Isolation and Characterization of Novel Bacteriophages to Target Carbapenem-Resistant Acinetobacter baumannii
Abstract
1. Introduction
2. Results
2.1. Bacteriophage Isolation, Host Spectrum Analysis, and RAPD–PCR Classification
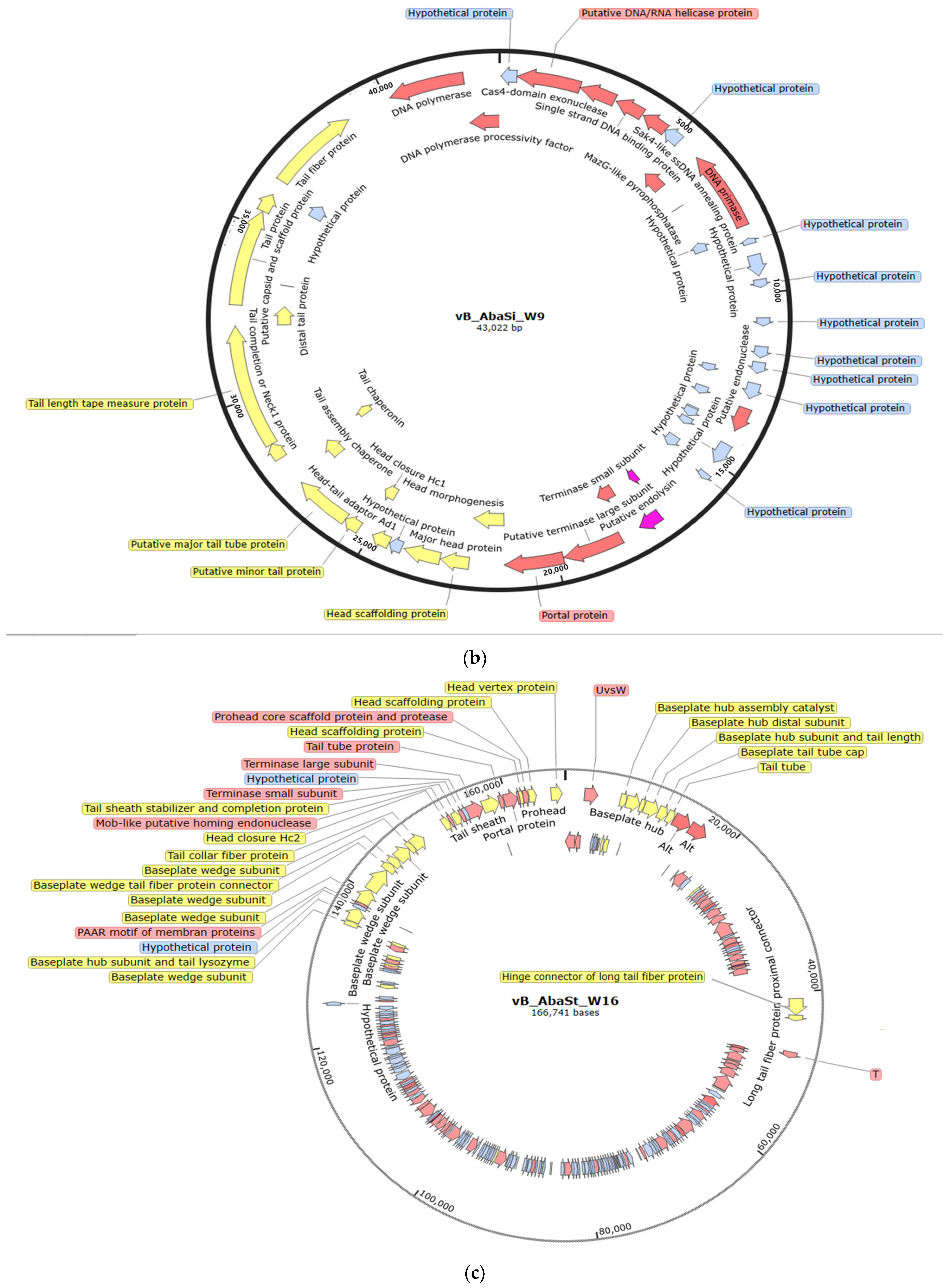
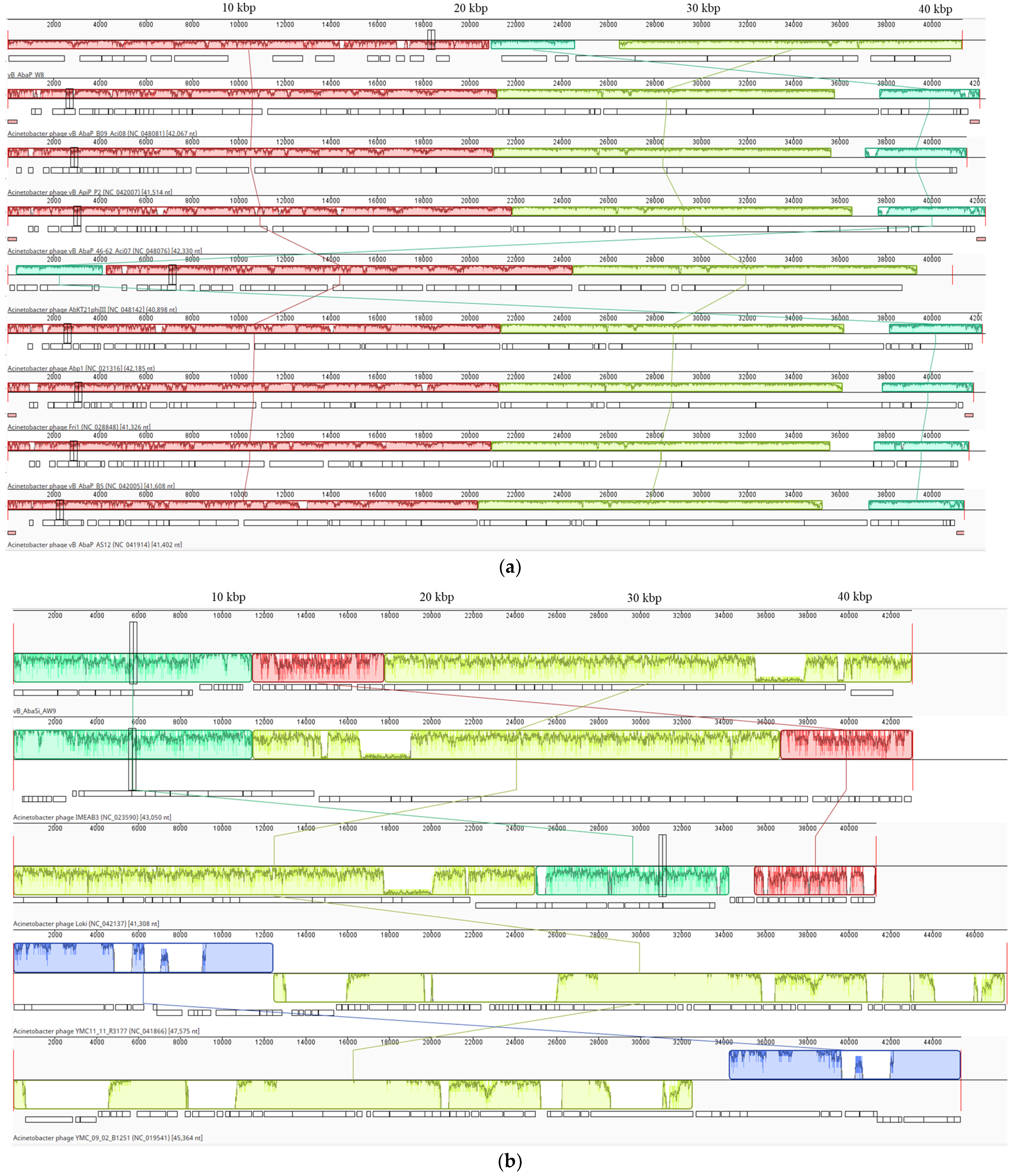
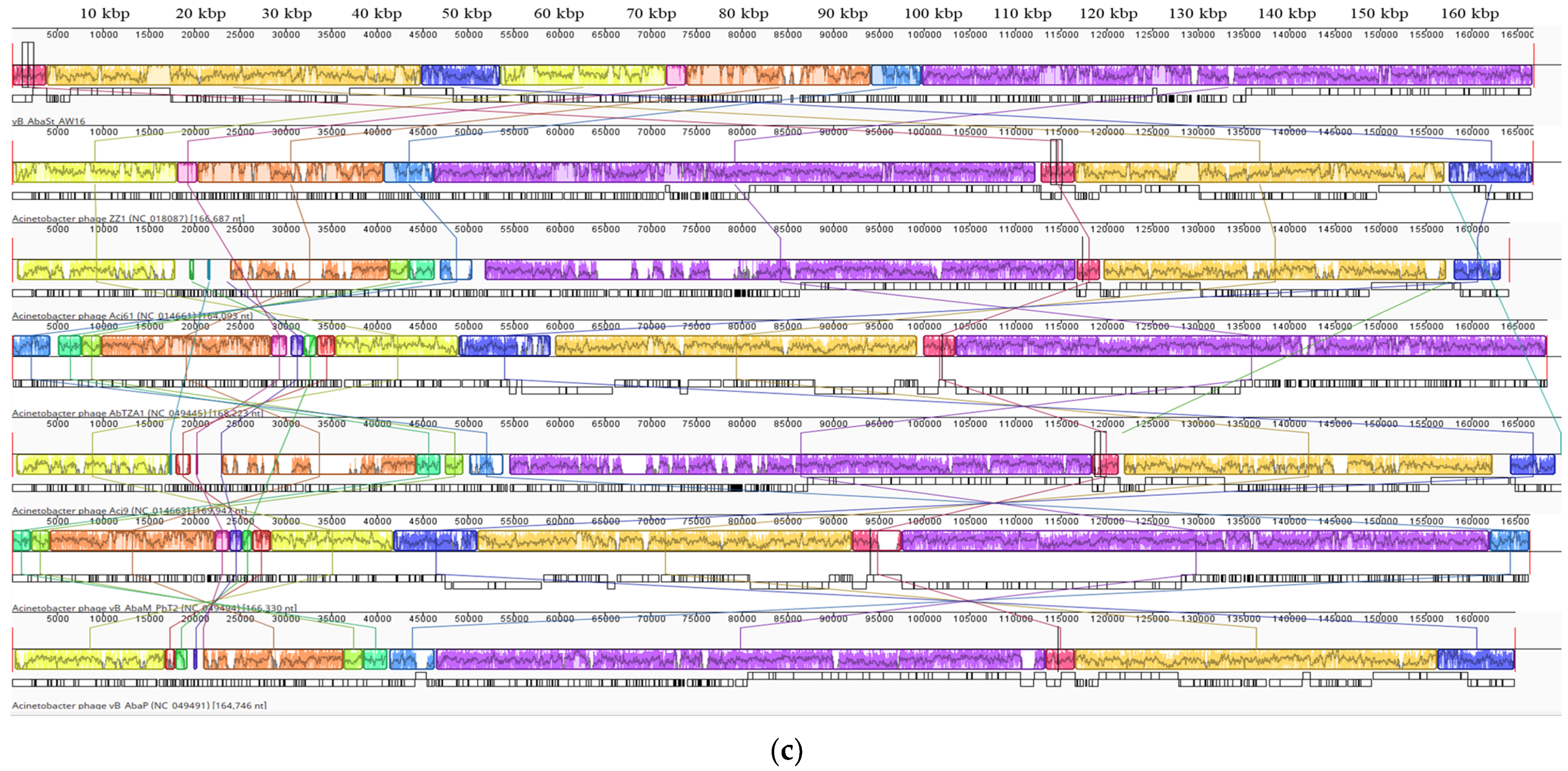
2.2. Phenotypic and Genotypic Characteristics of Three Novel Phages
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Processing
4.2. A. baumannii Strains Used in This Study
4.3. Isolation and Purification of Bacteriophages
4.4. Analysis of Host Spectrum Analysis and Efficiency of Plating (EOP)
4.5. Random Amplified Polymorphic DNA–Polymerase Chain Reaction
4.6. Plaque Formation and Morphology
4.7. Transmission Electron Microscopy (TEM)
4.8. Adsorption Assay and One-Step Growth Curve Assay
4.9. Genome Sequencing of Phages and Bioinformatics Analysis
4.10. Accession Numbers of the Genome Data
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Antunes, L.C.S.; Visca, P.; Towner, K.J. Acinetobacter baumannii: Evolution of a Global Pathogen. Pathog. Dis. 2014, 71, 292–301. [Google Scholar] [CrossRef] [PubMed]
- McConnell, M.J.; Actis, L.; Pachón, J. Acinetobacter baumannii: Human Infections, Factors Contributing to Pathogenesis and Animal Models. FEMS Microbiol. Rev. 2013, 37, 130–155. [Google Scholar] [CrossRef] [PubMed]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An Increasing Threat in Hospitals: Multidrug-Resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Pogue, J.M.; Mann, T.; Barber, K.E.; Kaye, K.S. Carbapenem-Resistant Acinetobacter baumannii: Epidemiology, Surveillance and Management. Expert Rev. Anti-Infect. Ther. 2013, 11, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.H.; Lee, D.E.; Hwang, H.R.; Kim, N.; Kim, H.J.; Lee, Y.C.; Kim, Y.K.; Lee, J.C. Clonal Change of Carbapenem-Resistant Acinetobacter baumannii Isolates in a Korean Hospital. Infect. Genet. Evol. 2021, 93, 104935. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.E.; Choi, S.M.; Yu, Y.; Shin, S.U.; Oh, T.H.; Kang, S.J.; Park, K.H.; Shin, J.H.; Kim, U.J.; Jung, S.I. Replacement of the Dominant ST191 Clone by ST369 among Carbapenem-Resistant Acinetobacter baumannii Bloodstream Isolates at a Tertiary Care Hospital in South Korea. Front. Microbiol. 2022, 13, 2580. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, J.N.; Putra, V.; Lodise, T.P. Treatment of Patients with Serious Infections Due to Carbapenem-Resistant Acinetobacter baumannii: How Viable Are the Current Options? Pharmacotherapy 2021, 41, 762–780. [Google Scholar] [CrossRef]
- Piperaki, E.T.; Tzouvelekis, L.S.; Miriagou, V.; Daikos, G.L. Carbapenem-Resistant Acinetobacter baumannii: In Pursuit of an Effective Treatment. Clin. Microbiol. Infect. 2019, 25, 951–957. [Google Scholar] [CrossRef]
- Gaiarsa, S.; Batisti Biffignandi, G.; Esposito, E.P.; Castelli, M.; Jolley, K.A.; Brisse, S.; Sassera, D.; Zarrilli, R. Comparative Analysis of the Two Acinetobacter baumannii Multilocus Sequence Typing (MLST) Schemes. Front. Microbiol. 2019, 10, 930. [Google Scholar] [CrossRef]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Luzon, M.A.D.; Wisplinghoff, H.; Rodríguez-Valera, F. Development of a Multilocus Sequence Typing Scheme for Characterization of Clinical Isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Cabezas, P.; Castro-Nallar, E.; Crandall, K.A. Pathogen Typing in the Genomics Era: MLST and the Future of Molecular Epidemiology. Infect. Genet. Evol. 2013, 16, 38–53. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, Y.; Yan, R.; Lan, P.; Liu, H.; Fu, Y.; Hua, X.; Jiang, Y.; Zhou, Z.; Yu, Y. Comparing Core-Genome MLST with PFGE and MLST for Cluster Analysis of Carbapenem-Resistant Acinetobacter baumannii. J. Glob. Antimicrob. Resist. 2022, 30, 148–151. [Google Scholar] [CrossRef] [PubMed]
- PubMLST. Primers Used for MLST of Acinetobacter baumannii Complex. Available online: https://pubmlst.org/primers-used-mlst-acinetobacter-baumannii-complex-pasteur-scheme (accessed on 16 November 2023).
- Niu, T.; Guo, L.; Kong, X.; He, F.; Ru, C.; Xiao, Y. Prevalent Dominant Acinetobacter baumannii ST191/195/208 Strains in Bloodstream Infections Have High Drug Resistance and Mortality. Infect. Drug Resist. 2023, 16, 2417–2427. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.J.; Kim, D.; Lee, H.; Lee, H.S.; Shin, J.H.; Uh, Y.; Shin, K.S.; Kim, Y.A.; Park, Y.S.; Shin, J.H.; et al. Counter Clinical Prognoses of Patients with Bloodstream Infections between Causative Acinetobacter baumannii Clones ST191 and ST451 Belonging to the International Clonal Lineage II. Front. Public Health 2019, 7, 233. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Marks, T.; Sharp, R. Bacteriophages and Biotechnology: A Review. Chem. Technol. Biotechnol. 2000, 75, 6–17. [Google Scholar] [CrossRef]
- Jamal, M.; Bukhari, S.M.A.U.S.; Andleeb, S.; Ali, M.; Raza, S.; Nawaz, M.A.; Hussain, T.; Rahman, S.U.; Shah, S.S.A. Bacteriophages: An Overview of the Control Strategies against Multiple Bacterial Infections in Different Fields. J. Basic Microbiol. 2019, 59, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Oechslin, F. Resistance Development to Bacteriophages Occurring during Bacteriophage Therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef]
- Hyman, P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef]
- Liu, M.; Hernandez-Morales, A.; Clark, J.; Le, T.; Biswas, B.; Bishop-Lilly, K.A.; Henry, M.; Quinones, J.; Voegtly, L.J.; Cer, R.Z.; et al. Comparative Genomics of Acinetobacter baumannii and Therapeutic Bacteriophages from a Patient Undergoing Phage Therapy. Nat. Commun. 2022, 13, 3776. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.L.; Barr, J.J. Unlocking the Next Generation of Phage Therapy: The Key is in the Receptors. Curr. Opin. Biotechnol. 2021, 68, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiao, S.; Huang, G. Acinetobacter baumannii Bacteriophage: Progress in Isolation, Genome Sequencing, Preclinical Research, and Clinical Application. Curr. Microbiol. 2023, 80, 199. [Google Scholar] [CrossRef] [PubMed]
- Tu, Q.; Pu, M.; Li, Y.; Wang, Y.; Li, M.; Song, L.; Li, M.; An, X.; Fan, H.; Tong, Y. Acinetobacter baumannii Phages: Past, Present and Future. Viruses 2023, 15, 673. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of Morphology-Based Taxa and Change to Binomial Species Names: 2022 Taxonomy Update of the ICTV Bacterial Viruses Subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, H.; Chen, H.; Zhang, J.; Wang, R.; Wang, Z.; Wang, H. Origin, Phylogeny, and Transmission of the Epidemic Clone ST208 of Carbapenem-Resistant Acinetobacter baumannii on a Global Scale. Microbiol. Spectr. 2022, 10, e02604-21. [Google Scholar] [CrossRef] [PubMed]
- Isler, B.; Doi, Y.; Bonomo, R.A.; Paterson, D.L. New Treatment Options against Carbapenem-Resistant Acinetobacter baumannii Infections. Antimicrob. Agents Chemother. 2018, 63. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.M.; Cho, H.W.; Kim, T.Y.; Park, J.S.; Jung, J.; Song, K.H.; Lee, H.; Kim, E.S.; Kim, H.B.; Park, K.U. Whole-Genome Sequencing for Investigating a Health Care-Associated Outbreak of Carbapenem-Resistant Acinetobacter baumannii. Diagnostics 2021, 11, 201. [Google Scholar] [CrossRef] [PubMed]
- Essoh, C.; Vernadet, J.-P.; Vergnaud, G.; Coulibaly, A.; Kakou-N’Douba, A.; N’Guetta, A.S.-P.; Resch, G.; Pourcel, C. Complete Genome Sequences of Five Acinetobacter baumannii Phages from Abidjan, Côte d’Ivoire. Microbiol. Resour. Announc. 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Ackermann, H.W.; Kropinski, A.M.; Lavigne, R.; Sutton, J.M.; Reynolds, D.M. Comparative Analysis of 37 Acinetobacter Bacteriophages. Viruses 2018, 10, 5. [Google Scholar] [CrossRef]
- Turner, D.; Wand, M.E.; Briers, Y.; Lavigne, R.; Sutton, J.M.; Reynolds, D.M. Characterisation and Genome Sequence of the Lytic Acinetobacter baumannii Bacteriophage VB_AbaS_Loki. PLoS ONE 2017, 12, e0172303. [Google Scholar] [CrossRef]
- Jin, J.; Li, Z.J.; Wang, S.W.; Wang, S.M.; Chen, S.J.; Huang, D.H.; Zhang, G.; Li, Y.H.; Wang, X.T.; Wang, J.; et al. Genome Organisation of the Acinetobacter Lytic Phage ZZ1 and Comparison with Other T4-like Acinetobacter Phages. BMC Genom. 2014, 15, 793. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.; Casjens, S.R.; Millard, A.D.; Harrison, C.; Gannon, L.; Chattaway, M.A. Genomic Analysis of Anderson Typing Phages of Salmonella typhimrium: Towards Understanding the Basis of Bacteria-Phage Interaction. Sci. Rep. 2023, 13, 10484. [Google Scholar] [CrossRef] [PubMed]
- Chaturongakul, S.; Ounjai, P. Phage-Host Interplay: Examples from Tailed Phages and Gram-Negative Bacterial Pathogens. Front. Microbiol. 2014, 5, 98825. [Google Scholar] [CrossRef] [PubMed]
- Stone, E.; Campbell, K.; Grant, I.; McAuliffe, O. Understanding and Exploiting Phage–Host Interactions. Viruses 2019, 11, 567. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Xie, L.; Liu, M.; Li, Q.; Wang, P.; Luo, C. Bactericidal Synergism Between Phage YC#06 and Antibiotics: A Combination Strategy to Target Multidrug-Resistant Acinetobacter baumannii In Vitro and In Vivo. Microbiol. Spectr. 2022, 10. [Google Scholar] [CrossRef]
- Badie, O.H.; Basyony, A.F.; Samir, R. Computer-Based Identification of Potential Druggable Targets in Multidrug-Resistant Acinetobacter baumannii: A Combined In Silico, In Vitro and In Vivo Study. Microorganisms 2022, 10, 1973. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T.; Danis-Wlodarczyk, K.M.; Wozniak, D.J. Phage Cocktail Development for Bacteriophage Therapy: Toward Improving Spectrum of Activity Breadth and Depth. Pharmaceuticals 2021, 14, 1019. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shen, W.; Zhong, Q.; Chen, Q.; He, X.; Baker, J.L.; Xiong, K.; Jin, X.; Wang, J.; Hu, F.; et al. Development of a Bacteriophage Cocktail to Constrain the Emergence of Phage-Resistant Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 327. [Google Scholar] [CrossRef] [PubMed]
- Cieplak, T.; Soffer, N.; Sulakvelidze, A.; Nielsen, D.S. A Bacteriophage Cocktail Targeting Escherichia coli Reduces E. coli in Simulated Gut Conditions, While Preserving a Non-Targeted Representative Commensal Normal Microbiota. Gut Microbes 2018, 9, 391–399. [Google Scholar] [CrossRef]
- Abdelrahman, F.; Easwaran, M.; Daramola, O.I.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E.; et al. Phage-Encoded Endolysins. Antibiotics 2021, 10, 124. [Google Scholar] [CrossRef]
- Heselpoth, R.D.; Swift, S.M.; Linden, S.B.; Mitchell, M.S.; Nelson, D.C. Enzybiotics: Endolysins and Bacteriocins. Bacteriophages 2021, 3, 989–1030. [Google Scholar] [CrossRef]
- Luong, T.; Salabarria, A.C.; Edwards, R.A.; Roach, D.R. Standardized Bacteriophage Purification for Personalized Phage Therapy. Nat. Protoc. 2020, 15, 2867–2890. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, X.; Kang, G.; Bai, L.; Wang, P.; Huang, H. Isolation and Characterization of AbTJ, an Acinetobacter baumannii Phage, and Functional Identification of Its Receptor-Binding Modules. Viruses 2020, 12, 205. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Joshi, H. Simple Two-Step, High Yield Protocol for Isolation and Amplification of Bacteriophages against Methicillin-Resistant Staphylococcus aureus (MRSA). Curr. Protoc. 2022, 2, e395. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, N.; Barr, J.J. Phage on Tap: A Quick and Efficient Protocol for the Preparation of Bacteriophage Laboratory Stocks. Methods Mol. Biol. 2018, 1838, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Kim, S.; Kim, S.M.; Seol, S.Y.; Kim, J. Characterization of Induced Staphylococcus aureus Bacteriophage SAP-26 and Its Anti-Biofilm Activity with Rifampicin. Biofouling 2011, 27, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Mardiana, M.; Teh, S.H.; Lin, L.C.; Lin, N.T. Isolation and Characterization of a Novel Siphoviridae Phage, VB_AbaS_TCUP2199, Infecting Multidrug-Resistant Acinetobacter baumannii. Viruses 2022, 14, 1240. [Google Scholar] [CrossRef]
- Mirzaei, M.K.; Nilsson, A.S. Isolation of Phages for Phage Therapy: A Comparison of Spot Tests and Efficiency of Plating Analyses for Determination of Host Range and Efficacy. PLoS ONE 2015, 10, e0127606. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, D.; Martín-Platero, A.M.; Rodríguez, A.; Martínez-Bueno, M.; García, P.; Martínez, B. Typing of Bacteriophages by Randomly Amplified Polymorphic DNA (RAPD)-PCR to Assess Genetic Diversity. FEMS Microbiol. Lett. 2011, 322, 90–97. [Google Scholar] [CrossRef]
- Abedon, S.T. Detection of Bacteriophages: Phage Plaques. Bacteriophages 2018, 2, 507–538. [Google Scholar] [CrossRef]
- Yang, H.; Liang, L.; Lin, S.; Jia, S. Isolation and Characterization of a Virulent Bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol. 2010, 10, 131. [Google Scholar] [CrossRef] [PubMed]
- Sisakhtpour, B.; Mirzaei, A.; Karbasizadeh, V.; Hosseini, N.; Shabani, M.; Moghim, S. The Characteristic and Potential Therapeutic Effect of Isolated Multidrug-Resistant Acinetobacter baumannii Lytic Phage. Ann. Clin. Microbiol. Antimicrob. 2022, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host Receptors for Bacteriophage Adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef] [PubMed]
- Letarov, A.V.; Kulikov, E.E. Adsorption of Bacteriophages on Bacterial Cells. Biochemistry 2017, 82, 1632–1658. [Google Scholar] [CrossRef] [PubMed]
- Kutter, E. Phage Host Range and Efficiency of Plating. Methods Mol. Biol. 2009, 501, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Philipson, C.W.; Voegtly, L.J.; Lueder, M.R.; Long, K.A.; Rice, G.K.; Frey, K.G.; Biswas, B.; Cer, R.Z.; Hamilton, T.; Bishop-Lilly, K.A. Characterizing Phage Genomes for Therapeutic Applications. Viruses 2018, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Harada, L.K.; Silva, E.C.; Campos, W.F.; Del Fiol, F.S.; Vila, M.; Dąbrowska, K.; Krylov, V.N.; Balcão, V.M. Biotechnological Applications of Bacteriophages: State of the Art. Microbiol. Res. 2018, 212–213, 38–58. [Google Scholar] [CrossRef] [PubMed]
- Grande, L.; Michelacci, V.; Tozzoli, R.; Ranieri, P.; Maugliani, A.; Caprioli, A.; Morabito, S. Whole Genome Sequence Comparison of Vtx2-Converting Phages from Enteroaggregative Haemorrhagic Escherichia coli Strains. BMC Genom. 2014, 15, 574. [Google Scholar] [CrossRef]
- Lood, R.; Collin, M. Characterization and Genome Sequencing of Two Propionibacterium acnes Phages Displaying Pseudolysogeny. BMC Genom. 2011, 12, 198. [Google Scholar] [CrossRef]
- de Paiva, V.A.; de Gomes, I.S.; Monteiro, C.R.; Mendonça, M.V.; Martins, P.M.; Santana, C.A.; Gonçalves-Almeida, V.; Izidoro, S.C.; de Melo-Minardi, R.C.; Silveira, S.d.A. Protein Structural Bioinformatics: An Overview. Comput. Biol. Med. 2022, 147, 105695. [Google Scholar] [CrossRef]


| 1 CRAB | Source | Date Collection | 2 CCs | 3 STs | Spot Test Results | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Φ AW8 | Φ AW9 | Φ AW15-1 | Φ AW15-2 | Φ AW15-3 | Φ AW15-4 | Φ AW16-1 | Φ AW16-2 | Φ AW16-3 | Φ AW16-4 | Φ AW16-5 | |||||
| KBN10P02782 | Blood | 2013 | 552 | 552 | - | - | CL | CL | CL | CL | CL | CL | CL | CL | CL |
| LIS20145805 | ND | 2014 | 110 | 229 | CL | OL | CL | CL | CL | CL | CL | CL | CL | CL | CL |
| LIS20145719 | ND | 2014 | CL | - | CL | CL | CL | CL | CL | CL | CL | CL | CL | ||
| LIS20144539 | ND | 2014 | CL | OL | CL | CL | CL | CL | CL | CL | CL | CL | CL | ||
| KBN10P04598 | Blood | 2016 | CL | - | CL | CL | CL | CL | CL | CL | CL | CL | CL | ||
| LIS20130976 | Sputum | 2013 | 92 | 357 | - | SCL | CL | CL | CL | CL | CL | CL | CL | CL | CL |
| LIS20130721 | Sputum | 2013 | - | SCL | CL | CL | CL | CL | CL | CL | CL | CL | CL | ||
| LIS20130567 | Urine | 2013 | - | SCL | CL | CL | CL | CL | CL | CL | CL | CL | CL | ||
| KBN10P04322 | Blood | 2016 | OL | SCL | SCL | SCL | SCL | SCL | SCL | SCL | SCL | SCL | SCL | ||
| KBN10P05102 | Blood | 2016 | 784 | CL | CL | CL | CL | CL | CL | CL | CL | CL | CL | CL | |
| KBN10P04703 | Blood | 2016 | CL | CL | CL | CL | CL | CL | CL | CL | CL | CL | CL | ||
| KBN10P04697 | Blood | 2017 | CL | CL | CL | CL | CL | CL | CL | CL | CL | CL | CL | ||
| KBN10P02972 | Pus | 2013 | 191 | - | SCL | - | - | - | - | - | - | - | - | - | |
| KBN10P02901 | ND | 2013 | - | SCL | - | - | - | - | - | - | - | - | - | ||
| KBN10P02755 | Blood | 2013 | - | OL | - | - | - | - | - | - | - | - | - | ||
| KBN10P04594 | Venous blood | 2016 | - | OL | - | - | - | - | - | - | - | - | - | ||
| KBN10P04627 | Central blood | 2016 | - | SCL | - | - | - | - | - | - | - | - | - | ||
| KBN10P04948 | Blood | 2017 | - | SCL | - | - | - | - | - | - | - | - | - | ||
| KBN10P02768 | Blood | 2013 | 208 | - | OL | - | - | - | - | - | - | - | - | - | |
| LIS20132370 | ND | 2013 | OL | OL | - | - | - | - | - | - | - | - | - | ||
| KBN10P04322 | Blood | 2016 | OL | SCL | - | - | - | - | - | - | - | - | - | ||
| LIS20140444 | ND | 2014 | 369 | OL | SCL | - | - | - | - | - | - | - | - | - | |
| LIS20138989 | ND | 2013 | - | SCL | - | - | - | - | - | - | - | - | - | ||
| LIS20137924 | Ascites | 2013 | - | SCL | - | - | - | - | - | - | - | - | - | ||
| KBN10P04633 | Blood | 2016 | - | SCL | - | - | - | - | - | - | - | - | - | ||
| KBN10P05663 | Blood | 2018 | - | SCL | - | - | - | - | - | - | - | - | - | ||
| KBN10P05982 | Blood | 2018 | - | SCL | - | - | - | - | - | - | - | - | - | ||
| KBN10P04600 | Blood | 2016 | 451 | - | SCL | - | - | - | - | - | - | - | - | - | |
| KBN10P05231 | Blood | 2016 | - | SCL | - | - | - | - | - | - | - | - | - | ||
| A. baumannii ATCC17978 | CL | CL | CL | CL | CL | CL | CL | CL | CL | CL | CL | ||||
| A. baumannii ATCC19606 | - | SCL | - | - | - | - | - | OL | OL | OL | OL | ||||
| Properties of Acinetobacter baumannii Phages | |||||
|---|---|---|---|---|---|
| Feature | Acinetobacter Phage | ||||
| AW8 (vB_AbaP_W8) | AW9 (vB_AbaSi_W9) | AW16-4 (vB_AbaSt_W16) | |||
| Phage characteristics | |||||
| Source | Hospital sewage | ||||
| Isolation date | 2022 | 2022 | 2022 | ||
| Isolation strain | A. baumannii ATCC17978 | A. baumannii clinical isolate KBN10P02782 | |||
| Summary of phage host spectra: no. lysed/no. tested (% lysed) | |||||
| 1 CC552 (n = 1) | 2 EOP > 0.1 | 0/1 (0%) | 0/1 (0%) | 1/1 (100%) | |
| EOP > 0.001 | 0/1 (0%) | 0/1 (0%) | 0/1 (0%) | ||
| CC110 (n = 4) | EOP > 0.1 | 4/4 (100%) | 0/4 (0%) | 4/4 (100%) | |
| EOP > 0.001 | 0/4 (0%) | 0/4 (0%) | 0/4 (0%) | ||
| CC92 (n = 24) | EOP > 0.1 | 3/24 (13%) | 3/24 (13%) | 7/24 (29%) | |
| EOP > 0.001 | 0/24 (0%) | 23/24 (96%) | 0/24 (0%) | ||
| Total clinical isolates (n = 29) | EOP > 0.1 | 7/29 (24%) | 3/29 (10%) | 12/29 (41%) | |
| EOP > 0.001 | 0/29 (0%) | 23/29 (79%) | 0/29 (0%) | ||
| Plaque morphology |  | 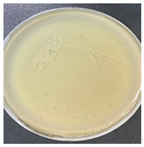 |  | ||
| Plaque size (cm) | 0.5–0.7 | 0.1–0.2 | 0.2–0.3 | ||
| Stock (PFU/mL) | 1.62 × 108 | 6.75 × 1014 | 2.84 × 1014 | ||
| Transmission electron microscopy (TEM) | Phage morphology |  | 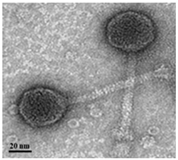 | 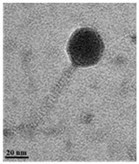 | |
| Podovirus | Myovirus | ||||
| Tail length | 2.75 ± 6.51 nm (n = 5) | 145.45 ± 4.17 nm (n = 15) | 112.5 ± 4.80 nm (n = 10) | ||
| Tail width | 8.74 ± 2.74 nm (n = 3) | 5.68 ± 4.44 nm (n = 15) | 5.6 ± 2.34 nm (n = 10) | ||
| Capsid dimeter | 68.54 ± 3.61 nm (n = 10) | 40.10 ± 2.65 nm (n = 15) | 38.2 ± 1.75 nm (n = 10) | ||
| Growth Properties | |||||
| Adsorption constant (PFU/mL) | 1 × 106 | ||||
| 90% adsorbed (min) | 1 | 1 | 3 | ||
| Latent period (min) | 10 | 20 | 5 | ||
| Burst size (PFU/cell) | 164 | 117 | 1102 | ||
| Attribute | Acinetobacter Phage vB_AbaP_W8 | Acinetobacter Phage vB_AbaSi_W9 | Acinetobacter Phage vB_AbaSt_W16 | |
|---|---|---|---|---|
| Accession no. | PP174318 | PP146379 | PP174317 | |
| Genome size (bp) | 41,326 | 43,022 | 166,741 | |
| GC content (%) | 39.2 | 45.6 | 34.4 | |
| Feature content | 48 | 56 | 242 | |
| Hypothetical proteins | 23 | 27 | 130 | |
| Length of direct terminal repeats (amino acids) | 25 | 29 | 112 | |
| tRNA genes | None | None | 10 | |
| tRNA genes | None | None | 10 | |
| Bacterial toxin genes | None | None | None | |
| Antibiotic resistance genes | None | None | None | |
| Genes indicating temperate lifecycle | None | None | None | |
| Family | Autographiviridae | Unclassified | Straboviridae | |
| Genus | Friunavirus | Lokivirus | Zedzedvirus | |
| Most similar phage sequence | Scientific name | Acinetobacter phage vB_AbaP_B09_Aci08 | Acinetobacter phage IMEAB3 | Acinetobacter phage ZZ1 |
| Query cover (%) | 90 | 99 | 94 | |
| Identity with (%) | 94.73 | 96.62 | 98.83 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, Y.-J.; Kim, S.; Shin, M.; Kim, J. Isolation and Characterization of Novel Bacteriophages to Target Carbapenem-Resistant Acinetobacter baumannii. Antibiotics 2024, 13, 610. https://doi.org/10.3390/antibiotics13070610
Choi Y-J, Kim S, Shin M, Kim J. Isolation and Characterization of Novel Bacteriophages to Target Carbapenem-Resistant Acinetobacter baumannii. Antibiotics. 2024; 13(7):610. https://doi.org/10.3390/antibiotics13070610
Chicago/Turabian StyleChoi, Yoon-Jung, Shukho Kim, Minsang Shin, and Jungmin Kim. 2024. "Isolation and Characterization of Novel Bacteriophages to Target Carbapenem-Resistant Acinetobacter baumannii" Antibiotics 13, no. 7: 610. https://doi.org/10.3390/antibiotics13070610
APA StyleChoi, Y.-J., Kim, S., Shin, M., & Kim, J. (2024). Isolation and Characterization of Novel Bacteriophages to Target Carbapenem-Resistant Acinetobacter baumannii. Antibiotics, 13(7), 610. https://doi.org/10.3390/antibiotics13070610






