Contrasting Dynamics of Intracellular and Extracellular Antibiotic Resistance Genes in Response to Nutrient Variations in Aquatic Environments
Abstract
1. Introduction
2. Results and Discussion
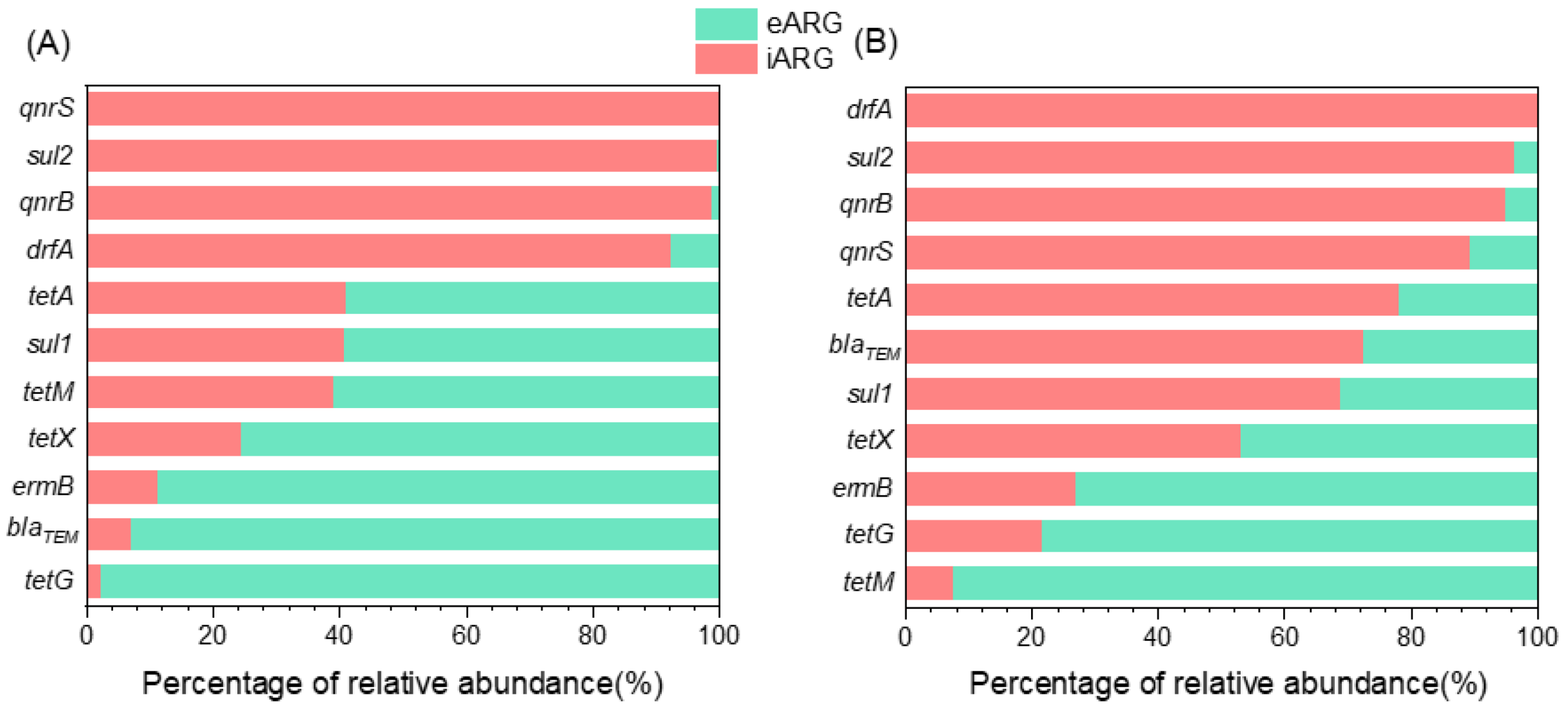
2.1. Dominance of eARGs in Water and iARGs in Biofilms
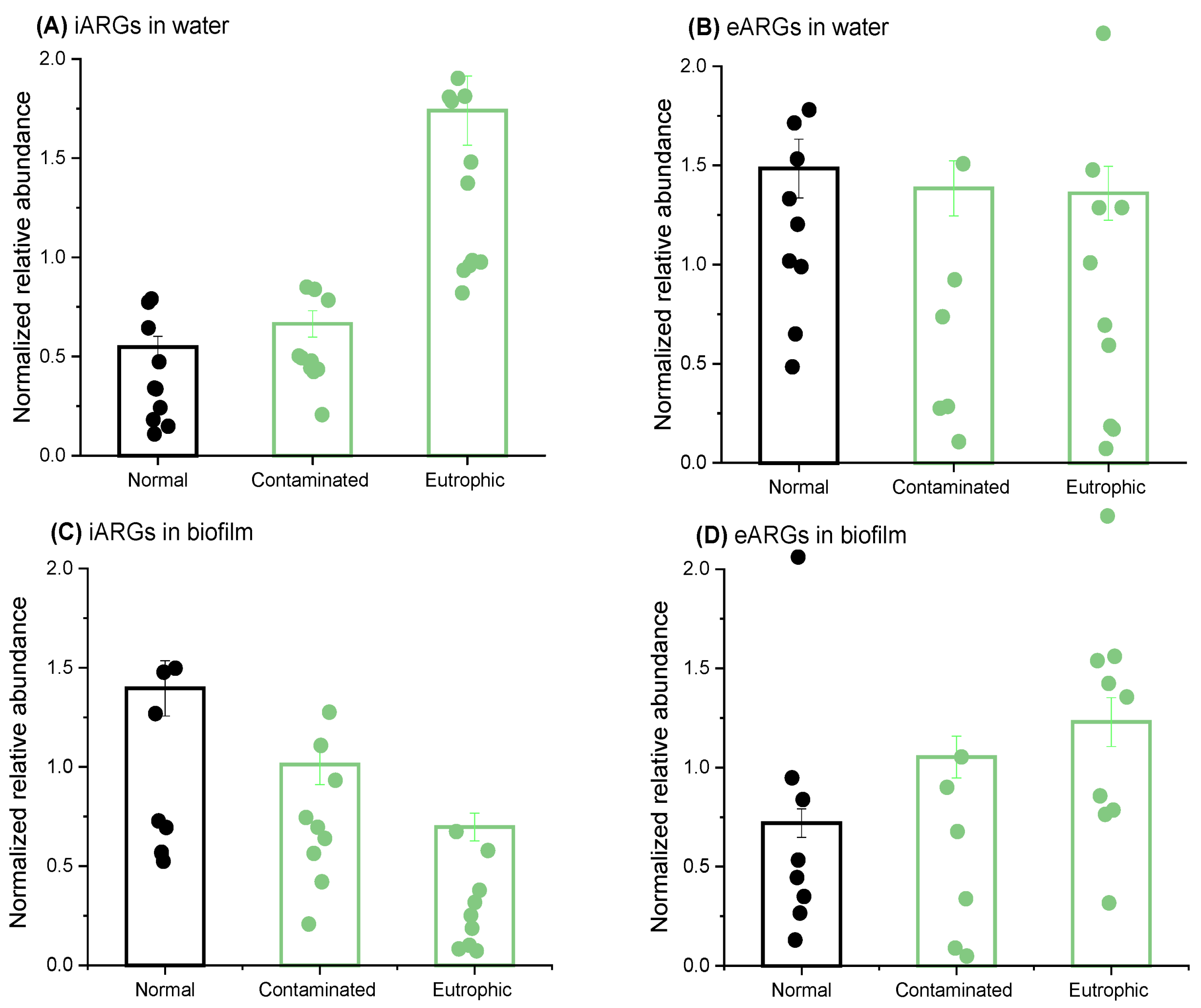
2.2. Nutrient Variations Induce Opposite Responses in iARGs and eARGs
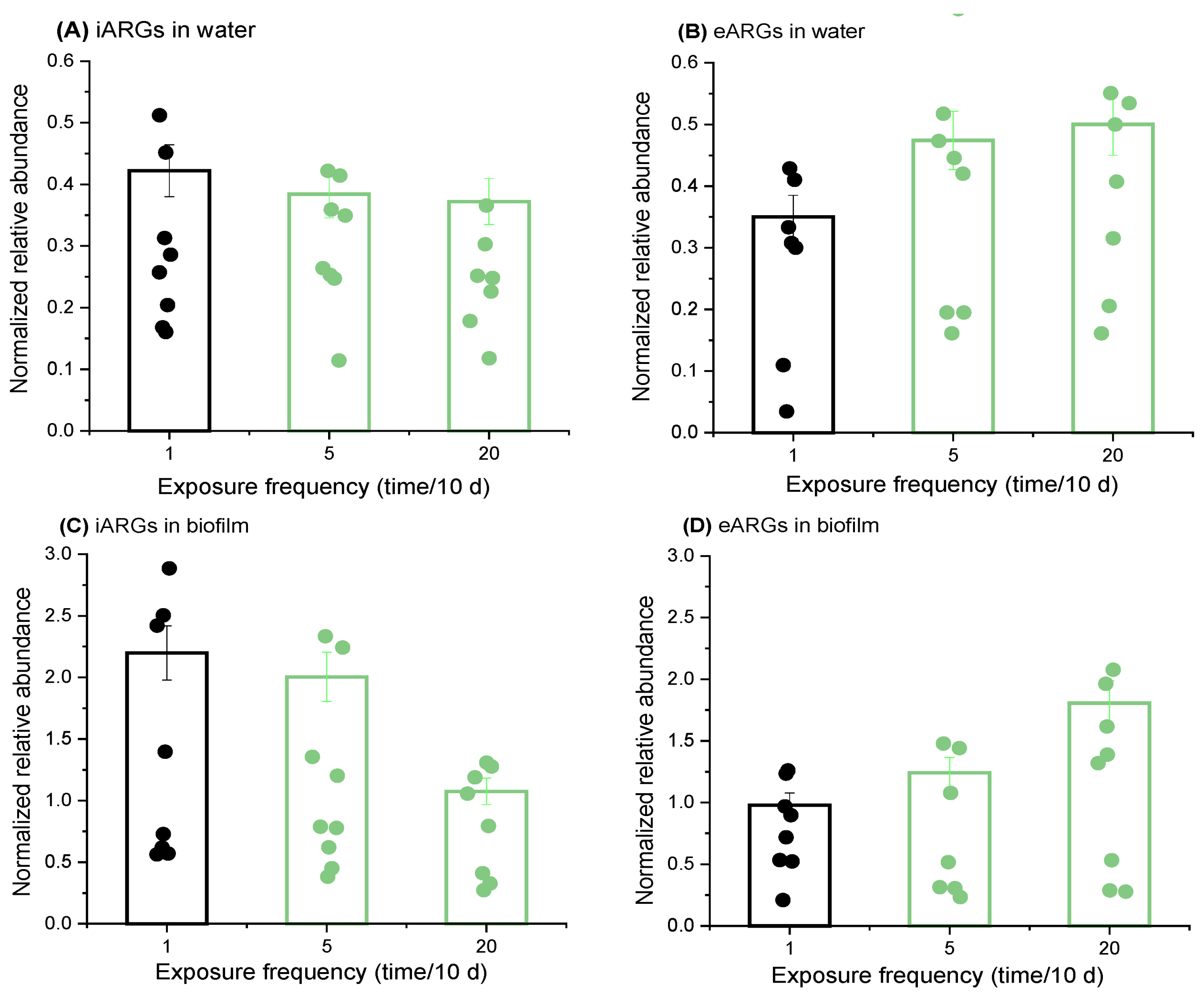
2.3. Increased Nutrient Delivery Frequencies Boost eARGs’ Abundance and Reduce iARGs
3. Materials and Methods
3.1. Sampling and Pretreatment
3.2. Construction of Microcosm
3.3. DNA Extraction
3.4. Quantitative PCR
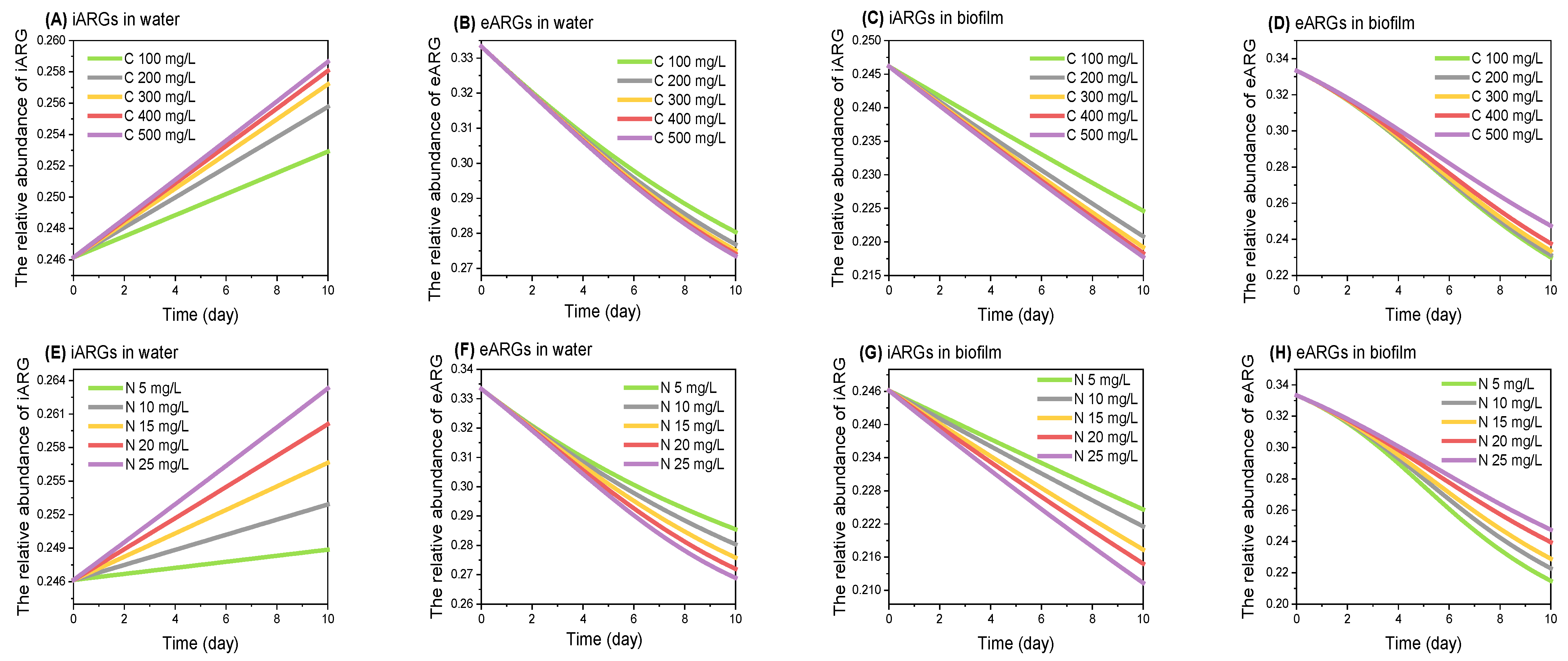
3.5. ARGs-Nutrient Model Construction
- r1 and r2—the maximum growth rates of antibiotic-resistant and antibiotic-sensitive strains, respectively;
- k1 and k2—the secretion rate of antibiotic-resistant strains and the transformation frequency of antibiotic-sensitive strains, respectively;
- a—the mortality rate of the strains;
- b—the degradation rate of the plasmids;
- C—the carbon concentration in the nutrients;
- N—the nitrogen concentration in the nutrients;
- M—the half-saturation constant at the maximum growth rate;
- n—the capacity of the strain in the microcosm.
3.6. Data Processing and Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Boeckel, T.P.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.T.; Levin, S.A.; Laxminarayan, R. Global antibiotic consumption 2000 to 2010: An analysis of national pharmaceutical sales data. Lancet Infect. Dis. 2014, 14, 742–750. [Google Scholar] [CrossRef]
- Zhang, Q.-Q.; Ying, G.-G.; Pan, C.-G.; Liu, Y.-S.; Zhao, J.-L. Comprehensive Evaluation of Antibiotics Emission and Fate in the River Basins of China: Source Analysis, Multimedia Modeling, and Linkage to Bacterial Resistance. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef] [PubMed]
- Qiao, M.; Ying, G.-G.; Singer, A.C.; Zhu, Y.-G. Review of antibiotic resistance in China and its environment. Environ. Int. 2018, 110, 160–172. [Google Scholar] [CrossRef]
- Li, S.; Shi, W.; Liu, W.; Li, H.; Zhang, W.; Hu, J.; Ke, Y.; Sun, W.; Ni, J. A duodecennial national synthesis of antibiotics in China’s major rivers and seas (2005–2016). Sci. Total Environ. 2018, 615, 906–917. [Google Scholar] [CrossRef]
- Gao, F.-Z.; He, L.-Y.; Hu, L.-X.; Chen, J.; Yang, Y.-Y.; He, L.-X.; Bai, H.; Liu, Y.-S.; Zhao, J.-L.; Ying, G.-G. The variations of antibiotics and antibiotic resistance genes in two subtropical large river basins of south China: Anthropogenic impacts and environmental risks. Environ. Pollut. 2022, 312, 119978. [Google Scholar] [CrossRef]
- Amarasiri, M.; Sano, D.; Suzuki, S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2020, 50, 2016–2059. [Google Scholar] [CrossRef]
- Gil, J. Antimicrobial use in livestock farming. Nat. Food 2023, 4, 138. [Google Scholar] [CrossRef]
- Sun, J.; Jin, L.; He, T.; Wei, Z.; Liu, X.; Zhu, L.; Li, X. Antibiotic resistance genes (ARGs) in agricultural soils from the Yangtze River Delta, China. Sci. Total Environ. 2020, 740, 140001. [Google Scholar] [CrossRef]
- Singh, R.; Singh, A.P.; Kumar, S.; Giri, B.S.; Kim, K.-H. Antibiotic resistance in major rivers in the world: A systematic review on occurrence, emergence, and management strategies. J. Clean. Prod. 2019, 234, 1484–1505. [Google Scholar] [CrossRef]
- Wang, C.; Liu, X.; Yang, Y.; Wang, Z. Antibiotic and antibiotic resistance genes in freshwater aquaculture ponds in China: A meta-analysis and assessment. J. Clean. Prod. 2021, 329, 129719. [Google Scholar] [CrossRef]
- Zhu, Y.-G.; Gillings, M.; Simonet, P.; Stekel, D.; Banwart, S.; Penuelas, J. Human dissemination of genes and microorganisms in Earth’s Critical Zone. Glob. Change Biol. 2018, 24, 1488–1499. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Gao, R.; Wei, Y.; Chen, T.; Fan, J.; Zhou, Z.; Makimilua, T.B.; Jiao, Y.; Chen, H. High-throughput profiling and analysis of antibiotic resistance genes in East Tiaoxi River, China. Environ. Pollut. 2017, 230, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Letten, A.D.; Hall, A.R.; Levine, J.M. Using ecological coexistence theory to understand antibiotic resistance and microbial competition. Nat. Ecol. Evol. 2021, 5, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.S.; Kam, Y.; Khin, Z.P.; Minton, S.E.; Gillies, R.J.; Gatenby, R.A. Evolutionary Approaches to Prolong Progression-Free Survival in Breast Cancer. Cancer Res. 2012, 72, 6362–6370. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.C.; Berry, D. Microbial nutrient niches in the gut. Environ. Microbiol. 2017, 19, 1366–1378. [Google Scholar] [CrossRef]
- Chowdhury, N.N.; Cox, A.R.; Wiesner, M.R. Nanoparticles as vectors for antibiotic resistance: The association of silica nanoparticles with environmentally relevant extracellular antibiotic resistance genes. Sci. Total Environ. 2021, 761, 143261. [Google Scholar] [CrossRef]
- Liu, S.-S.; Qu, H.-M.; Yang, D.; Hu, H.; Liu, W.-L.; Qiu, Z.-G.; Hou, A.-M.; Guo, J.; Li, J.-W.; Shen, Z.-Q.; et al. Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 2018, 136, 131–136. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, K.; Wu, N.; Li, W.; Xu, W.; Zhang, Y.; Niu, Z. Estuarine sediments are key hotspots of intracellular and extracellular antibiotic resistance genes: A high-throughput analysis in Haihe Estuary in China. Environ. Int. 2020, 135, 105385. [Google Scholar] [CrossRef]
- Mao, D.; Luo, Y.; Mathieu, J.; Wang, Q.; Feng, L.; Mu, Q.; Feng, C.; Alvarez, P.J.J. Persistence of Extracellular DNA in River Sediment Facilitates Antibiotic Resistance Gene Propagation. Environ. Sci. Technol. 2014, 48, 71–78. [Google Scholar] [CrossRef]
- Yu, W.; Xu, Y.; Wang, Y.; Sui, Q.; Xin, Y.; Wang, H.; Zhang, J.; Zhong, H.; Wei, Y. An extensive assessment of seasonal rainfall on intracellular and extracellular antibiotic resistance genes in Urban River systems. J. Hazard. Mater. 2023, 455, 131561. [Google Scholar] [CrossRef]
- Yu, J.; Wang, Z.; Wang, X.; Xu, J.; Jia, J. Study on Mechanism Experiments and Evaluation Methods for Water Eutrophication. J. Chem. 2017, 2017, 2036035. [Google Scholar] [CrossRef]
- Zhao, W.; Peng, Y.; Wang, M.; Huang, Y.; Li, X. Nutrient removal and microbial community structure variation in the two-sludge system treating low carbon/nitrogen domestic wastewater. Bioresour. Technol. 2019, 294, 122161. [Google Scholar] [CrossRef]
- Mao, J.; Lu, T. Population-Dynamic Modeling of Bacterial Horizontal Gene Transfer by Natural Transformation. Biophys. J. 2016, 110, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Elisabet, I.N.; Lena, E.F. Pharmacokinetic-Pharmacodynamic Modeling of Antibacterial Drugs. Pharmacol. Rev. 2013, 65, 1053. [Google Scholar] [CrossRef]
- Zhang, Y.; Niu, Z.; Zhang, Y.; Zhang, K. Occurrence of intracellular and extracellular antibiotic resistance genes in coastal areas of Bohai Bay (China) and the factors affecting them. Environ. Pollut. 2018, 236, 126–136. [Google Scholar] [CrossRef]
- Yuan, Q.; Wang, X.; Fang, H.; Cheng, Y.; Sun, R.; Luo, Y. Coastal mudflats as reservoirs of extracellular antibiotic resistance genes: Studies in Eastern China. J. Environ. Sci. 2023, 129, 58–68. [Google Scholar] [CrossRef]
- An, X.-L.; Su, J.-Q.; Li, B.; Ouyang, W.-Y.; Zhao, Y.; Chen, Q.-L.; Cui, L.; Chen, H.; Gillings, M.R.; Zhang, T.; et al. Tracking antibiotic resistome during wastewater treatment using high throughput quantitative PCR. Environ. Int. 2018, 117, 146–153. [Google Scholar] [CrossRef]
- Zhou, S.; Zhu, Y.; Yan, Y.; Wang, W.; Wang, Y. Deciphering extracellular antibiotic resistance genes (eARGs) in activated sludge by metagenome. Water Res. 2019, 161, 610–620. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, Y.; Yang, M.; Tian, Z.; Ren, L.; Zhang, S. Abundance and Distribution of Tetracycline Resistance Genes and Mobile Elements in an Oxytetracycline Production Wastewater Treatment System. Environ. Sci. Technol. 2012, 46, 7551–7557. [Google Scholar] [CrossRef]
- Pietramellara, G.; Ascher, J.; Borgogni, F.; Ceccherini, M.T.; Guerri, G.; Nannipieri, P. Extracellular DNA in soil and sediment: Fate and ecological relevance. Biol. Fertil. Soils 2009, 45, 219–235. [Google Scholar] [CrossRef]
- Schulz-Siegmund, M.; Aigner, A. Nucleic acid delivery with extracellular vesicles. Adv. Drug Deliv. Rev. 2021, 173, 89–111. [Google Scholar] [CrossRef] [PubMed]
- Zarei-Baygi, A.; Smith, A.L. Intracellular versus extracellular antibiotic resistance genes in the environment: Prevalence, horizontal transfer, and mitigation strategies. Bioresour. Technol. 2021, 319, 124181. [Google Scholar] [CrossRef]
- Rysz, M.; Mansfield, W.R.; Fortner, J.D.; Alvarez, P.J.J. Tetracycline Resistance Gene Maintenance under Varying Bacterial Growth Rate, Substrate and Oxygen Availability, and Tetracycline Concentration. Environ. Sci. Technol. 2013, 47, 6995–7001. [Google Scholar] [CrossRef]
- Vacca, I. Building up the matrix. Nat. Rev. Microbiol. 2017, 15, 513. [Google Scholar] [CrossRef]
- Guo, X.; Yang, Y.; Lu, D.; Niu, Z.; Feng, J.; Chen, Y.; Tou, F.; Garner, E.; Xu, J.; Liu, M.; et al. Biofilms as a sink for antibiotic resistance genes (ARGs) in the Yangtze Estuary. Water Res. 2018, 129, 277–286. [Google Scholar] [CrossRef]
- Janicka, E.; Kanclerz, J.; Wiatrowska, K.; Budka, A. Variability of Nitrogen and Phosphorus Content and Their Forms in Waters of a River-Lake System. Front. Environ. Sci. 2022, 10, 874754. [Google Scholar] [CrossRef]
- Danyang, W.; Xianqiang, T.; Rui, L.; Wenjun, Y. Spatial distribution patterns of nitrogen and phosphorus in water and bed sediment of the Three Gorges Reservoir. J. Clean. Prod. 2021, 322, 129026. [Google Scholar] [CrossRef]
- Dominiak, D.M.; Nielsen, J.L.; Nielsen, P.H. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ. Microbiol. 2011, 13, 710–721. [Google Scholar] [CrossRef]
- Ryu, K.H.; Sung, M.-G.; Kim, B.; Heo, S.; Chang, Y.K.; Lee, J.H. A mathematical model of intracellular behavior of microalgae for predicting growth and intracellular components syntheses under nutrient-replete and -deplete conditions. Biotechnol. Bioeng. 2018, 115, 2441–2455. [Google Scholar] [CrossRef] [PubMed]
- Letten, A.D.; Dhami, M.K.; Ke, P.-J.; Fukami, T. Species coexistence through simultaneous fluctuation-dependent mechanisms. Proc. Natl. Acad. Sci. USA 2018, 115, 6745–6750. [Google Scholar] [CrossRef] [PubMed]
- Yip, A.Y.G.; King, O.G.; Omelchenko, O.; Kurkimat, S.; Horrocks, V.; Mostyn, P.; Danckert, N.; Ghani, R.; Satta, G.; Jauneikaite, E.; et al. Antibiotics promote intestinal growth of carbapenem-resistant Enterobacteriaceae by enriching nutrients and depleting microbial metabolites. Nat. Commun. 2023, 14, 5094. [Google Scholar] [CrossRef]
- Côté, J.-P.; French, S.; Gehrke Sebastian, S.; MacNair Craig, R.; Mangat Chand, S.; Bharat, A.; Brown Eric, D. The Genome-Wide Interaction Network of Nutrient Stress Genes in Escherichia coli. mBio 2016, 7, e01714-16. [Google Scholar] [CrossRef]
- Brander, T.; Lesnic, D.; Cao, K. Inverse problems for a model of biofilm growth. IMA J. Appl. Math. 2023, 88, 258–281. [Google Scholar] [CrossRef]
- Hu, J.; Li, S.; Zhang, W.; Helbling, D.E.; Xu, N.; Sun, W.; Ni, J. Animal production predominantly contributes to antibiotic profiles in the Yangtze River. Water Res. 2023, 242, 120214. [Google Scholar] [CrossRef]
- Rajasekar, A.; Vadde, K.K.; Murava, R.T.; Qiu, M.; Guo, S.; Yu, T.; Wang, R.; Zhao, C. Occurrence of antibiotic resistance genes and potentially pathogenic bacteria in the Yangtze River tributary (Nanjing section) and their correlation with environmental factors. Environ. Res. Commun. 2023, 5, 035001. [Google Scholar] [CrossRef]
- Zhang, G.; Lu, S.; Wang, Y.; Liu, X.; Liu, Y.; Xu, J.; Zhang, T.; Wang, Z.; Yang, Y. Occurrence of antibiotics and antibiotic resistance genes and their correlations in lower Yangtze River, China. Environ. Pollut. 2020, 257, 113365. [Google Scholar] [CrossRef]
- Zou, Y.; Tu, W.; Wang, H.; Fang, T. Anaerobic digestion reduces extracellular antibiotic resistance genes in waste activated sludge: The effects of temperature and degradation mechanisms. Environ. Int. 2020, 143, 105980. [Google Scholar] [CrossRef]
- Wang, R.-N.; Zhang, Y.; Cao, Z.-H.; Wang, X.-Y.; Ma, B.; Wu, W.-B.; Hu, N.; Huo, Z.-Y.; Yuan, Q.-B. Occurrence of super antibiotic resistance genes in the downstream of the Yangtze River in China: Prevalence and antibiotic resistance profiles. Sci. Total Environ. 2019, 651, 1946–1957. [Google Scholar] [CrossRef]
- Yu, C.; Li, C.; Zhang, Y.; Du, X.; Wang, J.-H.; Chi, Z.-Y.; Zhang, Q. Effects of environment-relevant concentrations of antibiotics on seawater Chlorella sp. biofilm in artificial mariculture effluent. Algal Res. 2023, 70, 103008. [Google Scholar] [CrossRef]
- Zeeshan, M.; Pabst, S.; Sandyk, E.; Ruhl, A.S. Fates of selected pharmaceuticals in a large recirculated mesocosm with a pond and bank filtration. Sci. Total Environ. 2023, 892, 164575. [Google Scholar] [CrossRef]
- Yan, F.; An, L.; Xu, X.; Du, W.; Dai, R. A review of antibiotics in surface water and their removal by advanced electrocoagulation technologies. Sci. Total Environ. 2024, 906, 167737. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.-T.; Yuan, Q.-B.; Yang, J. Microbial selectivity of UV treatment on antibiotic-resistant heterotrophic bacteria in secondary effluents of a municipal wastewater treatment plant. Water Res. 2013, 47, 6388–6394. [Google Scholar] [CrossRef]
- Taylor, D.I.; Oviatt, C.A.; Giblin, A.E.; Tucker, J.; Diaz, R.J.; Keay, K. Wastewater input reductions reverse historic hypereutrophication of Boston Harbor, USA. Ambio 2020, 49, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Reverte, L.; Prieto-Simon, B.; Campas, M. New advances in electrochemical biosensors for the detection of toxins: Nanomaterials, magnetic beads and microfluidics systems. A review. Anal. Chim. Acta 2016, 908, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.-B.; Huang, Y.-M.; Wu, W.-B.; Zuo, P.; Hu, N.; Zhou, Y.-Z.; Alvarez, P.J.J. Redistribution of intracellular and extracellular free & adsorbed antibiotic resistance genes through a wastewater treatment plant by an enhanced extracellular DNA extraction method with magnetic beads. Environ. Int. 2019, 131, 104986. [Google Scholar] [CrossRef]
- Adams, N.M.; Bordelon, H.; Wang, K.-K.A.; Albert, L.E.; Wright, D.W.; Haselton, F.R. Comparison of Three Magnetic Bead Surface Functionalities for RNA Extraction and Detection. Acs Appl. Mater. Interfaces 2015, 7, 6062–6069. [Google Scholar] [CrossRef]
- Hao, H.; Shi, D.; Yang, D.; Yang, Z.; Qiu, Z.; Liu, W.; Shen, Z.; Yin, J.; Wang, H.; Li, J.; et al. Profiling of intracellular and extracellular antibiotic resistance genes in tap water. J. Hazard. Mater. 2019, 365, 340–345. [Google Scholar] [CrossRef]
- Yuan, Q.; Zhang, D.; Yu, P.; Sun, R.; Javed, H.; Wu, G.; Alvarez, P.J.J. Selective Adsorption and Photocatalytic Degradation of Extracellular Antibiotic Resistance Genes by Molecularly-Imprinted Graphitic Carbon Nitride. Environ. Sci. Technol. 2020, 54, 4621–4630. [Google Scholar] [CrossRef]
- Liu, L.; Zou, X.; Gao, Y.; Li, H.; Cheng, Y.; Zhang, X.; Yuan, Q. Differential dose-response patterns of intracellular and extracellular antibiotic resistance genes under sub-lethal antibiotic exposure. Ecotoxicol. Environ. Saf. 2023, 260, 115070. [Google Scholar] [CrossRef]
- Zeng, Y.-H.; Cai, Z.-H.; Zhu, J.-M.; Du, X.-P.; Zhou, J. Two hierarchical LuxR-LuxI type quorum sensing systems in Novosphingobium activate microcystin degradation through transcriptional regulation of the mlr pathway. Water Res. 2020, 183, 116092. [Google Scholar] [CrossRef]
- Guo, M.-T.; Yuan, Q.-B.; Yang, J. Ultraviolet reduction of erythromycin and tetracycline resistant heterotrophic bacteria and their resistance genes in municipal wastewater. Chemosphere 2013, 93, 2864–2868. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jin, M.; Qiu, Z.-G.; Guo, C.; Chen, Z.-L.; Shen, Z.-Q.; Wang, X.-W.; Li, J.-W. A Survey of Drug Resistance bla Genes Originating from Synthetic Plasmid Vectors in Six Chinese Rivers. Environ. Sci. Technol. 2012, 46, 13448–13454. [Google Scholar] [CrossRef]
- Maslov, S.; Sneppen, K. Population cycles and species diversity in dynamic Kill-the-Winner model of microbial ecosystems. Sci. Rep. 2017, 7, 39642. [Google Scholar] [CrossRef]
- Lax, S.; Abreu, C.I.; Gore, J. Higher temperatures generically favour slower-growing bacterial species in multispecies communities. Nat. Ecol. Evol. 2020, 4, 560–567. [Google Scholar] [CrossRef]
- Wang, Q. On a Lotka-Volterra competition-diffusion-advection model in general heterogeneous environments. J. Math. Anal. Appl. 2020, 489, 124127. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, J.; Zhang, Z.; Peng, Z.; Dai, X.; Ni, B.-J. Polyethylene terephthalate microplastic fibers increase the release of extracellular antibiotic resistance genes during sewage sludge anaerobic digestion. Water Res. 2022, 217, 118426. [Google Scholar] [CrossRef]
- Igler, C.; Rolff, J.; Regoes, R. Multi-step vs. single-step resistance evolution under different drugs, pharmacokinetics, and treatment regimens. eLife 2021, 10, e64116. [Google Scholar] [CrossRef]
- Yu, G.; Baeder, D.Y.; Regoes, R.R.; Rolff, J. Predicting drug resistance evolution: Insights from antimicrobial peptides and antibiotics. Proc. R. Soc. B Biol. Sci. 2018, 285, 20172687. [Google Scholar] [CrossRef]
- Jiang, X.; Zhu, Z.; Wu, J.; Lian, E.; Liu, D.; Yang, S.; Zhang, R. Bacterial and Protistan Community Variation across the Changjiang Estuary to the Ocean with Multiple Environmental Gradients. Microorganisms 2022, 10, 991. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Marshall, J.S.; Wargo, M.J. Hybrid Model of Bacterial Biofilm Growth. Bull. Math. Biol. 2020, 82, 27. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.-B.; Guo, M.-T.; Yang, J. Monitoring and assessing the impact of wastewater treatment on release of both antibiotic-resistant bacteria and their typical genes in a Chinese municipal wastewater treatment plant. Environ. Sci. Process. Impacts 2014, 16, 1930–1937. [Google Scholar] [CrossRef]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.C.; Oh, J.Y.; Cho, J.W.; Park, J.C.; Kim, J.M.; Seol, S.Y.; Cho, D.T. The prevalence of trimethoprim-resistance-conferring dihydrofolate reductase genes in urinary isolates of Escherichia coli in Korea. J. Antimicrob. Chemother. 2001, 47, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.; Nordmann, P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef]
- Marti, E.; Balcázar José, L. Real-Time PCR Assays for Quantification of qnr Genes in Environmental Water Samples and Chicken Feces. Appl. Environ. Microbiol. 2013, 79, 1743–1745. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Mao, D.; Rysz, M.; Zhang, H.; Xu, L.; Alvarez, P.J.J. Trends in Antibiotic Resistance Genes Occurrence in the Haihe River, China. Environ. Sci. Technol. 2010, 44, 7220–7225. [Google Scholar] [CrossRef] [PubMed]
- Pei, R.; Kim, S.-C.; Carlson, K.H.; Pruden, A. Effect of River Landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res. 2006, 40, 2427–2435. [Google Scholar] [CrossRef] [PubMed]
- Tamminen, M.; Karkman, A.; Lõhmus, A.; Muziasari, W.I.; Takasu, H.; Wada, S.; Suzuki, S.; Virta, M. Tetracycline Resistance Genes Persist at Aquaculture Farms in the Absence of Selection Pressure. Environ. Sci. Technol. 2011, 45, 386–391. [Google Scholar] [CrossRef]
- Zheng, G.; Lu, Y.; Wang, D.; Zhou, L. Importance of sludge conditioning in attenuating antibiotic resistance: Removal of antibiotic resistance genes by bioleaching and chemical conditioning with Fe[III]/CaO. Water Res. 2019, 152, 61–73. [Google Scholar] [CrossRef]
- Ghosh, S.; Ramsden, S.J.; LaPara, T.M. The role of anaerobic digestion in controlling the release of tetracycline resistance genes and class 1 integrons from municipal wastewater treatment plants. Appl. Microbiol. Biotechnol. 2009, 84, 791–796. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Zou, X.; Cheng, Y.; Li, H.; Zhang, X.; Yuan, Q. Contrasting Dynamics of Intracellular and Extracellular Antibiotic Resistance Genes in Response to Nutrient Variations in Aquatic Environments. Antibiotics 2024, 13, 817. https://doi.org/10.3390/antibiotics13090817
Liu L, Zou X, Cheng Y, Li H, Zhang X, Yuan Q. Contrasting Dynamics of Intracellular and Extracellular Antibiotic Resistance Genes in Response to Nutrient Variations in Aquatic Environments. Antibiotics. 2024; 13(9):817. https://doi.org/10.3390/antibiotics13090817
Chicago/Turabian StyleLiu, Lele, Xinyi Zou, Yuan Cheng, Huihui Li, Xueying Zhang, and Qingbin Yuan. 2024. "Contrasting Dynamics of Intracellular and Extracellular Antibiotic Resistance Genes in Response to Nutrient Variations in Aquatic Environments" Antibiotics 13, no. 9: 817. https://doi.org/10.3390/antibiotics13090817
APA StyleLiu, L., Zou, X., Cheng, Y., Li, H., Zhang, X., & Yuan, Q. (2024). Contrasting Dynamics of Intracellular and Extracellular Antibiotic Resistance Genes in Response to Nutrient Variations in Aquatic Environments. Antibiotics, 13(9), 817. https://doi.org/10.3390/antibiotics13090817





