrophoresis, and band patterns were analyzed.
4.3.4. Antimicrobial Susceptibility Testing
The susceptibility of CoNS isolates to antimicrobial agents was determined by the disk diffusion method, according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST). The following antimicrobial agents were used for the test: amoxicillin/clavulanic acid, chloramphenicol, ciprofloxacin, clindamycin, erythromycin, gentamicin, penicillin, tetracycline, trimethoprim/sulfamethoxazole (Oxoid, Basingstoke, UK). The susceptibility to vancomycin, daptomycin, and linezolid was determined by E-test. The multidrug-resistant (MDR) strains showed resistance to at least three classes of antibiotics. The inducible macrolide–lincosamide–streptogramin B (MLSB) resistance was detected by the D-test and interpreter according to the EUCAST. The penicillinase encoded by the blaZ gene was detected by the cefinase discs test (Becton, Dickinson and Company, Drogheda, Ireland).
Author Contributions
Writing—original draft preparation, M.K. and K.G.; writing—review and editing, M.K.-S., J.M. and K.G.; investigation, M.K., M.K.-S., M.W.-G., E.K. and M.W.; supervision, K.G. and J.M.; project administration, K.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors report there are no competing interests to declare.
References
- Kosecka-Strojek, M.; Buda, A.; Międzobrodzki, J. Staphylococcal Ecology and Epidemiology, in Pet-to-Man Travelling Staphylococci: A World in Progress; Savini, V., Ed.; Elsevier: Cambridge, MA, USA, 2018; pp. 11–24. [Google Scholar] [CrossRef]
- Garbacz, K.; Wierzbowska, M.; Kwapisz, E.; Kosecka-Strojek, M.; Bronk, M.; Saki, M.; Międzobrodzki, J. Distribution and antibiotic-resistance of different Staphylococcus species identified by matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) isolated from the oral cavity. J. Oral. Microbiol. 2021, 13, 1983322. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef] [PubMed]
- Pereira, V.C.; Romero, L.C.; Pinheiro-Hubinger, L.; Oliveira, A.; Martins, K.B.; Cunha, M.D.L.R.D.S.D. Coagulase-negative staphylococci: A 20-year study on the antimicrobial resistance profile of blood culture isolates from a teaching hospital. Braz. J. Infect. Dis. 2020, 24, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Casey, A.L.; Lambert, P.A.; Elliott, T.S. Staphylococci. Int. J. Antimicrob. Agents 2007, 29 (Suppl. S3), S23–S32. [Google Scholar] [CrossRef]
- García, A.; Martínez, C.; Juárez, R.I.; Téllez, R.; Paredes, M.A.; Herrera, M.d.R.; Giono, S. Methicillin resistance and biofilm production in clinical isolates of Staphylococcus aureus and coagulase-negative Staphylococcus in México. Biomedica 2019, 39, 513–523. [Google Scholar] [CrossRef]
- Mora-Ochomogo, M.; Lohans, C.T. β-Lactam antibiotic targets and resistance mechanisms: From covalent inhibitors to substrates. RSC Med. Chem. 2021, 12, 1623–1639. [Google Scholar] [CrossRef]
- Lade, H.; Kim, J.S. Molecular Determinants of β-Lactam Resistance in Methicillin-Resistant Staphylococcus aureus (MRSA): An Updated Review. Antibiotics 2023, 12, 1362. [Google Scholar] [CrossRef]
- Alghamdi, B.A.; Al-Johani, I.; Al-Shamrani, J.M.; Alshamrani, H.M.; Al-Otaibi, B.G.; Almazmomi, K.; Yusof, N.Y. Antimicrobial resistance in methicillin-resistant Staphylococcus aureus. Saudi J. Biol. Sci. 2023, 30, 103604. [Google Scholar] [CrossRef]
- García-Álvarez, L.; Holden, M.T.; Lindsay, H.; Webb, C.R.; Brown, D.F.; Curran, M.D.; Walpole, E.; Brooks, K.; Pickard, D.J.; Teale, C.; et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect. Dis. 2011, 11, 595–603. [Google Scholar] [CrossRef]
- Matsuhashi, M.; Song, M.D.; Ishino, F.; Wachi, M.; Doi, M.; Inoue, M.; Ubukata, K.; Yamashita, N.; Konno, M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J. Bacteriol. 1986, 167, 975–980. [Google Scholar] [CrossRef]
- Wolska-Gębarzewska, M.; Międzobrodzki, J.; Kosecka-Strojek, M. Current types of staphylococcal cassette chromosome mec (SCCmec) in clinically relevant coagulase-negative staphylococcal (CoNS) species. Crit. Rev. Microbiol. 2023, 50, 1020–1036. [Google Scholar] [CrossRef] [PubMed]
- Stańkowska, M.; Garbacz, K.; Piechowicz, L.; Bronk, M. Dissemination Of t437-SCCmecIV And Coagulase-Negative t037-SCCmecIII Types Among Borderline Oxacillin-Resistant Staphylococcus aureus Isolated From Skin Infections And Diabetic Foot Ulcers. Infect. Drug Resist. 2019, 12, 3197–3203. [Google Scholar] [CrossRef] [PubMed]
- Hryniewicz, M.M.; Garbacz, K. Borderline oxacillin-resistant Staphylococcus aureus (BORSA)—A more common problem than expected? J. Med. Microbiol. 2017, 66, 1367–1373. [Google Scholar] [CrossRef] [PubMed]
- The European Committee on Antimicrobial Susceptibility Testing. Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Expert_Rules/2018/Exprules_Staphylococcus_2018.pdf (accessed on 1 January 2018).
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Pinheiro, L.; Mello, P.L.; Abraão, L.M.; Corrente, J.E.; de Lourdes, R.S.C.M. Evaluation of reference values for phenotypic tests to detect oxacillin resistance in coagulase-negative staphylococci. Futur. Microbiol. 2018, 13, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Pefanis, A.; Thauvin-Eliopoulos, C.; Eliopoulos, G.M.; Moellering, R.C. Activity of ampicillin-sulbactam and oxacillin in experimental endocarditis caused by beta-lactamase-hyperproducing Staphylococcus aureus. Antimicrob. Agents Chemother. 1993, 37, 507–511. [Google Scholar] [CrossRef]
- Nelson, L.; Cockram, C.; Lui, G.; Lam, R.; Lam, E.; Lai, R.; Ip, M. Community case of methicillin-resistant Staphylococcus aureus infection. Emerg. Infect. Dis. 2006, 12, 172–174. [Google Scholar] [CrossRef]
- Sawhney, S.S.; Ransom, E.M.; Wallace, M.A.; Reich, P.J.; Dantas, G.; Burnham, C.-A.D. Comparative Genomics of Borderline Oxacillin-Resistant Staphylococcus aureus Detected during a Pseudo-outbreak of Methicillin-Resistant S. aureus in a Neonatal Intensive Care Unit. mBio 2022, 13, e0319621. [Google Scholar] [CrossRef]
- Conceição, T.; Coelho, C.; de Lencastre, H.; Aires-De-Sousa, M. Frequent occurrence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) strains in two African countries. J. Antimicrob. Chemother. 2015, 70, 3200–3204. [Google Scholar] [CrossRef][Green Version]
- Liu, J.-L.; Li, T.-M.; Zhong, N.; Wang, X.; Jiang, J.; Zhang, W.-X.; Tang, R.; Guo, Y.-J.; Liu, Y.; Hu, J.; et al. Current status of oxacillin-susceptible mecA-positive Staphylococcus aureus infection in Shanghai, China: A multicenter study. J. Microbiol. Immunol. Infect. 2021, 54, 1070–1077. [Google Scholar] [CrossRef]
- Ho, C.-M.; Lin, C.-Y.; Ho, M.-W.; Lin, H.-C.; Chen, C.-J.; Lin, L.-C.; Lu, J.-J. Methicillin-resistant Staphylococcus aureus isolates with SCCmec type V and spa types t437 or t1081 associated to discordant susceptibility results between oxacillin and cefoxitin, Central Taiwan. Diagn. Microbiol. Infect. Dis. 2016, 86, 405–411. [Google Scholar] [CrossRef]
- Hososaka, Y.; Hanaki, H.; Endo, H.; Suzuki, Y.; Nakae, T.; Nagasawa, Z.; Otsuka, Y.; Sunakawa, K. Characterization of oxacillin-susceptible mecA-positive Staphylococcus aureus: A new type of MRSA. J. Infect. Chemother. 2007, 13, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Petinaki, E.; Kontos, F.; Maniatis, A.N. Emergence of two oxacillin-susceptible mecA-positive Staphylococcus aureus clones in a Greek hospital. J. Antimicrob. Chemother. 2002, 50, 1090–1091. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wannet, W.J.B.; Spalburg, E.; Heck, M.E.O.C.; Pluister, G.N.; Willems, R.J.L.; de Neeling, A.J. Widespread dissemination in the Netherlands of the epidemic berlin methicillin-resistant Staphylococcus aureus clone with low-level resistance to oxacillin. J. Clin. Microbiol. 2004, 42, 3077–3082. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Harrison, E.M.; Ba, X.; Coll, F.; Blane, B.; Restif, O.; Carvell, H.; Köser, C.U.; Jamrozy, D.; Reuter, S.; Lovering, A.; et al. Genomic identification of cryptic susceptibility to penicillins and beta-lactamase inhibitors in methicillin resistant Staphylococcus aureus. Nat. Microbiol. 2019, 4, 1680–1691. [Google Scholar] [CrossRef]
- Ba, X.; Harrison, E.M.; Lovering, A.L.; Gleadall, N.; Zadoks, R.; Parkhill, J.; Peacock, S.J.; Holden, M.T.G.; Paterson, G.K.; Holmes, M.A. Old drugs to treat resistant bugs: Methicillin-resistant Staphylococcus aureus isolates with mecC are susceptible to a combination of Penicillin and Clavulanic acid. Antimicrob. Agents Chemother. 2015, 59, 7396–7404. [Google Scholar] [CrossRef]
- Gostev, V.; Sabinova, K.; Sopova, J.; Kalinogorskaya, O.; Sulian, O.; Chulkova, P.; Velizhanina, M.; Pavlova, P.; Danilov, L.; Kraeva, L.; et al. Phenotypic and genomic characteristics of oxacillin-susceptible mecA-positive Staphylococcus aureus, rapid selection of high-level resistance to beta-lactams. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 1125–1133. [Google Scholar] [CrossRef]
- Maalej, S.M.; Rhimi, F.M.; Fines, M.; Mnif, B.; Leclercq, R.; Hammami, A. Analysis of borderline oxacillin-resistant Staphylococcus aureus (BORSA) strains isolated in Tunisia. J. Clin. Microbiol. 2012, 50, 3345–3348. [Google Scholar] [CrossRef]
- Chambers, H.F. Methicillin resistance in staphylococci: Molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 1997, 10, 781–791. [Google Scholar] [CrossRef]
- Balslev, U.; Bremmelgaard, A.; Svejgaard, E.; Havstreym, J.; Westh, H. An outbreak of borderline oxacillin-resistant Staphylococcus aureus (BORSA) in a dermatological unit. Microb. Drug Resist. 2005, 11, 78–81. [Google Scholar] [CrossRef]
- Khorvash, F.; Mostafavizadeh, K.; Mobasherizadeh, S. Frequency of mecA gene and borderline oxacillin resistant Staphylococcus aureus in nosocomial acquired methicillin resistance Staphylococcus aureus infections. Pak. J. Biol. Sci. 2008, 11, 1282–1285. [Google Scholar] [CrossRef]
- Leahy, T.R.; Yau, Y.C.; Atenafu, E.; Corey, M.; Ratjen, F.; Waters, V. Epidemiology of borderline oxacillin-resistant Staphylococcus aureus in pediatric cystic fibrosis. Pediatr. Pulmonol. 2011, 46, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Konstantinovski, M.M.; Veldkamp, K.E.; Lavrijsen, A.P.M.; Bosch, T.; Kraakman, M.E.M.; Nooij, S.; Claas, E.C.J.; Gooskens, J. Hospital transmission of borderline oxacillin-resistant Staphylococcus aureus evaluated by whole-genome sequencing. J. Med. Microbiol. 2021, 70, 001384. [Google Scholar] [CrossRef] [PubMed]
- Skinner, S.; Murray, M.; Walus, T.; Karlowsky, J.A. Failure of cloxacillin in treatment of a patient with borderline oxacillin-resistant Staphylococcus aureus endocarditis. J. Clin. Microbiol. 2009, 47, 859–861. [Google Scholar] [CrossRef]
- Szczuka, E.; Koznowski, A. Zróżnicowanie kaset SCCmec u metycylinoopornych gronkowców koagulazo-ujemnych. Post. Mikrobiol. 2014, 53, 223–228. [Google Scholar]
- Secchi, C.; Antunes, A.L.S.; Perez, L.R.R.; Cantarelli, V.V.; D’Azevedo, P.A. Identification and detection of methicillin resistance in non-epidermidis coagulase-negative staphylococci. Braz. J. Infect. Dis. 2008, 12, 316–320. [Google Scholar] [CrossRef]
- Swenson, J.M.; Tenover, F.C.; Cefoxitin Disk Study Group. Results of disk diffusion testing with cefoxitin correlate with presence of mecA in Staphylococcus spp. J. Clin. Microbiol. 2005, 43, 3818–3823. [Google Scholar] [CrossRef]
- Nair, D.; Shashindran, N.; Kumar, A.; Vinodh, V.; Biswas, L.; Biswas, R. Comparison of Phenotypic MRSA Detection Methods with PCR for mecA Gene in the Background of Emergence of Oxacillin-Susceptible MRSA. Microb. Drug Resist. 2021, 27, 1190–1194. [Google Scholar] [CrossRef]
- Ito, T.; Kuwahara-Arai, K.; Katayama, Y.; Uehara, Y.; Han, X.; Kondo, Y.; Hiramatsu, K. Staphylococcal Cassette Chromosome mec (SCCmec) analysis of MRSA. Methods Mol. Biol. 2014, 1085, 131–148. [Google Scholar] [CrossRef]
- Becker, K.; van Alen, S.; Idelevich, E.A.; Schleimer, N.; Seggewiß, J.; Mellmann, A.; Kaspar, U.; Peters, G. Plasmid-Encoded Transferable mecB-Mediated Methicillin Resistance in Staphylococcus aureus. Emerg. Infect. Dis. 2018, 24, 242–248. [Google Scholar] [CrossRef]
- Milheiriço, C.; Oliveira, D.C.; de Lencastre, H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 3374–3377. [Google Scholar] [CrossRef]
- Kondo, Y.; Ito, T.; Ma, X.X.; Watanabe, S.; Kreiswirth, B.N.; Etienne, J.; Hiramatsu, K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 2007, 51, 264–274. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).