Exploring Weissella confusa W1 and W2 Strains Isolated from Khao-Mahk as Probiotic Candidates: From Phenotypic Traits to Genomic Insights
Abstract
1. Introduction
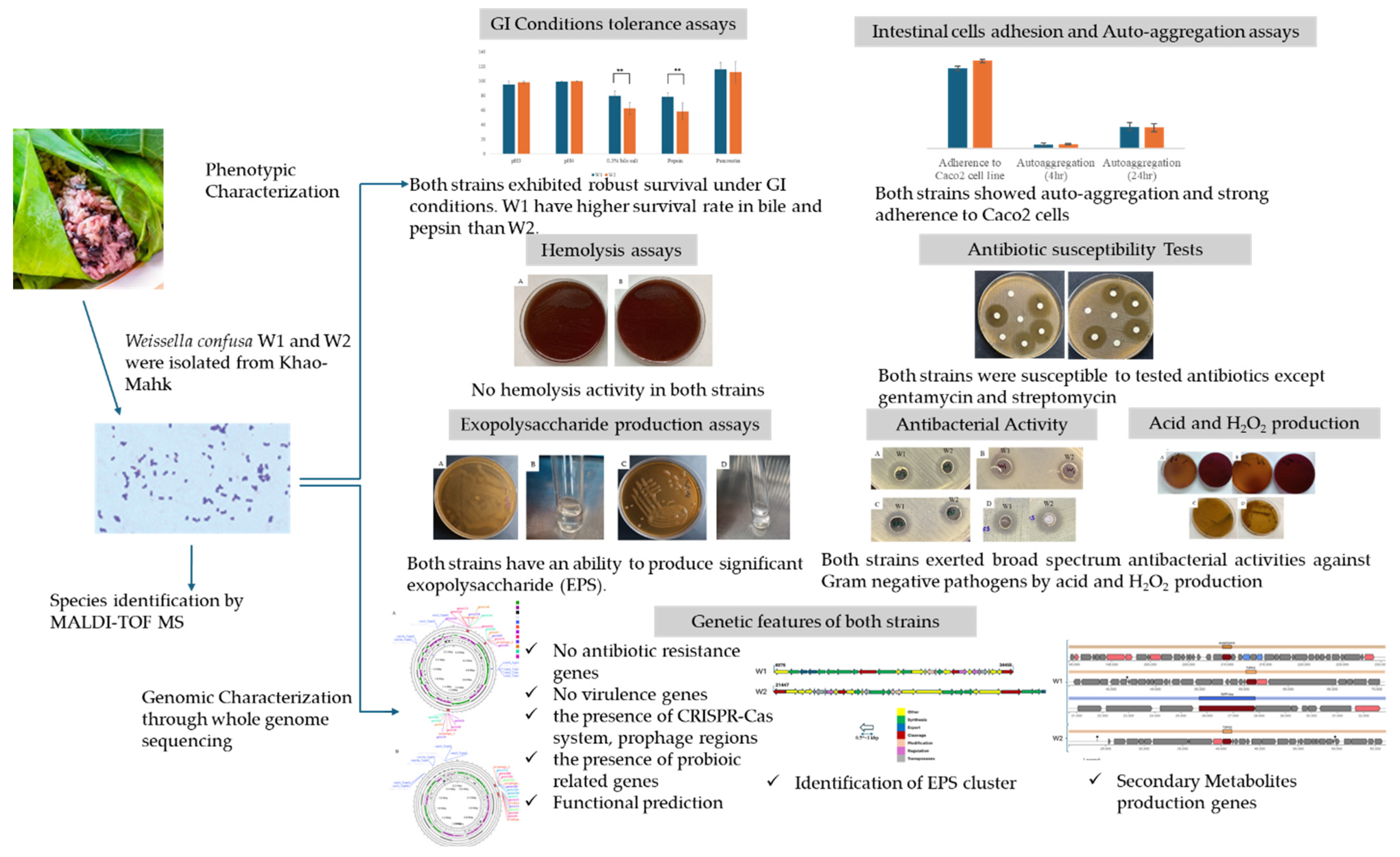
2. Results and Discussion
2.1. Bacterial Isolation and Identification
2.2. Phenotypic Assays
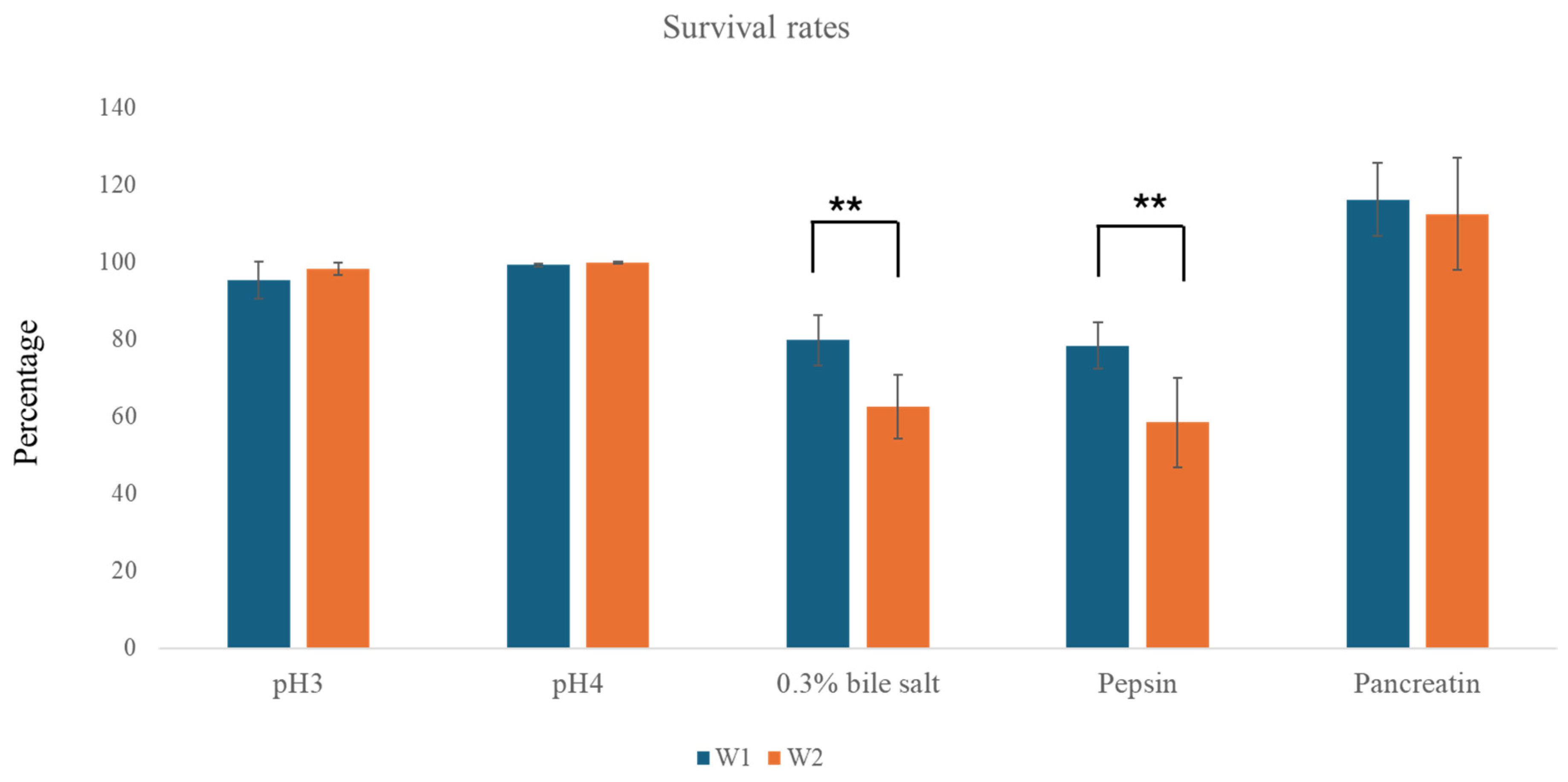
2.2.1. Tolerance Tests to Gastric and Intestinal Conditions
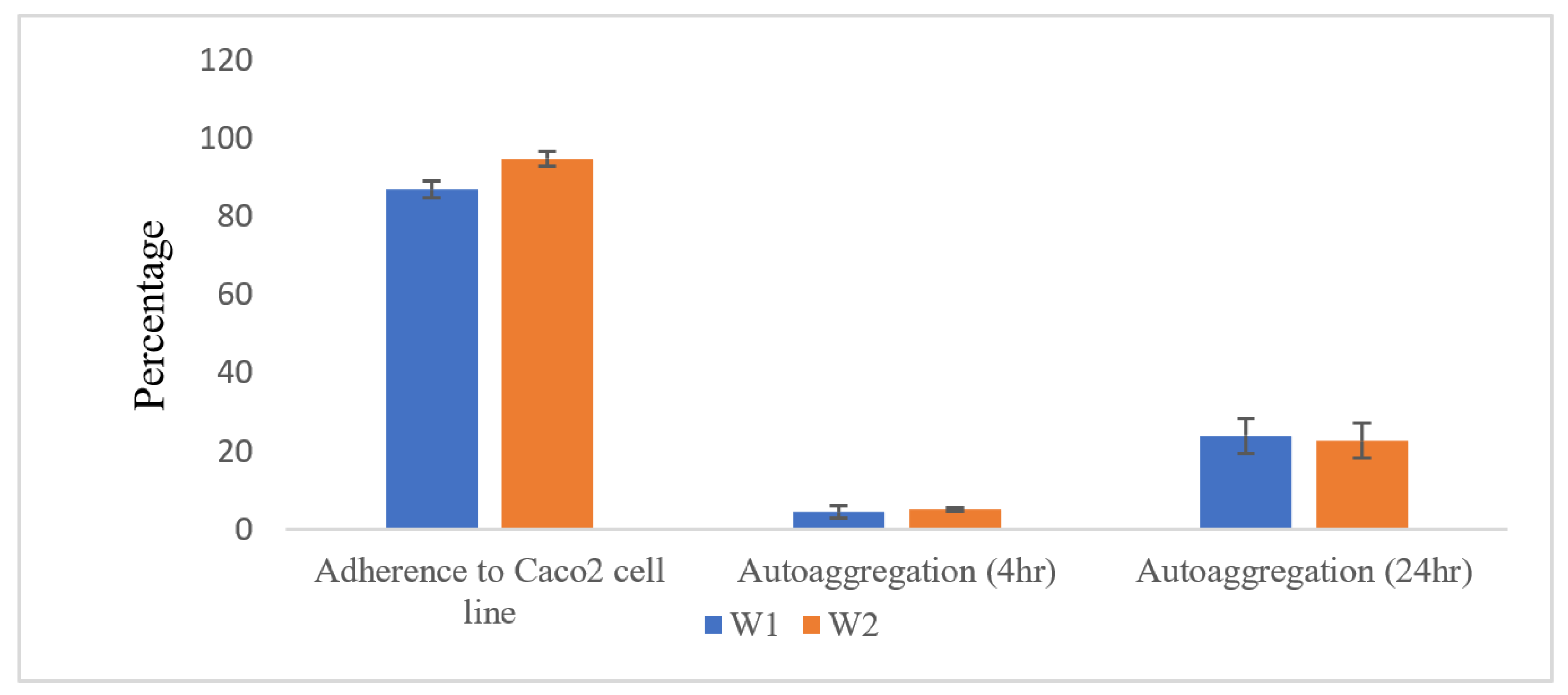
2.2.2. Auto-Aggregation and Adherence to Caco2 Intestinal Cell Line
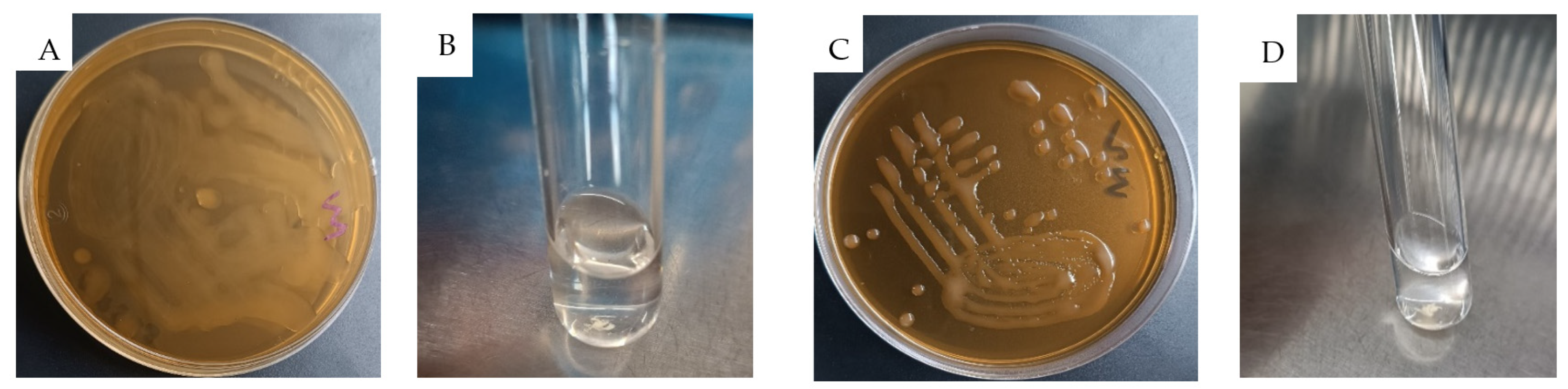
2.2.3. Exopolysaccharide (EPS) Production
2.2.4. Safety Assessment
- (a)
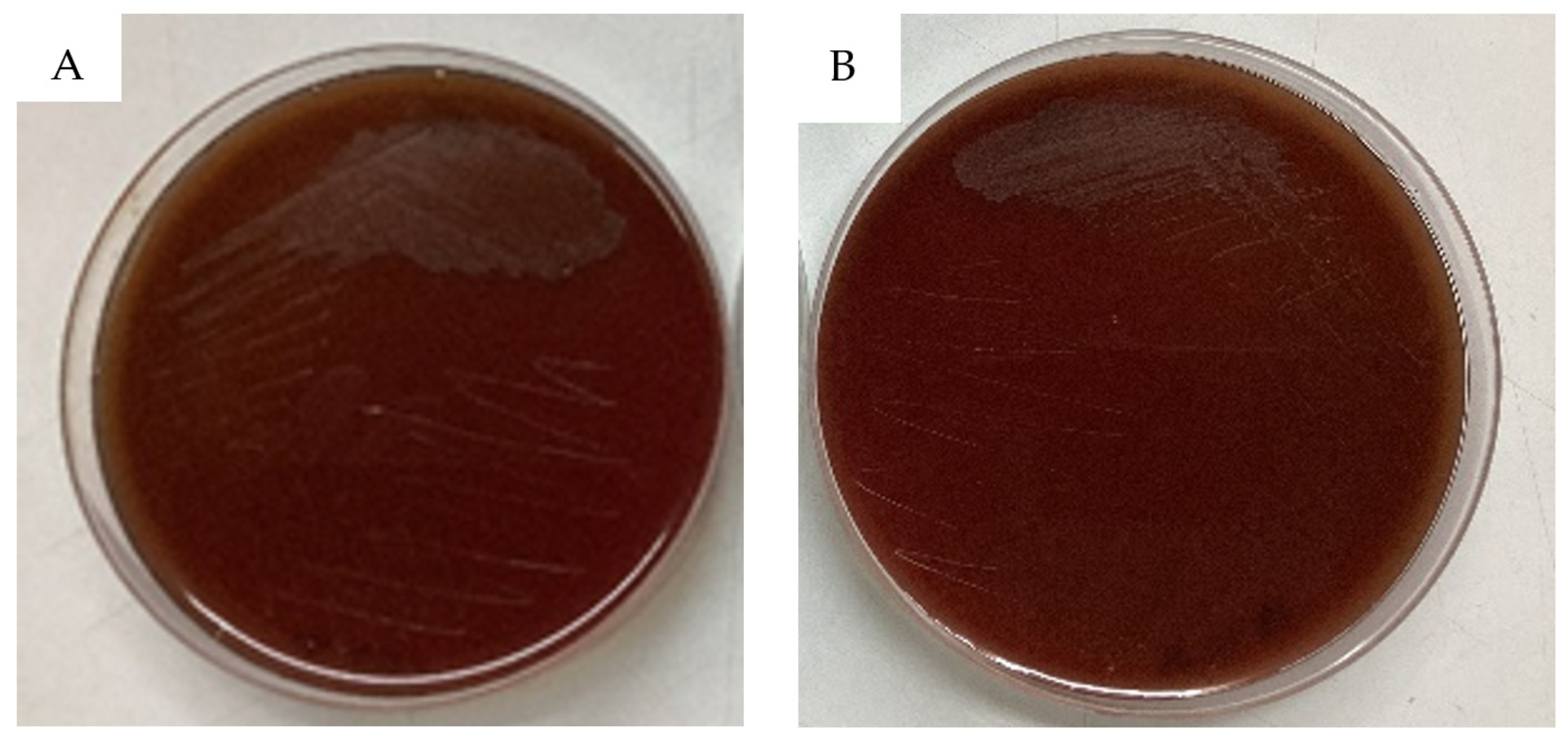
- Hemolysis Test
- (b)
- Antibiotic Susceptibility Test
2.2.5. Production of Antimicrobial Substances
2.2.6. Antibacterial Activity
2.3. Genomic Analysis
2.3.1. Genome Features
2.3.2. Genomic Safety Assessment
2.3.3. Prophage and CRISPR-Cas
2.3.4. Functional Prediction
2.3.5. Genes Related to Probiotic Properties
2.3.6. EPS Production-Related Gene Cluster
2.3.7. Secondary Metabolic Products
2.3.8. Data Summarization
3. Material and Methods
3.1. Bacteria Isolation and Identification
3.2. Determination of Probiotic Properties
3.2.1. Determination of Bile Tolerance
3.2.2. Determination of Acid Tolerance
3.2.3. Detection of Pepsin and Pancreatin Tolerance
3.2.4. Auto-Aggregation
3.2.5. Exopolysaccharide Production Activity
3.2.6. Determination for Adherence to Intestinal Epithelial Cells
3.2.7. Acid and H2O2 Production
3.3. Phenotypic Safety Assessment
3.3.1. Hemolytic Potential
3.3.2. Susceptibility to Antibiotics
3.4. Determination of Antimicrobial Activities of Probiotic Isolates (Agar-Well Diffusion Method)
3.5. Genomic DNA Extraction and Bioinformatics Analysis
3.6. Statiscal Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tananuwong, K.; Tewaruth, W. Extraction and application of antioxidants from black glutinous rice. LWT 2010, 43, 476–481. [Google Scholar] [CrossRef]
- Nuraida, L. A review: Health promoting lactic acid bacteria in traditional Indonesian fermented foods. Food Sci. Hum. Wellness 2015, 4, 47–55. [Google Scholar] [CrossRef]
- Fuller, R. Probiotics in human medicine. Gut 1991, 32, 439. [Google Scholar] [CrossRef]
- Tongyai, A.; Riebroy, S.; Maneerat, S.; Siriwong, N.; Chulakarungka, S. Changes in Physicochemical and Sensory Characteristics during Fermentation of Khaow-Maak from Black Glutinous Rice. 2012. Available online: https://kukr.lib.ku.ac.th/kukr_es/rice/search_detail/dowload_digital_file/13167/87849 (accessed on 12 December 2012).
- Sornsenee, P.; Singkhamanan, K.; Sangkhathat, S.; Saengsuwan, P.; Romyasamit, C. Probiotic Properties of Lactobacillus Species Isolated from Fermented Palm Sap in Thailand. Probiotics Antimicrob. Proteins 2021, 13, 957–969. [Google Scholar] [CrossRef]
- Sulistiani; Novarina, I.; Inawati; Dinoto, A.; Julistiono, H.; Handayani, R.; Saputra, S. Assessment of Potential Probiotic Lactic Acid Bacteria from Tempe and Tape. IOP Conf. Ser. Earth Environ. Sci. 2020, 572, 012026. [CrossRef]
- Collins, M.D.; Samelis, J.; Metaxopoulos, J.; Wallbanks, S. Taxonomic studies on some leuconostoc-like organisms from fermented sausages: Description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J. Appl. Bacteriol. 1993, 75, 595–603. [Google Scholar] [CrossRef]
- Fusco, V.; Quero, G.M.; Cho, G.S.; Kabisch, J.; Meske, D.; Neve, H.; Bockelmann, W.; Franz, C.M.A.P. The genus Weissella: Taxonomy, ecology and biotechnological potential. Front. Microbiol. 2015, 6, 130276. [Google Scholar] [CrossRef]
- Bjo $ Rkroth, K.J.; Schillinger, U.; Geisen, R.; Weiss, N.; Hoste, B.; Holzapfel, W.H.; Korkeala, H.J.; Vandamme, P. Taxonomic study of Weissella confusa and description of Weissella cibaria sp. nov., detected in food and clinical samples. Int. J. Syst. Evol. Microbiol. 2002, 52, 141–148. [Google Scholar] [CrossRef]
- Ayeni, F.A.; Sánchez, B.; Adeniyi, B.A.; de los Reyes-Gavilán, C.G.; Margolles, A.; Ruas-Madiedo, P. Evaluation of the functional potential of Weissella and Lactobacillus isolates obtained from Nigerian traditional fermented foods and cow’s intestine. Int. J. Food Microbiol. 2011, 147, 97–104. [Google Scholar] [CrossRef]
- Lee, K.W.; Park, J.Y.; Jeong, H.R.; Heo, H.J.; Han, N.S.; Kim, J.H. Probiotic properties of Weissella strains isolated from human faeces. Anaerobe 2012, 18, 96–102. [Google Scholar] [CrossRef]
- Teixeira, C.G.; Fusieger, A.; Milião, G.L.; Martins, E.; Drider, D.; Nero, L.A.; de Carvalho, A.F. Weissella: An Emerging Bacterium with Promising Health Benefits. Probiotics Antimicrob. Proteins 2021, 13, 915–925. [Google Scholar] [CrossRef]
- Yu, H.S.; Jang, H.J.; Lee, N.K.; Paik, H.D. Evaluation of the probiotic characteristics and prophylactic potential of Weissella cibaria strains isolated from kimchi. LWT 2019, 112, 108229. [Google Scholar] [CrossRef]
- Nami, Y.; Panahi, B.; MohammadZadeh Jalaly, H.; Hejazi, M.A. Isolation and assessment of novel exopolysaccharide-producing Weissella confusa ABRIIFBI-96 isolated from an Iranian homemade dairy fermented food “Tof” as a main starter culture for probiotic fermented milk. LWT 2024, 197, 115910. [Google Scholar] [CrossRef]
- Sadeghi, M.; Panahi, B.; Mazlumi, A.; Hejazi, M.A.; Nami, Y. Screening of potential probiotic lactic acid bacteria with antimicrobial properties and selection of superior bacteria for application as biocontrol using machine learning models. LWT 2022, 162, 113471. [Google Scholar] [CrossRef]
- Jurášková, D.; Ribeiro, S.C.; Silva, C.C.G. Exopolysaccharides Produced by Lactic Acid Bacteria: From Biosynthesis to Health-Promoting Properties. Foods 2022, 11, 156. [Google Scholar] [CrossRef]
- Verdenelli, M.C.; Ghelfi, F.; Silvi, S.; Orpianesi, C.; Cecchini, C.; Cresci, A. Probiotic properties of Lactobacillus rhamnosus and Lactobacillus paracasei isolated from human faeces. Eur. J. Nutr. 2009, 48, 355–363. [Google Scholar] [CrossRef]
- Sornplang, P.; Piyadeatsoontorn, S. Probiotic isolates from unconventional sources: A review. J. Anim. Sci. Technol. 2016, 58, 26. [Google Scholar] [CrossRef]
- Rodríguez-Sánchez, S.; Fernández-Pacheco, P.; Seseña, S.; Pintado, C.; Ll Palop, M. Selection of probiotic Lactobacillus strains with antimicrobial activity to be used as biocontrol agents in food industry. LWT 2021, 143, 111142. [Google Scholar] [CrossRef]
- Kandasamy, S.; Yoo, J.; Yun, J.; Lee, K.H.; Kang, H.B.; Kim, J.E.; Oh, M.H.; Ham, J.S. Probiogenomic In-Silico Analysis and Safety Assessment of Lactiplantibacillus plantarum DJF10 Strain Isolated from Korean Raw Milk. Int. J. Mol. Sci. 2022, 23, 14494. [Google Scholar] [CrossRef]
- Patrone, V.; Al-Surrayai, T.; Romaniello, F.; Fontana, A.; Milani, G.; Sagheddu, V.; Puglisi, E.; Callegari, M.L.; Al-Mansour, H.; Kishk, M.W.; et al. Integrated Phenotypic-Genotypic Analysis of Candidate Probiotic Weissella cibaria Strains Isolated from Dairy Cows in Kuwait. Probiotics Antimicrob. Proteins 2021, 13, 809–823. [Google Scholar] [CrossRef]
- Paulo, E.M.; Vasconcelos, M.P.; Oliveira, I.S.; Affe, H.M.d.J.; Nascimento, R.; de Melo, I.S.; Roque, M.R.d.A.; de Assis, S.A. An alternative method for screening lactic acid bacteria for the production of exopolysaccharides with rapid confirmation. Food Sci. Technol. 2012, 32, 710–714. [Google Scholar] [CrossRef]
- Kumari, M.; Kumar, R.; Singh, D.; Bhatt, S.; Gupta, M. Physiological and genomic characterization of an exopolysaccharide-producing Weissella cibaria CH2 from cheese of the western Himalayas. Food Biosci. 2020, 35, 100570. [Google Scholar] [CrossRef]
- Pradhan, D.; Mallappa, R.H.; Grover, S. Comprehensive approaches for assessing the safety of probiotic bacteria. Food Control 2020, 108, 106872. [Google Scholar] [CrossRef]
- Lakra, A.K.; Domdi, L.; Hanjon, G.; Tilwani, Y.M.; Arul, V. Some probiotic potential of Weissella confusa MD1 and Weissella cibaria MD2 isolated from fermented batter. LWT 2020, 125, 109261. [Google Scholar] [CrossRef]
- Schellenberg, J.J.; Dumonceaux, T.J.; Hill, J.E.; Kimani, J.; Jaoko, W.; Wachihi, C.; Mungai, J.N.; Lane, M.; Fowke, K.R.; Ball, T.B.; et al. Selection, Phenotyping and Identification of Acid and Hydrogen Peroxide Producing Bacteria from Vaginal Samples of Canadian and East African Women. PLoS ONE 2012, 7, e41217. [Google Scholar] [CrossRef]
- de Moreno de LeBlanc, A.; LeBlanc, J.G. Effect of probiotic administration on the intestinal microbiota, current knowledge and potential applications. World J. Gastroenterol. 2014, 20, 16518. [Google Scholar] [CrossRef]
- Pridmore, R.D.; Pittet, A.C.; Praplan, F.; Cavadini, C. Hydrogen peroxide production by Lactobacillus johnsonii NCC 533 and its role in anti-Salmonella activity. FEMS Microbiol. Lett. 2008, 283, 210–215. [Google Scholar] [CrossRef]
- Ho Sui, S.J.; Fedynak, A.; Hsiao, W.W.L.; Langille, M.G.I.; Brinkman, F.S.L. The Association of Virulence Factors with Genomic Islands. PLoS ONE 2009, 4, e8094. [Google Scholar] [CrossRef]
- Sharma, V.; Harjai, K.; Shukla, G. Effect of bacteriocin and exopolysaccharides isolated from probiotic on P. aeruginosa PAO1 biofilm. Folia Microbiol. 2018, 63, 181–190. [Google Scholar] [CrossRef]
- Colautti, A.; Arnoldi, M.; Comi, G.; Iacumin, L. Antibiotic resistance and virulence factors in lactobacilli: Something to carefully consider. Food Microbiol. 2022, 103, 103934. [Google Scholar] [CrossRef]
- Canchaya, C.; Proux, C.; Fournous, G.; Bruttin, A.; Brüssow, H. Prophage Genomics. Microbiol. Mol. Biol. Rev. 2003, 67, 238–276. [Google Scholar] [CrossRef]
- Yuan, S.; Wang, Y.; Zhao, F.; Kang, L. Complete Genome Sequence of Weissella confusa LM1 and Comparative Genomic Analysis. Front. Microbiol. 2021, 12, 749218. [Google Scholar] [CrossRef]
- Kleerebezem, M.; Boekhorst, J.; Van Kranenburg, R.; Molenaar, D.; Kuipers, O.P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.E.J.; et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 2003, 100, 1990–1995. [Google Scholar] [CrossRef]
- Liu, Z.; Dong, H.; Cui, Y.; Cong, L.; Zhang, D. Application of different types of CRISPR/Cas-based systems in bacteria. Microb. Cell Factories 2020, 19, 172. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Van Pijkeren, J.P.; Canchaya, C.; Ryan, K.A.; Li, Y.; Claesson, M.J.; Sheil, B.; Steidler, L.; O’Mahony, L.; Fitzgerald, G.F.; Van Sinderen, D.; et al. Comparative and functional analysis of sortase-dependent proteins in the predicted secretome of Lactobacillus salivarius UCC118. Appl. Environ. Microbiol. 2006, 72, 4143–4153. [Google Scholar] [CrossRef]
- Vankerckhoven, V.; Moreillon, P.; Phane Piu, S.; Giddey, M.; Huys, G.; Vancanneyt, M.; Goossens, H.; Entenza, J.M. Infectivity of Lactobacillus rhamnosus and Lactobacillus paracasei isolates in a rat model of experimental endocarditis. J. Med. Microbiol. 2007, 56, 1017–1024. [Google Scholar] [CrossRef][Green Version]
- Deo, D.; Davray, D.; Kulkarni, R. A Diverse Repertoire of Exopolysaccharide Biosynthesis Gene Clusters in Lactobacillus Revealed by Comparative Analysis in 106 Sequenced Genomes. Microorganisms 2019, 7, 444. [Google Scholar] [CrossRef]
- Pérez-Ramos, A.; Nácher-Vázquez, M.; López, P. Current and Future Applications of Bacterial Extracellular Polysaccharides. In Probiotics Prebiotics Synbiotics; Elsevier: Amsterdam, The Netherlands, 2016; pp. 329–344. [Google Scholar] [CrossRef]
- Nourikyan, J.; Kjos, M.; Mercy, C.; Cluzel, C.; Morlot, C.; Noirot-Gros, M.F.; Guiral, S.; Lavergne, J.P.; Veening, J.W.; Grangeasse, C. Autophosphorylation of the Bacterial Tyrosine-Kinase CpsD Connects Capsule Synthesis with the Cell Cycle in Streptococcus pneumoniae. PLoS Genet. 2015, 11, e1005518. [Google Scholar] [CrossRef]
- Vijayakumar, R.; Raja, S.S. Secondary Metabolites: Sources and Applications; InTech, 2018; Available online: https://www.intechopen.com/books/6670 (accessed on 5 September 2018). [CrossRef]
- Chávarri, M.; Diez-Gutiérrez, L.; Marañón, I.; Javier, R.; Barron, L. Secondary Metabolites from Probiotic Metabolism. In Advances in Probiotics; Elsevier: Amsterdam, The Netherlands, 2021; pp. 259–276. [Google Scholar] [CrossRef]
- Schöner, T.A.; Gassel, S.; Osawa, A.; Tobias, N.J.; Okuno, Y.; Sakakibara, Y.; Shindo, K.; Sandmann, G.; Bode, H.B. Aryl Polyenes, a Highly Abundant Class of Bacterial Natural Products, Are Functionally Related to Antioxidative Carotenoids. Chembiochem A Eur. J. Chem. Biol. 2016, 17, 247–253. [Google Scholar] [CrossRef]
- Naughton, L.M.; Romano, S.; O’Gara, F.; Dobson, A.D.W. Identification of secondary metabolite gene clusters in the Pseudovibrio Genus reveals encouraging biosynthetic potential toward the production of novel bioactive compounds. Front. Microbiol. 2017, 8, 1494. [Google Scholar] [CrossRef]
- Li, Y.; Rebuffat, S. The manifold roles of microbial ribosomal peptide–based natural products in physiology and ecology. J. Biol. Chem. 2020, 295, 34–54. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Yanagida, F.; Shinohara, T. Isolation and identification of lactic acid bacteria from soil using an enrichment procedure. Lett. Appl. Microbiol. 2005, 40, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Chukamnerd, A.; Singkhamanan, K.; Chongsuvivatwong, V.; Palittapongarnpim, P.; Doi, Y.; Pomwised, R.; Sakunrang, C.; Jeenkeawpiam, K.; Yingkajorn, M.; Chusri, S.; et al. Whole-genome analysis of carbapenem-resistant Acinetobacter baumannii from clinical isolates in Southern Thailand. Comput. Struct. Biotechnol. J. 2022, 20, 545–558. [Google Scholar] [CrossRef]
- Rodríguez, E.; Arqués, J.L.; Rodríguez, R.; Peirotén, Á.; Landete, J.M.; Medina, M. Antimicrobial properties of probiotic strains isolated from breast-fed infants. J. Funct. Foods 2012, 4, 542–551. [Google Scholar] [CrossRef]
- Mulaw, G.; Sisay Tessema, T.; Muleta, D.; Tesfaye, A. In vitro evaluation of probiotic properties of lactic acid bacteria isolated from some traditionally fermented Ethiopian food products. Int. J. Microbiol. 2019, 2019, 7179514. [Google Scholar] [CrossRef] [PubMed]
- Romyasamit, C.; Thatrimontrichai, A.; Aroonkesorn, A.; Chanket, W.; Ingviya, N.; Saengsuwan, P.; Singkhamanan, K. Enterococcus faecalis Isolated from Infant Feces Inhibits Toxigenic Clostridioides (Clostridium) difficile. Front. Pediatr. 2020, 8, 612. [Google Scholar] [CrossRef] [PubMed]
- Somashekaraiah, R.; Shruthi, B.; Deepthi, B.V.; Sreenivasa, M.Y. Probiotic Properties of Lactic Acid Bacteria Isolated from Neera: A Naturally Fermenting Coconut Palm Nectar. Front. Microbiol. 2019, 10, 1382. [Google Scholar] [CrossRef] [PubMed]
- Ruas-Madiedo, P.; De Los Reyes-Gavilán, C. Invited review: Methods for the screening, isolation, and characterization of exopolysaccharides produced by lactic acid bacteria. J. Dairy Sci. 2005, 88, 843–856. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rodríguez-Aparicio, L.; Rúa, J.; Martínez-Blanco, H.; Navasa, N.; García-Armesto, M.R.; Ferrero, M.Á. In vitro evaluation of physiological probiotic properties of different lactic acid bacteria strains of dairy and human origin. J. Funct. Foods 2012, 4, 531–541. [Google Scholar] [CrossRef]
- Song, Y.L.; Kato, N.; Matsumiya, Y.; Liu, C.X.; Kato, H.; Watanabe, K. Identification of and Hydrogen Peroxide Production by Fecal and Vaginal Lactobacilli Isolated from Japanese Women and Newborn Infants. J. Clin. Microbiol. 1999, 37, 3062. [Google Scholar] [CrossRef] [PubMed]
- Foulquié Moreno, M.; Callewaert, R.; Devreese, B.; Van Beeumen, J.; De Vuyst, L. Isolation and biochemical characterisation of enterocins produced by enterococci from different sources. J. Appl. Microbiol. 2003, 94, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Nigam, A.; Kumar, A.; Madhusuda, H.; Bhola, N. In-vitro Screening of antibacterial activity of lactic acid bacteria against common enteric pathogens Astha Nigam, Madhusudan HV, and Neelam Bhola Departmaent of Microbiology, CloneGen Biotechnology (P) Ltd. J. Biomed. Sci. 2012, 1. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Zdobnov, E.M. BUSCO: Assessing Genomic Data Quality and Beyond. Curr. Protoc. 2021, 1, e323. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Coudert, E.; Gehant, S.; de Castro, E.; Pozzato, M.; Baratin, D.; Neto, T.; Sigrist, C.J.A.; Redaschi, N.; Bridge, A.; Consortium, T.U. Annotation of biologically relevant ligands in UniProtKB using ChEBI. Bioinformatics 2022, 39, btac793. [Google Scholar] [CrossRef]
- Biomatters. Geneious Prime, 2023.2.1; Biomatters: Auckland, New Zealand, 2024; Available online: https://www.geneious.com/updates/geneious-prime-2023-2 (accessed on 1 June 2024).
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024. [Google Scholar]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.T.L.C.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder–an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb. Genom. 2022, 8, 000748. [Google Scholar] [CrossRef] [PubMed]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xiong, Z.; Sun, L.; Yang, J.; Jin, Q. VFDB 2012 update: Toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2012, 40, D641–D645. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kautsar, S.A.; Medema, M.H.; Weber, T. The antiSMASH database version 3: Increased taxonomic coverage and new query features for modular enzymes. Nucleic Acids Res. 2021, 49, D639–D643. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Starikova, E.V.; Tikhonova, P.O.; Prianichnikov, N.A.; Rands, C.M.; Zdobnov, E.M.; Ilina, E.N.; Govorun, V.M. Phigaro: High-throughput prophage sequence annotation. Bioinformatics 2020, 36, 3882–3884. [Google Scholar] [CrossRef]
- Pourcel, C.; Touchon, M.; Villeriot, N.; Vernadet, J.-P.; Couvin, D.; Toffano-Nioche, C.; Vergnaud, G. CRISPRCasdb a successor of CRISPRdb containing CRISPR arrays and cas genes from complete genome sequences, and tools to download and query lists of repeats and spacers. Nucleic Acids Res. 2020, 48, D535–D544. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
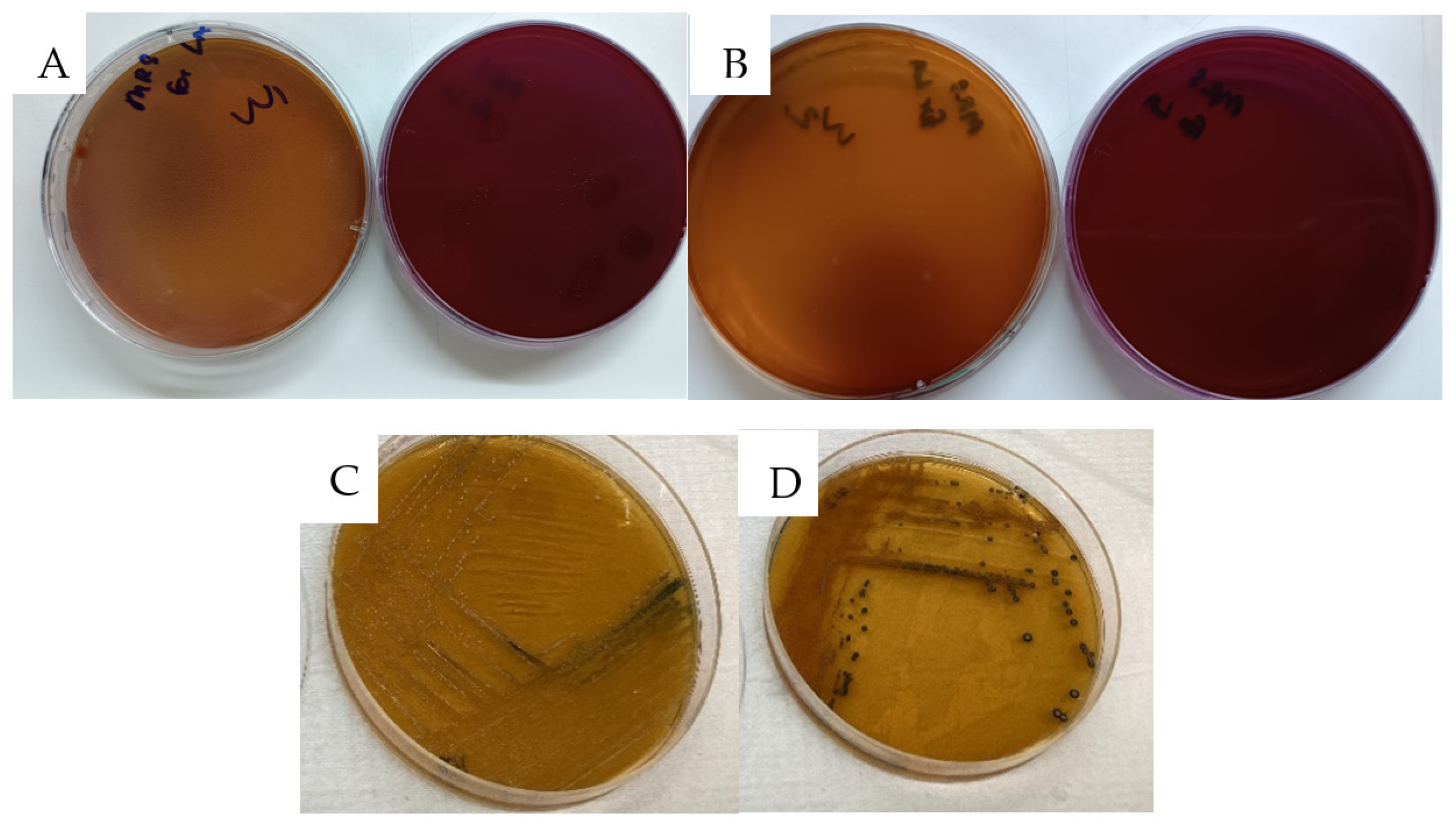
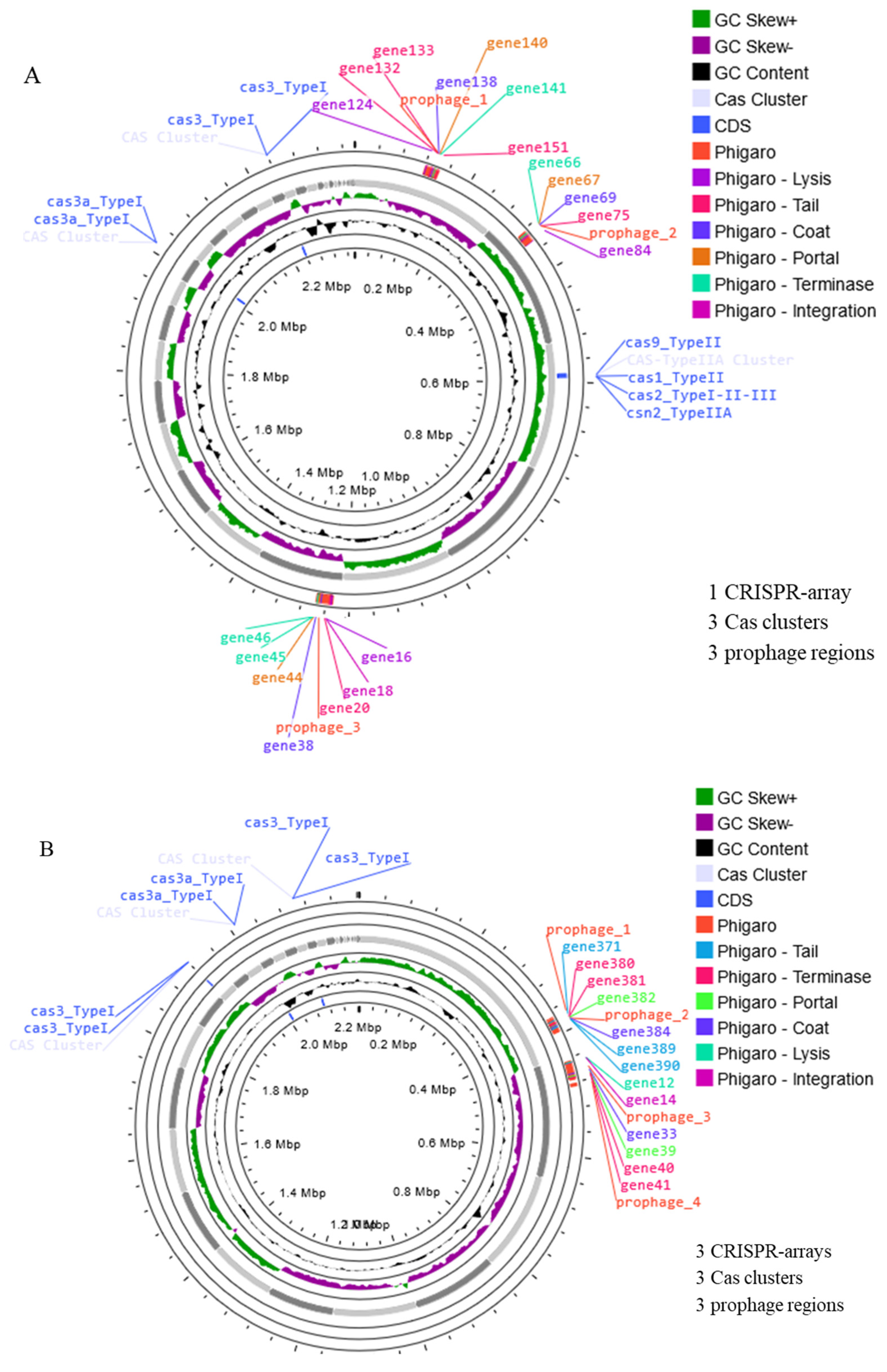

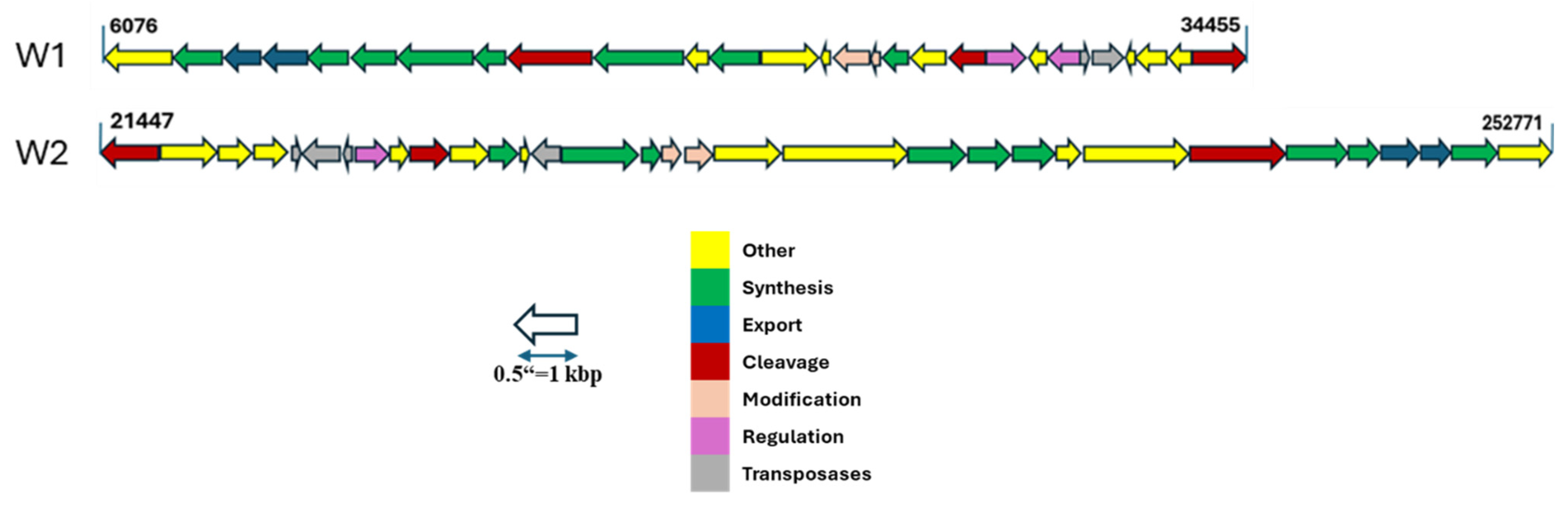
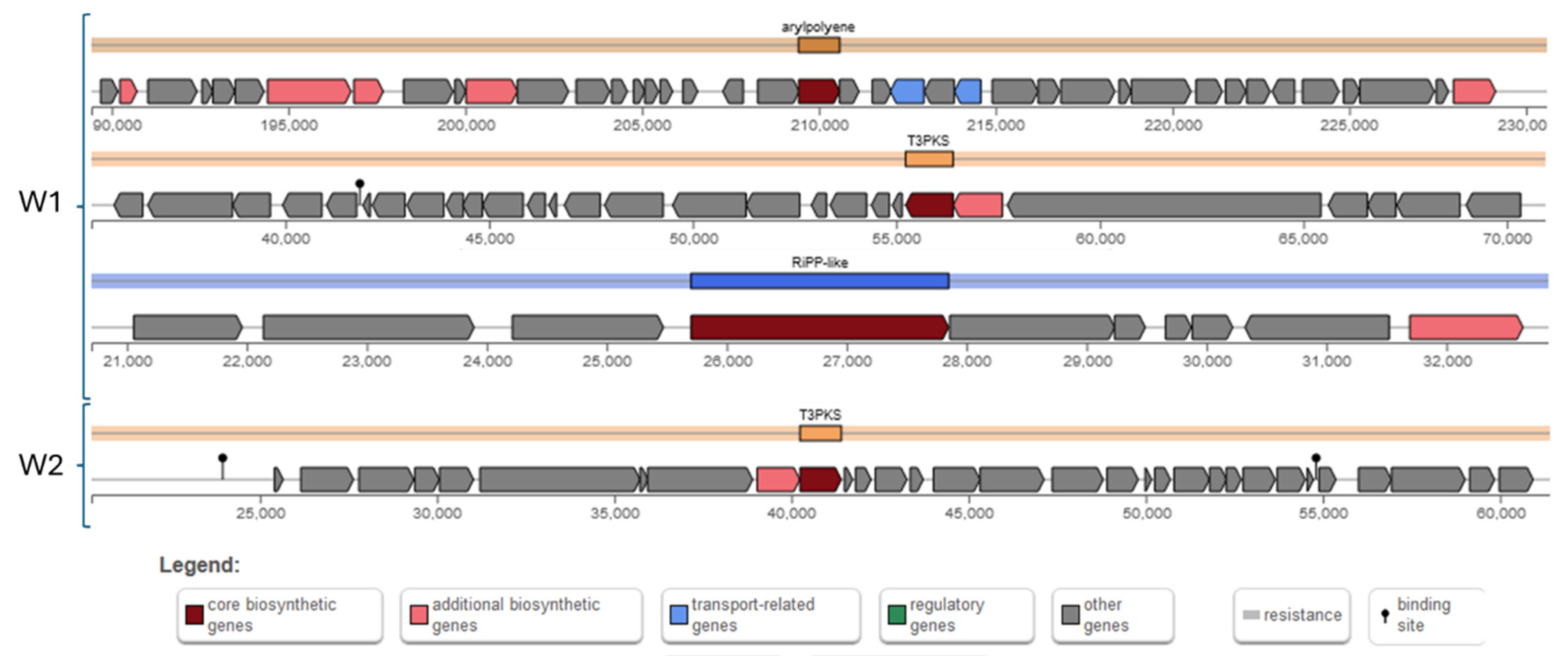
| Antibiotics | W1 | W2 |
|---|---|---|
| Inhibition Zone (mm) | Inhibition Zone (mm) | |
| Ampicillin (10 µg) | 21.1 ± 1.5 (S) | 20.8 ± 1.2 (S) |
| Erythromycin (15 µg) | 23.3 ± 1.8 (S) | 23.3 ± 1.8 (S) |
| Chloramphenicol (30 µg) | 24.8 ± 2.0 (S) | 24.5 ± 1.6 (S) |
| Gentamycin (10 µg) | 9.8 ± 0.6 (R) | 9.3 ± 0.8 (R) |
| Tetracycline (30 µg) | 23.2 ± 2.0 (S) | 22 ± 1.3 (S) |
| Streptomycin (10 µg) | 0 (R) | 0 (R) |
| Clindamycin (2 µg) | 25.8 ± 2.1 (S) | 24.8 ± 2.0 (S) |
| Zone of Inhibition (mm) ± SD | ||||
|---|---|---|---|---|
| Strains | S. maltophilia DMST 19079 | Sa. enterica Serotype Typhi DMST 22842 | V. cholerae DMST 2873 | CRAB SK005 |
| W1 | 13 ± 1.7 mm | 11.3 ± 1.5 mm | 10.7 ± 2.3 mm | 8.6 ± 1.2 mm |
| W2 | 12.3 ± 2.3 mm | 11.0 ± 1.7 mm | 10.0 ± 1.7 mm | 9.7 ± 1.5 mm |
| W1 | W2 | |
|---|---|---|
| Total genome size (bp) | 2,380,646 | 2,203,909 |
| Total contigs (≥200 bp) | 51 | 42 |
| G + C (%) | 44.62 | 44.78 |
| Largest contig (bp) | 277,000 | 438,208 |
| N50 | 209,762 | 157,099 |
| N90 | 37,460 | 31,601 |
| L50 | 5 | 5 |
| L90 | 17 | 14 |
| CDS | 2265 | 2131 |
| rRNA | 5 | 4 |
| tRNA | 81 | 74 |
| ANI value with W. confusa VTT E-133279 from NCBI (%) | 97.73 | 97.95 |
| Genes | Function/Description | W1 (No. of Gene) | W2 (No. of Gene) |
|---|---|---|---|
| Stress Response | |||
| lepA2 | Accurate and efficient protein synthesis under certain stress conditions | 1 | 2 |
| lepA | Accurate and efficient protein synthesis under certain stress conditions | 1 | 1 |
| uspA | Universal stress protein UspA and related nucleotide-binding proteins | 4 | 4 |
| uspA3 | Universal stress protein UspA and related nucleotide-binding proteins | 1 | 1 |
| ctsR | Firmicute transcriptional repressor of Class III stress genes | 1 | 1 |
| typA | Membrane GTPase involved in stress response | 1 | 1 |
| obg | An essential GTPase that plays a role in control of the cell cycle, stress response, ribosome biogenesis, differentiation, and morphogenesis | 1 | 1 |
| hslO | Redox regulated molecular chaperone for oxidative stress | 1 | 1 |
| groL | Prevents misfolding and promotes the refolding and proper assembly of unfolded polypeptides generated under stress conditions | 1 | 1 |
| Heat Shock | |||
| dnaK | Heat-shock 70 kDa protein | 1 | 1 |
| grpE | Participates actively in the response to hyperosmotic and heat shock by preventing the aggregation of stress-denatured proteins, in association with DnaK and GrpE | 1 | 1 |
| hrcA | Negative regulator of Class I heat-shock genes (grpE-dnaK-dnaJ and groELS operons). Prevents heat-shock induction of these operons | 1 | 1 |
| yabO | Ribosome-associated heat-shock protein implicated in the recycling of the 50S subunit | 1 | 1 |
| Cold Shock | |||
| cspB | Cold-shock protein | 1 | 1 |
| cshB | Conjunction with the cold-shock proteins to ensure proper initiation of transcription at low and optimal temperatures | 1 | 1 |
| cspC | Cold-shock proteins | 1 | 1 |
| Bile Tolerance | |||
| mleP | Sodium Bile acid symporter family | 1 | 1 |
| recU | Endonuclease for DNA repair, homologous recombination, and chromosome segregation | 1 | 1 |
| Acid Stress | |||
| nhaC | Sodium/hydrogen antiporter | 1 | 1 |
| nhaP3 | NhaP-type Sodium/hydrogen and Potassium/hydrogen antiporters | 0 | 2 |
| atpC | Produces ATP from ADP in the presence of a proton gradient across the membrane | 1 | 1 |
| atpD | Produces ATP from ADP in the presence of a proton gradient across the membrane | 1 | 1 |
| atpG | Produces ATP from ADP in the presence of a proton gradient across the membrane | 1 | 1 |
| atpA | Produces ATP from ADP in the presence of a proton gradient across the membrane | 1 | 1 |
| atpH | Produces ATP from ADP in the presence of a proton gradient across the membrane | 1 | 1 |
| atpF | Produces ATP from ADP in the presence of a proton gradient across the membrane | 1 | 1 |
| atpE | Produces ATP from ADP in the presence of a proton gradient across the membrane | 1 | 1 |
| atpB | Produces ATP from ADP in the presence of a proton gradient across the membrane | 1 | 1 |
| nhaK22 | Sodium/hydrogen exchanger family | 1 | 1 |
| Adherence | |||
| gap | Glyceraldehyde-3-phosphate dehydrogenase | 1 | 1 |
| brpA | Biofilm regulatory protein A | 1 | 0 |
| cps2I | Psort location Cytoplasmic Membrane, score | 1 | 0 |
| srtC/srtA | Sortase family | 2 | 2 |
| fbpA | fibronectin binding protein | 1 | 1 |
| tuf | elongation factor Tu | 1 | 1 |
| pgaC1, pgaC2 | Poly-beta-1/2,6-N-acetyl-D-glucosamine synthase | 2 | 2 |
| eno | Enolase | 1 | 1 |
| epsH | Putative glycosyltransferase epsH | 1 | 1 |
| epsL | Putative sugar transferase | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thant, E.P.; Surachat, K.; Chusri, S.; Romyasamit, C.; Pomwised, R.; Wonglapsuwan, M.; Yaikhan, T.; Suwannasin, S.; Singkhamanan, K. Exploring Weissella confusa W1 and W2 Strains Isolated from Khao-Mahk as Probiotic Candidates: From Phenotypic Traits to Genomic Insights. Antibiotics 2024, 13, 604. https://doi.org/10.3390/antibiotics13070604
Thant EP, Surachat K, Chusri S, Romyasamit C, Pomwised R, Wonglapsuwan M, Yaikhan T, Suwannasin S, Singkhamanan K. Exploring Weissella confusa W1 and W2 Strains Isolated from Khao-Mahk as Probiotic Candidates: From Phenotypic Traits to Genomic Insights. Antibiotics. 2024; 13(7):604. https://doi.org/10.3390/antibiotics13070604
Chicago/Turabian StyleThant, Ei Phway, Komwit Surachat, Sarunyou Chusri, Chonticha Romyasamit, Rattanaruji Pomwised, Monwadee Wonglapsuwan, Thunchanok Yaikhan, Sirikan Suwannasin, and Kamonnut Singkhamanan. 2024. "Exploring Weissella confusa W1 and W2 Strains Isolated from Khao-Mahk as Probiotic Candidates: From Phenotypic Traits to Genomic Insights" Antibiotics 13, no. 7: 604. https://doi.org/10.3390/antibiotics13070604
APA StyleThant, E. P., Surachat, K., Chusri, S., Romyasamit, C., Pomwised, R., Wonglapsuwan, M., Yaikhan, T., Suwannasin, S., & Singkhamanan, K. (2024). Exploring Weissella confusa W1 and W2 Strains Isolated from Khao-Mahk as Probiotic Candidates: From Phenotypic Traits to Genomic Insights. Antibiotics, 13(7), 604. https://doi.org/10.3390/antibiotics13070604







