Abstract
Knowledge of the microbiota present in food processing environments is a significant advance that will allow for better evaluation of the risk of food contamination and a better design of the procedures for sanitization. The levels of microbial group indicators of hygienic quality were determined in different areas of the slaughter lines of two poultry slaughterhouses in the northwest of Spain (22 surfaces in each slaughterhouse were studied). The average microbial levels (cfu/cm2) were 2.15 × 102 ± 4.26 × 102 (total aerobic counts, TAC), 1.99 × 102 ± 5.00 × 102 (psychrotrophic microorganisms), 3.10 × 100 ± 1.37 × 101 (enterobacteria), 3.96 × 100 ± 2.55 × 101 (coliforms), 1.80 × 10−1 ± 7.79 × 10−1 (enterococci), and 1.12 × 10−1 ± 3.35 × 10−1 (vancomycin-resistant enterococci, VRE). TAC and psychrotrophic microorganisms were the most abundant groups in all samples (p < 0.05). The counts of both microbial groups were higher (p < 0.05) in samples of Slaughterhouse A than in those of Slaughterhouse B. Microbial loads for the rest of the bacteria were not influenced by the slaughterhouse sampled (p > 0.05). All 44 samples showed TAC and psychrotrophic microorganisms. Colonies of the rest of the microbial groups were only found in 26 samples (59.1% of the total). The isolates (one from each sample) were identified with MALDI-TOF and PCR. Gram-negative bacteria (all Enterobacteriaceae) were isolated in 23 samples, and Gram-positive bacteria were isolated in 16 (9 Enterococcus spp., 2 Enterococcus spp. and VRE, 3 VRE, 1 Enterococcus spp. and Listeria spp., and 1 Listeria spp.). The resistance of the strains to 11 (Enterococcus spp.) or 17 (Enterobacteriaceae) antibiotics was determined (disk diffusion, CLSI), finding an average of 2.05 ± 2.06 resistances per strain (3.46 ± 2.27 if reduced susceptibility reactions are included). A total of 37.3% of the Enterobacteriaceae isolates had a gene for resistance to beta-lactam antibiotics (blaTEM, blaCTX-M-15, blaKPC, blaCMY-2 or blaNDM). The high prevalence of resistant bacteria and resistance genes highlights the need to establish measures to control the spread of antibiotic resistance in poultry slaughterhouses. The findings of this work could contribute to the design of more effective sanitation procedures.
1. Introduction
The high consumption (10.1 kg per capita worldwide and 23.2 kg per capita in Europe) [1] and production (139.2 million tons worldwide and 12.8 million tons in Europe) [2] of poultry meat justifies the importance of guaranteeing that this food is safe for the consumer and has a long shelf life [3,4]. To this end, numerous control measures have been implemented in recent years throughout the entire poultry meat production chain, such as feeding farm animals with prebiotics or vaccinating them against some pathogens [5,6]. However, the degree of contamination of this food with pathogenic and spoilage microorganisms remains high [7]. An important part of this contamination can occur during the dressing of the carcasses in the slaughterhouse through contact with the different facility surfaces [8,9,10]. The degree of food contamination is directly related to the microbial load on surfaces and equipment in processing plants [11]. Cross-contamination is especially high when correct cleaning and disinfection protocols are not followed [12,13].
The problem of microbial contamination of poultry meat is aggravated when microorganisms present antimicrobial resistance (AMR). The spread of AMR poses a serious threat to public health, as the effective treatment of infections is hampered, leading to increased costs for the health system and increased mortality [14,15]. Beta-lactams are the antibiotics of choice for the treatment of many food-borne infections, so it is essential to know and monitor the prevalence of extended-spectrum beta-lactamases (ESBL) in bacteria of food origin [16]. The prevalence of these enzymes in Enterobacteriaceae isolates from broilers has been studied in many European countries at the abattoir level and has provided further evidence of chicken meat as a potential zoonotic source of ESBL-producing bacteria [17]. Although there are a large number of genes that encode ESBL enzymes, in Europe, the CTX-M and TEM groups are some of the most frequently detected in food-producing animals [18]. However, recent studies carried out in Spain and Portugal have reported the presence of other resistance genes, such as blaNDM-1 (61.2% of isolates) [19], blaCMY-2 (76.2%) [20]), or blaKPC (91.0%) [21], in some EBSL-producing Enterobacteriaceae isolates.
Given that the presence of high microbial levels in processing environments directly influences the microbiological quality of meat [22,23], comprehensive monitoring of this microbiota is crucial. Total aerobic counts (TAC), psychrotrophs, Enterobacteriaceae, coliforms, and enterococci are among the most common microbial groups used in meat and poultry industries as general indicators of processing hygiene, storage quality, and potential shelf life, both in the oxygen atmosphere and in vacuum-packed meat [3]. For the detection and quantification of microorganisms on surfaces and equipment in food industries, analysis techniques have traditionally been used that involve their cultivation in different media (selective and non-selective), with the subsequent isolation and identification of the colonies performed using biochemical or molecular methods such as polymerase chain reaction (PCR) [24]. In the last decade, matrix-assisted laser desorption/ionization (MALDI-TOF) has gained popularity in microbiology laboratories, as it allows for the identification of bacteria in a quick and easy way [25].
One of the strategies to reduce the contamination of poultry meat is to reduce microbial levels on slaughterhouse surfaces and equipment. To achieve this, in the first stage, it is necessary to expand current knowledge about the microbiota of the different areas of the slaughter line, from stunning to cutting. This research has been proposed in this context, the objective of which was to determine the microbial groups present on the surfaces and equipment of two poultry slaughterhouses and to characterize their resistance to antibiotics, emphasizing the presence of extended-spectrum beta-lactamases.
2. Results
2.1. Microbial Counts
The levels of the different microbial groups studied (total aerobic counts (TAC), psychrotrophs, enterobacteria, coliforms, enterococci, and vancomycin-resistant enterococci (VRE)) are shown in Table 1. The TAC and psychrotrophic microorganisms were the most abundant bacterial groups in all samples (p < 0.05), with average values (cfu/cm2) of 2.15 × 102 ± 4.26 × 102 and 1.99 × 102 ± 5.00 × 102, respectively. The lowest plate counts were found for VRE, with an average value (cfu/cm2) of 1.12 × 10−1 ± 3.35 × 10−1. No significant differences (p > 0.05) were found in terms of the plate counts between the rest of the microbial groups studied. The mean values were 3.10 × 100 ± 1.37 × 101 for enterobacteria, 3.96 × 100 ± 2.55 × 101 for coliforms, and 1.80 × 10−1 ± 7.79 × 10−1 for enterococci. The TAC and psychrotrophic loads were higher (p < 0.05) in samples from Slaughterhouse A (2.90 × 102 ± 5.25 × 102 and 2.14 × 102 ± 3.59 × 102, respectively) than they were in samples from Slaughterhouse B (1.41 × 102 ± 2.91 × 102 and 1.84 × 102 ± 6.18 × 102, respectively). Microbial loads for the rest of the bacterial groups were not influenced by the slaughterhouse sampled (p > 0.05).

Table 1.
Load (cfu/cm2) of the different microbial groups analyzed.
2.2. Isolation and Identification of Microorganisms
One colony from each microbial group, except for TAC and psychrotrophic microorganisms, was isolated from each sample for subsequent identification using MALDI-TOF and PCR. Microorganisms could be isolated from 26 samples (Table 2), representing 59.1% of the total. In 23 of these samples (88.5% of the positive ones and 52.3% of the total), Gram-negative bacteria (all Enterobacteriaceae) were isolated, while only Gram-positive bacterial genera were isolated in 16 samples (61.5% of the positive ones and 36.4% of the total). Strains of Enterococcus spp. were isolated in 15 of the samples with Gram-positive bacteria, and VRE were found in 5 of them. Colonies with the typical morphology of Listeria spp. were found in two samples (A13 and A22; 7.7% of the positive samples and 4.5% of the total).

Table 2.
Surfaces and culture media from which the colonies analyzed were isolated in the two slaughterhouses (A and B).
2.3. Phenotypic Resistance to Antibiotics
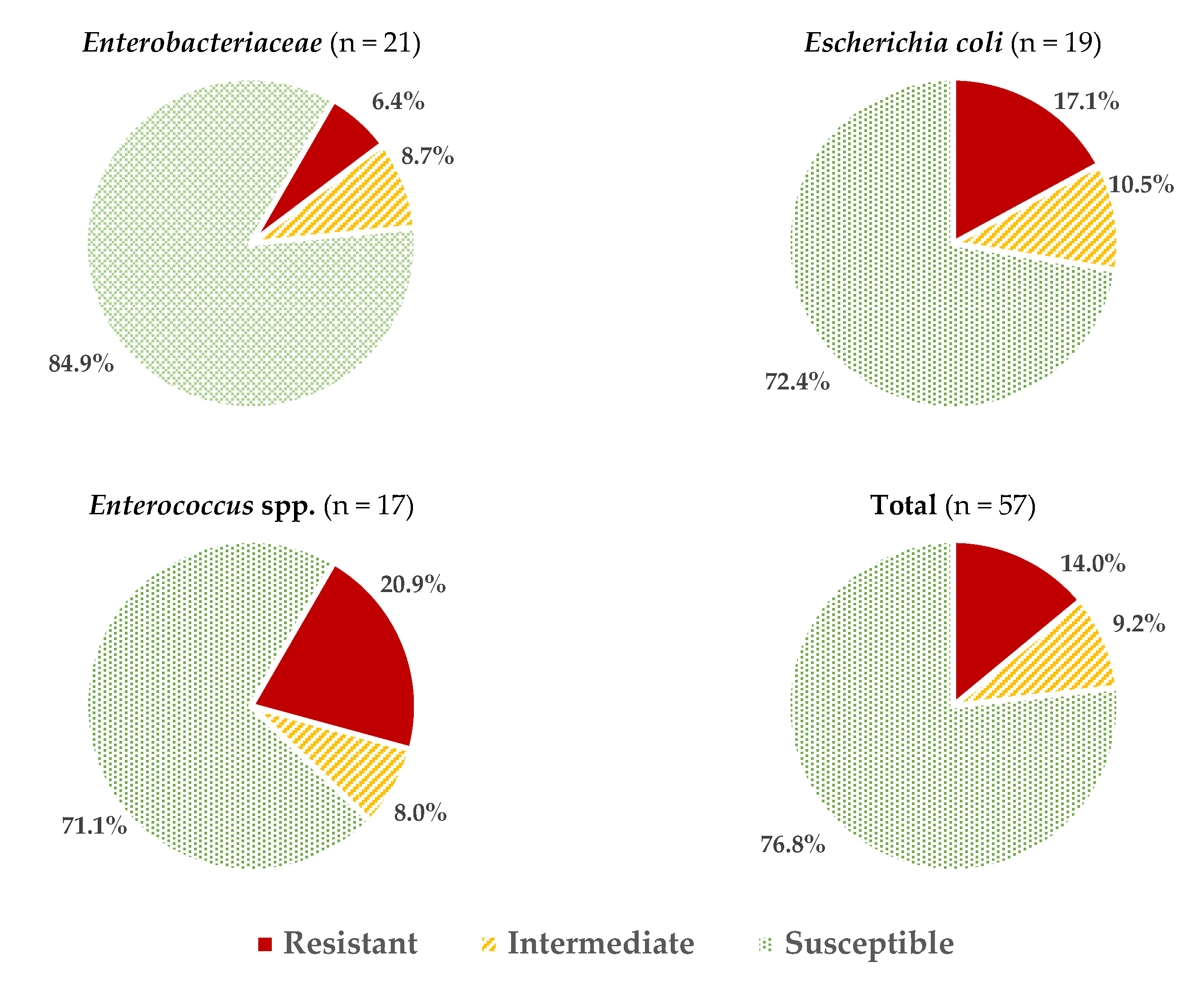
Considering all the data obtained for all the microbial groups studied, an average of 2.05 ± 2.06 resistances per strain were found, or 3.46 ± 2.27 if reduced susceptibility reactions are also included. These values vary depending on the microorganism considered (Figure 1), with the isolates of Enterococcus spp. and E. coli having the highest percentage of resistance (20.9 ± 11.6% and 17.0 ± 15.5%, respectively; these percentages were calculated considering the number of positive reactions (resistance) over the total number of tests carried out (strains × antibiotics). The resistance values (%) for other genera of the family Enterobacteriaceae (6.4 ± 6.5%) were significantly lower (p > 0.05).
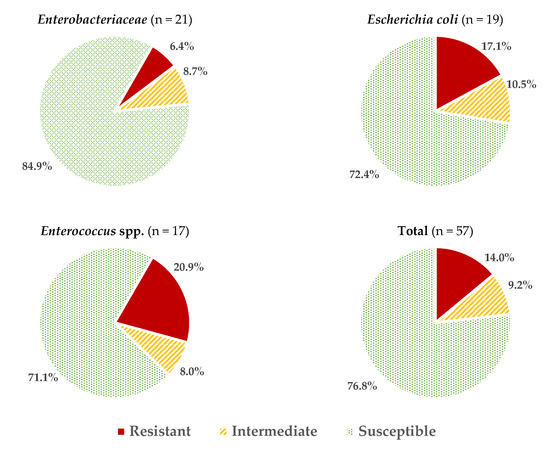
Figure 1.
Average percentages of resistant, intermediate (with reduced susceptibility), and susceptible strains in the different microbial groups studied.
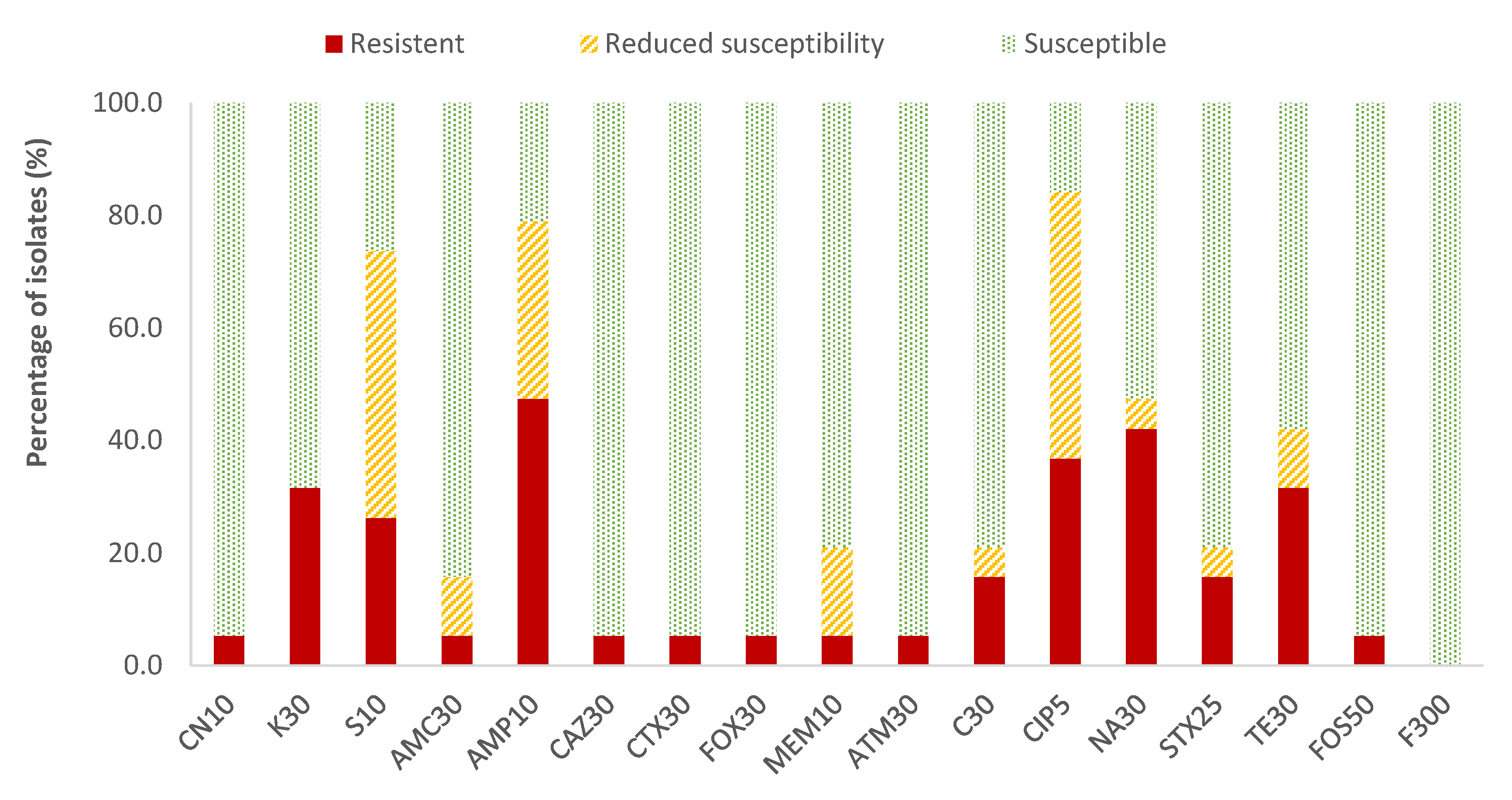
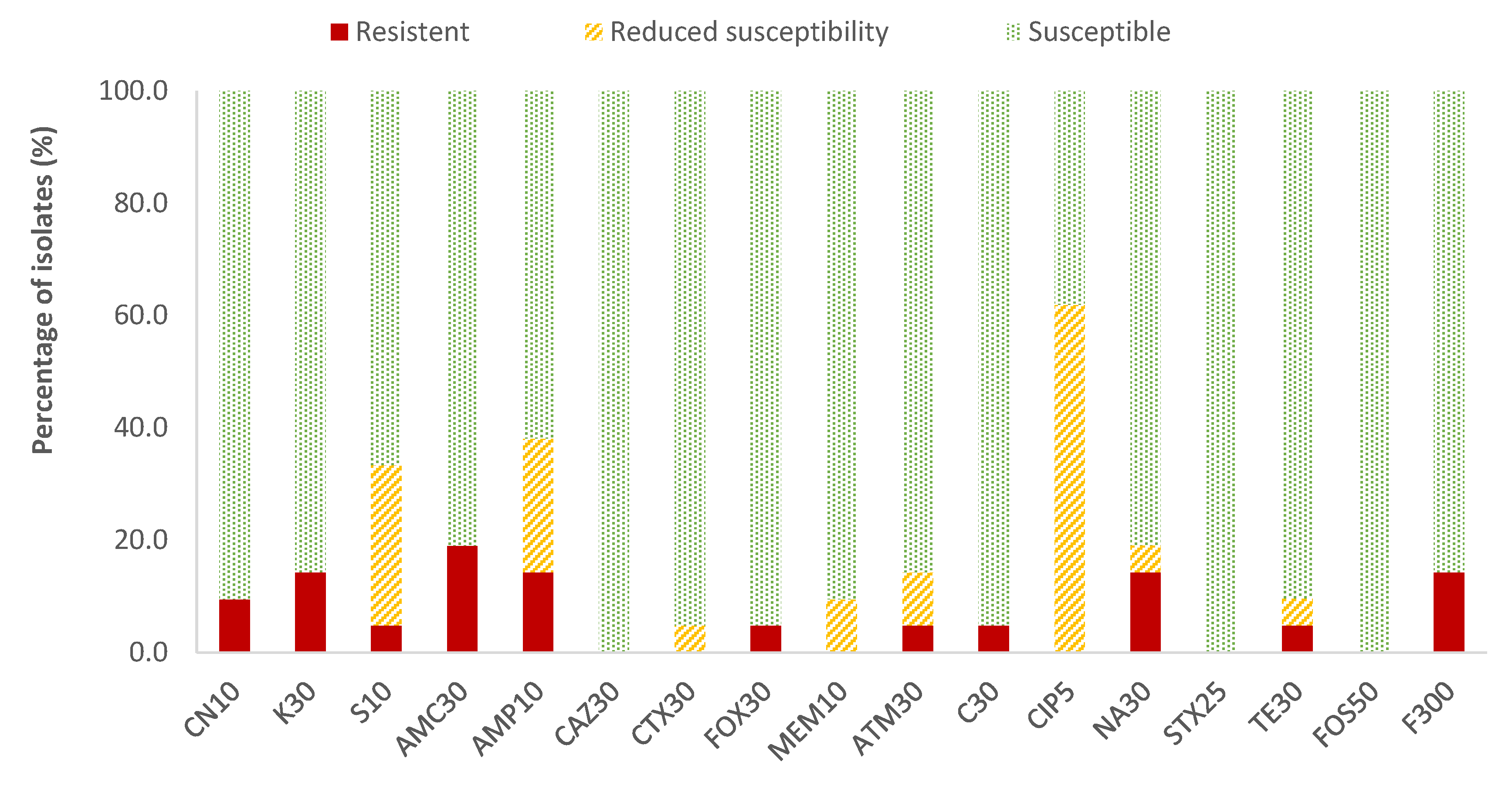
In the case of Enterobacteriaceae, 100% susceptibility to ceftazidime (CAZ) sulfamethoxazole/trimethoprim (SXT) and fosfomycin (FOS) was observed, with the exception of E. coli, for which there was just one antibiotic (nitrofurantoin, F) to which all strains were susceptible; the resistance percentages of E. coli to ceftazidime (CAZ), fosfomycin (FOS) and sulfamethoxazole/trimethoprim (SXT) were 5.3%, 5.3% and 15.8%, respectively. There were no antibiotics for which all Enterobacteriaceae strains were resistant to it. For the rest of the antibiotics, in E. coli the highest prevalence of resistance was obtained for ampicillin (AMP; 47.4%) and nalidixic acid (NA; 42.1%), while for the rest of the enterobacteria, both antibiotics showed 14.3% of resistances. Considering both resistance and reduced susceptibility, the highest values were obtained in the case of ciprofloxacin (CIP), with resistance prevalences of 84.2% and 61.9% for E. coli and for the rest of enterobacteria, respectively, and for ampicillin (AMP), with 78.9% resistance for E. coli and 38.1% for the rest of the enterobacteria (Figure 2 and Figure 3).
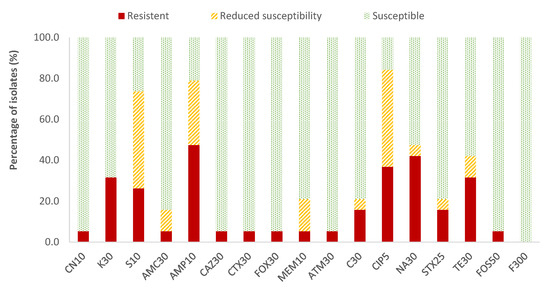
Figure 2.
Percentage of resistant E. coli strains, with reduced susceptibility or susceptibility to each of the antibiotics tested. Gentamicin, 10 µg (CN10), kanamycin, 30 µg (K30), streptomycin, 10 µg (S10), amoxicillin/clavulanic acid, 30 µg (AMC30), ampicillin, 10 µg (AMP10), ceftazidime, 30 µg (CAZ30), cefotaxime, 30 µg (CTX30), cefoxitin, 30 µg (FOX30), meropenem, 10 µg (MEM10), aztreonam, 30 µg (ATM30), chloramphenicol, 30 µg (C30), ciprofloxacin, 5 µg (CIP5), nalidixic acid, 30 µg (NA30), sulfamethoxazole/trimethoprim, 25 µg (SXT25), tetracycline, 30 µg (TE30), fosfomycin, 50 µg (FOS50), nitrofurantoin, 300 µg (F300).
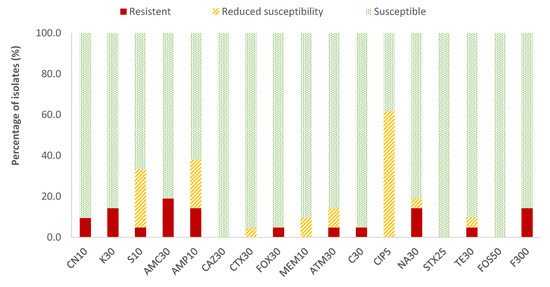
Figure 3.
Percentage of resistant Enterobacteriaceae strains other than E. coli, with reduced susceptibility or susceptibility to each of the antibiotics tested. Gentamicin, 10 µg (CN10), kanamycin, 30 µg (K30), streptomycin, 10 µg (S10), amoxicillin/clavulanic acid, 30 µg (AMC30), ampicillin, 10 µg (AMP10), ceftazidime, 30 µg (CAZ30), cefotaxime, 30 µg (CTX30), cefoxitin, 30 µg (FOX30), meropenem, 10 µg (MEM10), aztreonam, 30 µg (ATM30), chloramphenicol, 30 µg (C30), ciprofloxacin, 5 µg (CIP5), nalidixic acid, 30 µg (NA30), sulfamethoxazole/trimethoprim, 25 µg (SXT25), tetracycline, 30 µg (TE30), fosfomycin, 50 µg (FOS50), nitrofurantoin, 300 µg (F300).
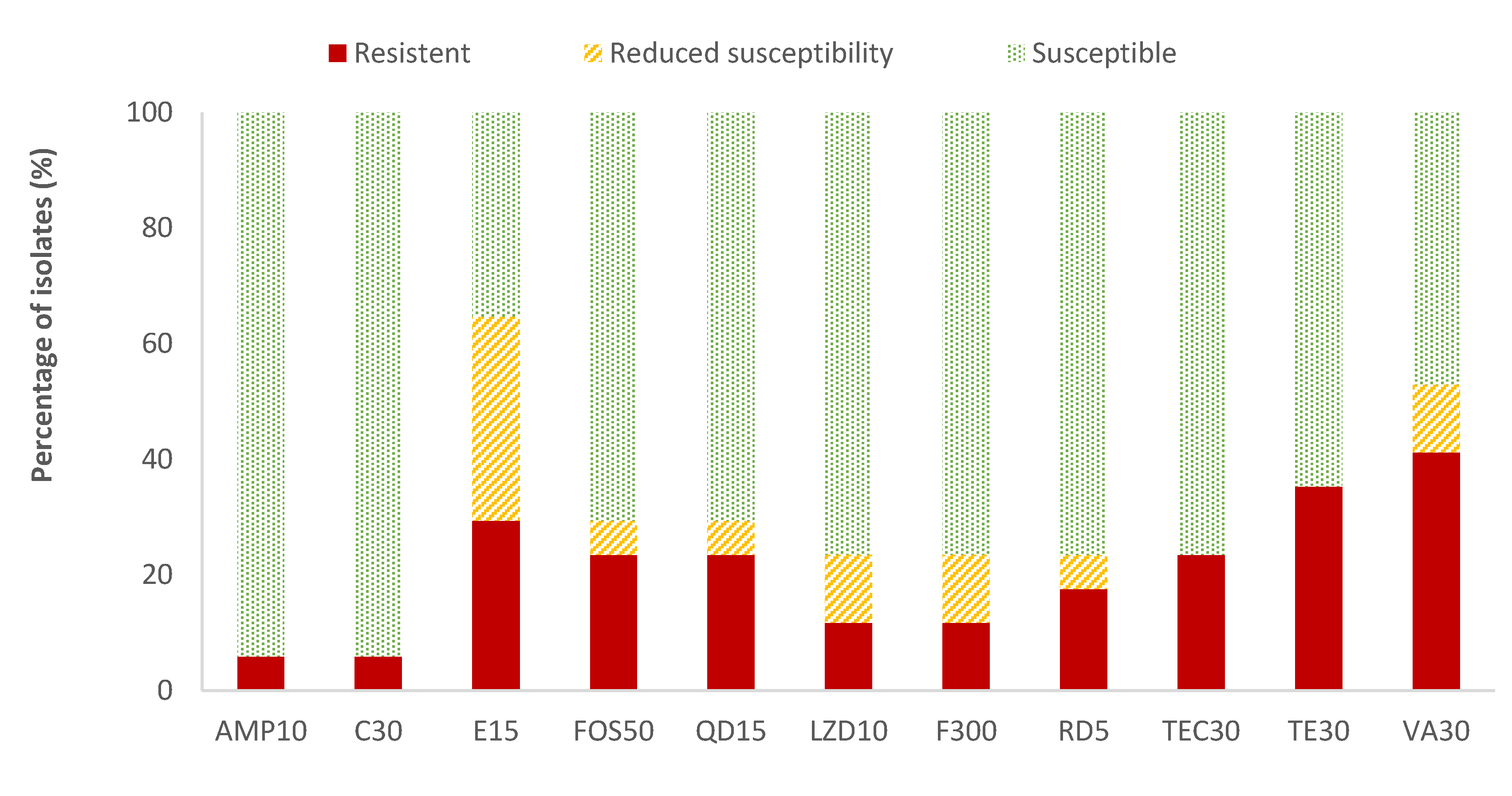
The highest resistance percentages of Enterococcus spp. were found for vancomycin (41.2%) and tetracycline (35.3%), while ampicillin (AMP) and chloramphenicol (C) showed the lowest resistant percentages (5.9%). The resistance percentages of the rest of the antibiotics evaluated ranged between 11.8% (for linezolid and nitrofurantoin) and 29.4% (for erythromycin). Considering all the data on the resistance and reduced susceptibility of Enterococcus spp., the high values observed for erythromycin (64.7%), vancomycin (52.9%), and tetracycline (35.3%) are notable (Figure 4). Listeria spp. was only detected in two samples. Therefore, as not average data could be obtained, the susceptibility testing to antibiotics was not performed to this microorganism.
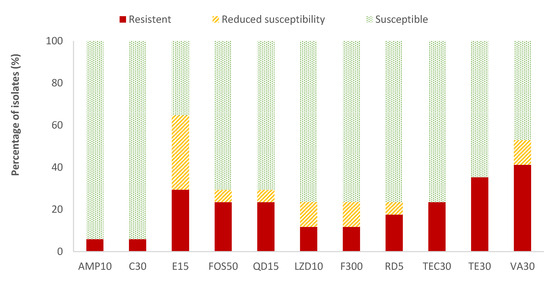
Figure 4.
Percentage of Enterococcus spp. strains resistant, intermediate (with reduced susceptibility), or susceptible to each of the antibiotics tested. Ampicillin, 10 µg (AMP10), chloramphenicol, 30 µg (C30), erythromycin, 15 µg (E15), fosfomycin, 50 µg (FOS50), quinupristin/dalfopristin, 15 µg (QD15), linezolid, 10 µg (LZD10), nitrofurantoin, 300 µg (F300), rifampicin, 5 µg (RD5), teicoplanin, 30 µg (TEC30), tetracycline, 30 µg (TE30), and vancomycin, 30 µg (VA).
2.4. Detection of Resistance Genes to Beta-Lactam Antibiotics in Enterobacteriaceae
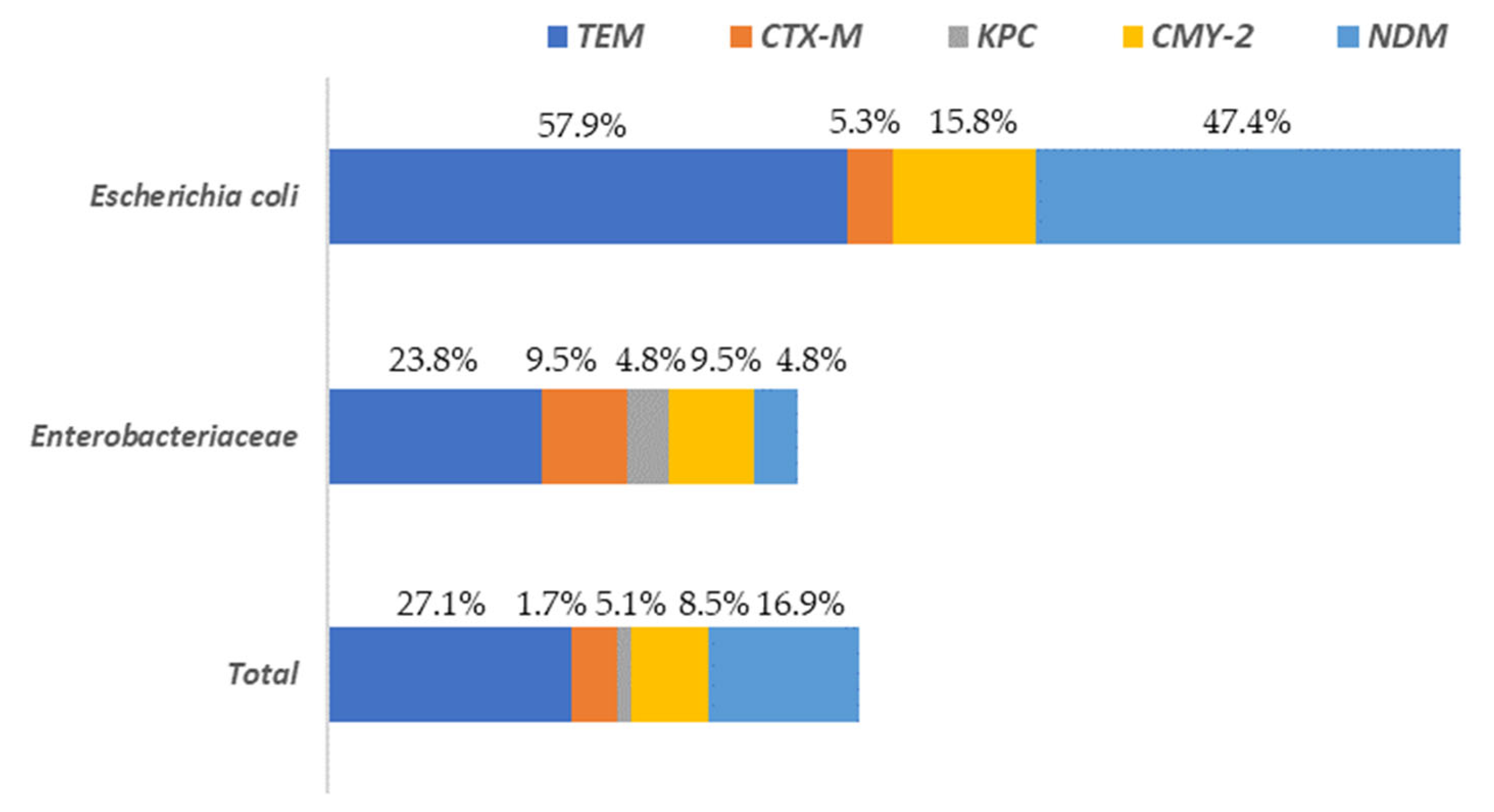
A total of 37.3% of the isolates presented at least one of the resistance genes to the beta-lactam antibiotics analyzed (blaTEM, blaCTX-M, blaKPC, blaCMY-2 or blaNDM). These percentages varied depending on the microbial group considered, being 78.9% for E. coli and 33.3% for the rest of the enterobacteria. The gene with the greatest presence in all Enterobacteriaceae isolates, was blaTEM, followed by blaNDM and blaCMY-2 (Figure 5).
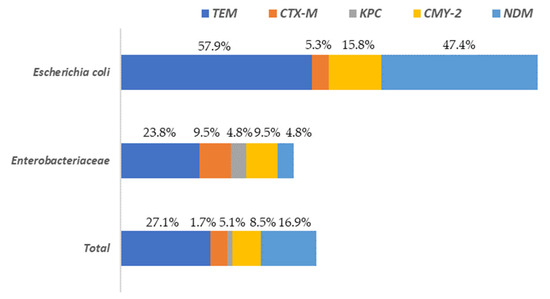
Figure 5.
Distribution of beta-lactam antibiotic resistance genes among Escherichia and the rest of Enterobacteriaceae (other than E. coli) isolated on surfaces in both slaughterhouses.
The highest levels of blaTEM were found in E. coli (57.9%) isolates while for the rest of Enterobacteriaceae its prevalence was notably lower (23.8%). The genes blaCTX-M and blaKPC were present in a very low percentage of the isolates (5.1% and 1.7%, respectively), with the highest values (9.5% and 4.8%, respectively) observed for the Enterobacteriaceae isolates, without taking into account E. coli. Regarding to the rest of the genes studied (blaCMY-2 and blaNDM) the highest values were found in E. coli isolates, ranging from 15.8% (blaCMY-2) to 47.4% (blaNDM). In the Supplementary Materials (Table S1) the results for all isolates tested for AST and the presence of AMR are shown. A column is also included to identify the sample origin (in which slaughterhouse the surface was found), as well as another for genus identification.
3. Discussion
3.1. Microbial Counts
The average TAC (2.15 × 102 ± 4.26 × 102 cfu/cm2) is similar to those observed by Whyte et al. [26] in three poultry meat production plants in Ireland, who found values between 2.8 × 101 and 1.8 × 103 cfu/cm2. Notably, in the present study, 72.7% of the surfaces analyzed had counts greater than 10 cfu/cm2 for this microbial group, a percentage that is considered high. The microbiological criteria for inert surfaces in meat industries established in Decision 2001/471/EC indicated that the number of bacteria present on a surface is acceptable if it does not exceed the value of 10 cfu/cm2 (TAC) or 1 cfu/cm2 (Enterobacteriaceae). With values higher than those indicated, the aforementioned standard established the need to implement corrective actions. Although this decision was repealed with effect from 1 January 2006, these microbiological criteria can still be used to evaluate hygiene in meat industries. The TAC values in the present study are considerably higher than those previously obtained by Alonso-Calleja et al. [27] in lamb slaughterhouses, where the percentages of samples with values greater than 10 cfu/cm2 varied between 3.1 and 17.1%. In this regard, it should be noted that the differences between slaughterhouses must be interpreted with caution due to the fundamental particularities of one, with regard to the species slaughtered, which present different levels and types of microorganisms.
The mean counts of psychrotrophic microorganisms observed in this study (1.99 × 102 ± 5.0 × 102 cfu/cm2) were also similar to those previously described by Whyte et al. [26], who obtained values between 1.4 × 101 and 3.6 × 102 cfu/cm2. However, other authors found much higher levels of psychrotrophs as follows: between 2.5 × 101 and 5.0 × 107 cfu/cm2 [28] and between 1.9 × 103 and 6.3 × 106 cfu/cm2 [29] on surfaces in a bovine slaughterhouse.
At this point in the production chain, it is essential to prevent psychrotrophic microorganisms from adhering to the carcass to produce meat with a long shelf life [20]. The presence of high counts of these microorganisms on food contact surfaces may pose a health risk to consumers due to the possibility of the cross-contamination of meat and, subsequently, their ability to grow and multiply under refrigerated conditions during storage and transportation [30]. Furthermore, psychrotrophic microorganisms include some spoilage bacteria (i.e., Pseudomonas or Brochothrix), so high counts of these microorganisms can be associated with important economic losses [3].
Enterobacteriaceae counts (3.10 × 100 ± 1.37 × 101 cfu/cm2) were also similar to those obtained by other authors (between 3.9 × 10−1 and 4.2 × 101 cfu/cm2) [26] on poultry slaughterhouse surfaces. Furthermore, 84.1% of the samples had values less than or equal to 1 cfu/cm2 (value established as a limit in Decision 2001/471/EC, now repealed). These data are similar to those previously obtained by Alonso-Calleja et al. [27], in which only one of the samples from one of the two slaughterhouses studied (assuming 1.4% of the samples from that slaughterhouse) presented a value greater than 1 cfu/cm2.
The counts of coliforms (3.96 × 100 ± 2.55 × 101 cfu/cm2) were similar to those previously obtained by other authors in samples from poultry slaughterhouse surfaces (between 3.9 × 10−1 and 4.2 × 101 cfu/cm2) [26]. The multiplication of these microorganisms may be affected by the storage at 4 °C of the samples, implying a lower value of their counts. However, some studies of raw milk [31] and poultry meat [32] suggested that this storage should be prolonged to a time above 24 h to be truly representative. In the study being presented here, microbiological analysis took place within a maximum period of 18 h from sample collection. In the case of enterococci, the average values in the present study ranged from 1.8 × 10−1 ± 7.79 × 10−1 cfu/cm2 (enterococci) to 1.12 × 10−1 ± 3.35 × 10−1 cfu/cm2 (VRE), which are much lower figures than those obtained by Nortjé et al. [29] from bovine slaughterhouse surfaces (between 7.9 × 103 and 5.0 × 104 cfu/cm2).
3.2. Isolation and Identification of Microorganisms
Among the isolations made from VRBGA (characteristic colonies of Enterobacteriaceae grew in 38.6% of the samples analyzed), E. coli was identified by MALDI-TOF and PCR in 47.1% of the positive samples (assuming 18.2% of the total samples analyzed), while the presence of Salmonella spp. was detected in none of them. The prevalence data for E. coli are lower than those obtained by other authors in pig (35.1%) [33] and bovine (41.6%) [34] slaughterhouses but higher than those found in lamb slaughterhouse surfaces (4.2%) [35]. Other authors obtained data similar to those of the present study, both for E. coli (20.2%) [36] and Salmonella spp. (0.34%) [37], but higher as for Enterobacteriaceae in general (60%) [38], on different surfaces of slaughterhouses and retail establishments. Monitoring the presence of different genera of Enterobacteriaceae, including the ones reported in this study, along the slaughter line enables the verification of slaughter operation hygiene and the use of good manufacturing practices by slaughter operators [39].
The prevalences of Enterococcus spp. obtained in this study (36.4%) are very different from those found by other authors consulted. Soares-Santos et al. [40] identified E. faecium strains in 75.0% of the swine slaughterhouse surface samples analyzed; Lavilla Lerma et al. [35] also detected strains of Enterococcus spp. on lamb slaughterhouse surfaces, but in a much smaller percentage (0.8%), and Wambui et al. [41] isolated strains of several Enterococcus species (E. faecalis, E. mundtii, E. thailandicus, E. faecium, E. hirae, E. casseliflavus, and E. devriesei) from slaughterhouse surfaces of small- and medium-sized enterprises in Kenya (assuming 26.6% of the samples analyzed).
It should be noted that vancomycin-resistant enterococci (VRE), confirmed by PCR at the genus level, were detected in five samples. VRE are associated with increasing frequency in both nosocomial and community-acquired human infections. Although there is little evidence that these infections are directly related to the consumption or handling of contaminated food, it is a proven fact that the presence of these microorganisms in food presents a clear risk of transferring resistance genes to other bacterial cells throughout the food chain, thereby contributing to the spread and persistence of antibiotic resistance in the food production chain [42,43].
Meanwhile, Listeria spp. was only detected in two samples (representing 4.6% of the total). The prevalence of Listeria is similar to the values obtained by other authors in samples collected from beef slaughterhouse surfaces, as follows: 6.0% [44], 5.6% (identified as L. monocytogenes, L. innocua and L. ivanovii) [45], and 4.9% of the samples in poultry slaughterhouses [37]. The contamination with these pathogenic bacteria when slaughtering animals may occur by means of direct fecal material, contaminated skin during the removal of viscera, and the contact of carcasses with one another, but also with the equipment in the slaughterhouse. For this reason, food of animal origin has become an important vehicle for the transmission of Listeria to humans [46].
3.3. Phenotypic Resistance to Antibiotics
The antibiotic resistance data obtained for Enterobacteriaceae were different, depending on whether it was E. coli or another Enterobacteriaceae. The E. coli isolates tested in the present study showed ranges of resistance (if taking in count both resistance and reduced susceptibility) to antibiotics similar to those previously obtained by Gregova et al. [47] in strains of this microbial species isolated in poultry slaughterhouses, as follows: 89.0% of the strains were resistant to ampicillin and 10.0% were resistant to chloramphenicol. Our results for ampicillin are also like those obtained by Elabbasy et al. [48] in strains isolated in beef slaughterhouse, (where 60% were resistant) but lower for other antibiotics: 65.0% were resistant to nalidixic acid, 55.0% were resistant to sulfamethoxazole/trimethoprim, and 75.0% were resistant to tetracycline.
For the rest of the enterobacteria, the results obtained in the present study were very different from those of E. coli, with lower resistance percentages. Contrarily, some authors have found very high resistance values for other species of Enterobacteriaceae from inert slaughterhouse surfaces. In this regard, Savin et al. [49] resistance in 96.7% (ciprofloxacin and cefotaxime) of Klebsiella spp. strains isolated from poultry slaughterhouse samples. Homeier-Bachmann et al. [50] obtained enterobacteria strains with high percentages of resistance to second and third generation cephalosporins (89% to cefotaxime and 95.0% to ceftazidime), and ciprofloxacin (53.0%).
In general, the enterococcal isolates analyzed showed a degree of resistance to antibiotics similar to that observed in other studies. Soares-Santos et al. [40] showed that all enterococcal isolates had sensitivity to ampicillin, ciprofloxacin, gentamicin, linezolid, nitrofurantoin, and teicoplanin, while 58% of the strains were resistant to quinupristin-dafopristin, 55% were resistant to tetracycline, 38% were resistant to rifampicin, 31% were resistant to erythromycin, 7% were resistant to streptomycin, 4% were resistant to chloramphenicol, and 1% were resistant to vancomycin. Similar results were obtained by Wambui et al. [41], who observed that all enterococcal isolates were sensitive to ciprofloxacin, penicillin, ampicillin, vancomycin, nitrofurantoin, teicoplanin, linezolid, and levofloxacin while presenting resistance to rifampicin (46.3%), erythromycin (23.9%), tetracycline (20.9%), and chloramphenicol (7.5%). Furthermore, it should be noted that the colonies isolated from the SBV medium, all of which were resistant to vancomycin (VRE), presented a higher percentage of resistance to antibiotics than those from the KAE medium.
Some of the antibiotics associated with the highest percentage of resistance or with reduced susceptibility strains, such as ampicillin, ciprofloxacin, streptomycin, cefoxitin, gentamycin, nalidixic acid, or tetracycline for Enterobacteriaceae, and streptomycin, gentamycin, kanamycin, tetracycline, or vancomycin for Enterococcus, are classified as “critically important” or “highly important” antimicrobial agents in human and veterinary medicine [51,52].
3.4. Detection of Resistance Genes to Beta-Lactam Antibiotics in Enterobacteriaceae
In this study, the presence of five resistance genes related to beta-lactam antibiotics (blaTEM, blaCTX-M, blaKPC, blaCYM-2, and blaNDM) was studied. The resistance mechanisms associated with these genes are based on the synthesis of extended-spectrum beta-lactamases capable of hydrolyzing all penicillins, cephalosporins, and carbapenems (with the exception of aztreonam). Furthermore, microorganisms carrying these genes usually have associated resistance mechanisms to other antibiotics, such as aminoglycosides or fluoroquinolones, leaving few therapeutic options available [53]. The use of carbapenems in veterinary medicine is off-label and should be reserved for cases with limited therapeutic alternatives and in which animals have a high likelihood of survival with appropriate therapy [54]. In Spain and the European Union, these drugs are not commonly used in agriculture or at the farm level to treat animals, but the localization of these genes in mobile genetic elements, together with others (conferring resistance to other beta-lactams, fluoroquinolones, or aminoglycosides), contributes significantly to its spread [55].
The prevalence of some of these genes was very high in E. coli agreeing with the fact that poultry meat is the meat that usually presents the greatest contamination with bacteria carrying extended-spectrum beta-lactamases [56]. The genes with the greatest presence in all the Enterobacteriaceae strains studied were blaTEM and blaNDM. However, their percentages of appearance were much lower than those found in poultry farms in China, with the detection ranges of these genes somewhere between 53.2% and 98.6% of the samples [57]. Although the prevalence of ESBLs tends to differ among countries, blaTEM, blaSHV, blaCTX-M, and blaOXA genes are the main classes of ESBLs and blaKPC, blaIMP, blaVIM, and blaNDM genes are the most common carbapenemases found in Gram-negative bacteria [55].
The presence of some of these genes has been previously reported by other authors in food-producing environments, particularly in poultry slaughterhouses. Lim et al. [58] characterized strains of E. coli-producing ESBL isolated from chicken slaughterhouses in South Korea. Wei et al. [59] studied the presence of plasmid-mediated ESBL in strains of E. coli that were also isolated from a chicken slaughterhouse in Korea. Adel et al. [60] found a high prevalence of ESBL resistance genes in Salmonella strains isolated from retail meats and slaughterhouses in Egypt. There have also been many studies about the presence of ESBL-producing strains isolated from slaughterhouses carried out in Spain and Europe, but most of them are focused on Enterobacteriaceae strains isolated from fresh meat or carcasses [61,62,63,64] instead of from strains isolated directly from the processing surfaces.
The frequency of CTX-M type beta-lactamases was considerably low (only 5.1% in the total of the isolates studied) compared to the data obtained by other authors, who have detected this type of gene in much higher percentages, as follows: 72.5% [65] and 100% [66]. This fact is also striking because some authors have shown that poultry meat is the origin of the plasmids that code for beta-lactamases of this type [67]. The CTX-M is a plasmid-encoded plasmid carrying resistance genes for other antibiotics, such as tetracycline, aminoglycosides, and sulfamethoxazole/trimethoprim [68]. This is a fact that can be observed in some of the isolates found in this study. One of the CTX-M-positive strains are sensitive to cefotaxime but resistant or with reduced susceptibility to other antibiotics from those groups, such as kanamycin, streptomycin, or tetracycline (Table S1).
Something similar occurs with the presence of TEM-type beta-lactamases, which have been detected in 27.1% of the isolates in the present study. Other authors have described this type of beta-lactamase as one of the most predominant in strains of Salmonella spp. isolated from poultry, poultry meat, and clinical samples from patients [69]. Furthermore, Gundran et al. [65] detected blaTEM genes in 58.0% of samples obtained from Philippine poultry farms.
In this study, carbapenemases genes were found throughout almost the entire slaughter chain. The blaTEM and blaNDM genes were the most common, being found in approximately at 50.0% strains of Escherichia isolated from different surfaces, such as the slaughter area, the stunning tank, different cutters, or aspiration equipment (Table S1). On the other hand, the blaKPC was the least predominant of the carbapenemases genes, being found in just some of the strains isolated in the cutting areas of the chain. This can occur as a result of the intense protocols of cleaning and disinfection applied throughout the first steps of the slaughter process and as a result of disinfectants effective against co- and cross-resistance, since corresponding genes are often located on the same plasmid [70].
4. Material and Methods
4.1. Sampling
Samples were taken in two poultry slaughterhouses (A and B) located in the northwest of Spain. Sampling was carried out on 22 different surfaces along the slaughter chain of each slaughterhouse (44 in total), from the animal stunning area to the cutting room (Table 3).

Table 3.
Surfaces analyzed and area (cm2) of each of them.
Sampling was carried out within a maximum of one hour after cleaning and disinfection and consisted of vigorously rubbing the surface with three swabs moistened with phosphate-buffered saline (PBS). The swabs were subsequently placed in tubes with 10 mL of PBS and stored at 4 °C until they were processed in the laboratory (which took place within a maximum period of 18 h from sample collection).
4.2. Microbiological Analysis
The levels of viable aerobic microbiota (total viable counts, TAC), psychrotrophic microorganisms, enterobacteria, coliforms, enterococci, and vancomycin-resistant enterococci (VRE) were determined through direct plate counts using specific media for each microbial group. Data were expressed as cfu/cm2. Because large areas were sampled, and bacteria levels were sometimes low, the results (cfu/cm2) in some cases are expressed as 10 to the power of a negative number. The prevalence of Listeria spp. (UNE-EN ISO 11290-1:2018) [71] and Salmonella spp. (UNE-EN ISO 6579-1:2017) [72] were determined, carrying out two stages of the enrichment of the samples in a liquid medium and subsequent seeding in a chromogenic medium. All culture media used (Oxoid Ltd., Basingstoke, United Kingdom), along with their incubation conditions, are listed in Table 4.

Table 4.
Culture media and incubation conditions used for the determination of each microbial group.
Colonies were isolated from all the samples presumed positive for any of the microbial groups studied (except TAC and psychrotrophs) with morphology and characteristics typical of the microorganism in question (enterobacteria, coliforms, enterococci, VRE, Salmonella spp. or Listeria spp.) for subsequent identification using MALDI-TOF and PCR. The isolated colonies were cultured at 37–42 °C (depending on the microbial group) for 24 h in test tubes with 9 mL of tryptone soya broth (TSB, Oxoid) and subsequently stored at −30 °C in TSB with 20% of glycerol.
4.3. Identification of Isolates Using MALDI-TOF
All isolates were streaked on tryptone soya agar (TSA, Oxoid) plates and cultured at 37 °C for 24 h for subsequent identification using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF). An isolated colony was taken from each sample using a sterile toothpick and distributed in a thin film on a well of a stainless-steel plate (Bruker Daltonics GmbH & Co. KG, Bremen, Germany). It was then air dried for 5 min and covered with 1 μL of 70% formic acid (Panreac, Barcelona, Spain). Once the acid had evaporated (approximately 5 min), the film was covered with 1 μL of HCCA matrix (α-Cyano-4-hydroxycinnamic acid; Bruker Daltonics) and allowed to dry for another 5 min at room temperature. Spectra were acquired with the MALDI Biotyper system and compared to the reference database (Bruker Daltonics).
4.4. Confirmation of Isolates Using PCR
The identification of the isolates (up to family, genus, or species level) was confirmed using polymerase chain reaction (PCR). To do so, genomic DNA was extracted from isolates grown in TSB (Oxoid) following the previously described protocol [77].
For the amplification reaction, 5 µL of the sample and 20 µL of the master mix, prepared with 1X NH4 Buffer, 2 mM MgCl2, 0.2 mM dNTPs (each), 0.5 mM primers (each one), 1U of Taq-DNA polymerase, and 14.25 µL of MiliQ water, were used. All reagents were supplied by BIORON GmbH (Ludwigshafen, Germany), except for dNTPs, which were purchased from EURX Sp. z o.o (Gdansk, Poland), and primers purchased from Macrogen (Beotkkot, Geumcheon-gu, Seoul, South Korea). The reactions were carried out in a ProFlex PCR System thermocycler (Waltham, MA, United States) programmed at 94 °C for 5 min, followed by the amplification cycles necessary for each pair of primers (Table 5) and 10 min of final elongation at 72 °C.

Table 5.
Primers used for each microorganism with its nucleotide sequence, the size of the amplified fragment (bp), and the thermocycling program used.
4.5. Phenotypic Resistance to Antibiotics
The susceptibility of the strains to some of the families of antibiotics most used in clinical practice was determined. These included aminoglycosides, beta-lactams, macrolides, glycopeptides, sulfonamides, rifamycins, tetracyclines, phenicols, fluoroquinolones, and nitrofurans. For this, the isolates were seeded in tubes with 9 mL of Mueller Hinton Broth (MHB, Oxoid) and incubated at 37 °C for 8–10 h, until they were in the exponential growth phase. Subsequently, antibiograms of each strain were performed (disk diffusion technique), sowing the grass cultures on Mueller Hinton Agar (MHA, Oxoid) plates. The plates were incubated at 37 °C for 18–20 h, after which time the inhibition zones were measured, allowing the strains to be classified as susceptible, with reduced susceptibility (intermediate), or resistant according to the established criteria [82].
The following antibiotic disks (Oxoid) were used for Enterobacteriaceae isolates: gentamicin (CN, 10 µg), kanamycin (K, 30 µg), streptomycin (S, 10 µg), amoxoxycillin/clavulanic acid (AMC, 30 µg), ampicillin (AMP, 10 µg), ceftazidime (CAZ, 30 µg), cefotaxime (CTX, 30 µg), cefoxitin (FOX, 30 µg), meropenem (MEM, 10 µg), aztreonam (ATM, 30 µg), chloramphenicol (C, 30 µg), ciprofloxacin (CIP, 5 µg), nalidixic acid (NA, 30 µg), sulfamethaxazole/trimethoprim (SXT, 25 µg), fosfomycin (FOS, 50 µg) and nitrofurantoin (F, 300 µg). While for Enterococcus spp. isolates the used disks were: ampicillin (AMP, 10 µg), chloramphenicol (C, 30 µg), erythromycin (E, 15 µg), fosfomycin (FOS, 50 µg), quinupristin/dalfopristin (15 µg), linezolid (LZD, 10 µg), nitrofurantoin (F, 300 µg), rifampicin (RD, 5 µg), teicoplanin (TEC, 30 µg), tetracycline (TE, 30 µg), and vancomycin (VA, 30 µg).
4.6. Detection of Resistance Genes to Beta-Lactam Antibiotics in Enterobacteriaceae
The production capacity of five of the most common extended-spectrum beta-lactamases was determined using real-time PCR (qPCR) detection of the blaTEM, blaCTX-M-15, blaKPC, blaCYM-2, and blaNDM genes (Table 6). For the amplification reaction, 6.5 µL of the previously extracted genomic DNA (10 ng/µL), 10 µL of the commercial Forget-Me-Not™ EvaGreen® qPCR master mix (2×; Biotium, Landing Parkway, Fremont, CA, USA), 3 µL of Rox (1:10; Biotium), and 0.25 µL of each primer (25 µM; Macrogen) were used. The reactions were carried out in a StepOne™ thermocycler (Applied Biosystems, Foster City, CA, USA) programmed with an initial denaturation at 95 °C for 10 min, followed by 40 cycles made up of 5 s at 95 °C and 30 s at 60 °C. At the end of the amplification, a melting curve was performed using the following cycling parameters: 57 °C for 30 s and temperature changes of 5 °C to a final temperature of 99 °C. Each PCR reaction included a negative control (without DNA).

Table 6.
Primers used for qPCR detection of the beta-lactam resistance genes studied.
4.7. Statistical Analysis
After verifying that the data did not meet the normality (Shapiro–Wilk test) and homogeneity of variances (Levene test) criteria, microbial counts were analyzed using the Kruskal–Wallis test. To compare microbial groups (pairwise) and slaughterhouses (A and B), Mann–Whitney tests were performed. All analyses were carried out using RStudio 4.1.3 software [84], establishing a probability level of 95% to determine significant differences (p < 0.05).
5. Conclusions
The results obtained in this research allow us to describe the microbiota present on the surfaces and equipment of different areas of the slaughter line (from stunning to cutting) of two poultry slaughterhouses. The average levels of microorganisms indicating hygienic quality (TAC, psychrotrophs, enterobacteria, coliforms, and enterococci) are in the range of values obtained by other authors, although important differences have been found between surfaces. The high presence of strains with resistance to some antibiotics and, in some cases, the high prevalence of genes for resistance to beta-lactams is striking. This fact highlights the importance of establishing appropriate measures to control the spread of microorganisms resistant to antibiotics in poultry slaughterhouses. However, the low prevalence of potentially pathogenic species, such as Salmonella spp. and Listeria spp., and their low level of resistance to the antibiotics most used for the treatment of these infections is reassuring. These findings are of great practical importance since they can serve as a basis for improving cleaning and disinfection procedures in poultry slaughterhouses and other similar meat industries. It should be noted, however, that data from only two slaughterhouses are presented, so additional studies should be conducted to obtain more general conclusions.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/antibiotics13070587/s1: Table S1: Results of antimicrobial susceptibility tests (disk diffusion) and detection of antibiotic resistance genes (PCR).
Author Contributions
Conceptualization, S.P.-M., C.R.-M., R.C. and C.A.-C.; methodology, S.P.-M., C.R.-M., R.C. and C.A.-C.; software, S.P.-M. and C.R.-M.; validation, R.C. and C.A.-C.; formal analysis, S.P.-M., C.R.-M., D.R.-C. and N.P.-E.; investigation, S.P.-M., C.R.-M., D.R.-C. and N.P.-E.; resources, R.C. and C.A.-C.; data curation, S.P.-M., C.R.-M., R.C. and C.A.-C.; writing—original draft preparation, S.P.-M., C.R.-M., D.R.-C., N.P.-E., R.C. and C.A.-C.; writing—review and editing, S.P.-M., C.R.-M., R.C. and C.A.-C.; supervision, R.C. and C.A.-C.; project administration, R.C. and C.A.-C.; funding acquisition, R.C. and C.A.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the MINISTERIO DE CIENCIA E INNOVACIÓN, grant numbers RTI2018-098267-R-C33 and PID2022-142329OB-C31, and the JUNTA DE CASTILLA Y LEÓN (CONSEJERÍA DE EDUCACIÓN), grant number LE018P20. The APC was funded by the MINISTERIO DE CIENCIA E INNOVACIÓN, grant number PID2022-142329OB-C31. Sarah Panera-Martínez is recipient of a predoctoral research fellowship from the Junta de Castilla y León (Consejería de Educación, Spain), which is co-financed by the European Social Fund. Cristina Rodríguez-Melcón is recipient of a postdoctoral research fellowship “Margarita Salas”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- MAPA. Indicadores Económicos del Sector Avícola de Carne Ministerio de Agricultura, Pesca y Alimentación. 2022. Available online: https://www.mapa.gob.es/es/ganaderia/temas/produccion-y-mercados-ganaderos/indicadorescompleto_tcm30-623991.pdf (accessed on 14 May 2024).
- OECD-FAO. Agricultural Outlook 2023–2032. 2023. Available online: https://www.oecd-ilibrary.org/agriculture-and-food/oecd-fao-agricultural-outlook-2023-2032_08801ab7-en (accessed on 14 May 2024).
- Del Río, E.; Panizo-Morán, M.; Prieto, M.; Alonso-Calleja, C.; Capita, R. Effect of various chemical decontamination treatments on natural microflora and sensory characteristics of poultry. Int. J. Food Microbiol. 2007, 115, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, H.; Saghaian, S.; Boccia, F. Antibiotic-free poultry meat consumption and its determinants. Foods 2023, 12, 1776. [Google Scholar] [CrossRef]
- Ricke, S.C. Impact of prebiotics on poultry production and food safety. Yale J. Biol. Med. 2018, 91, 151. [Google Scholar]
- Śmiałek, M.; Kowalczyk, J.; Koncicki, A. Influence of vaccination of broiler chickens against Escherichia coli with live attenuated vaccine on general properties of E. coli population, IBV vaccination efficiency, and production parameters—A field experiment. Poultry Sci. 2020, 99, 5452–5460. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Diarra, M.S.; Rehman, M.A.; Li, L.; Yu, H.; Yin, X.; Aslam, M.; Carrillo, C.D.; Yang, C.; Gong, J. Virulence potential of antimicrobial-resistant extraintestinal pathogenic Escherichia coli from retail poultry meat in a Caenorhabditis elegans model. J. Food Prot. 2023, 86, 100008. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Ding, X.; Zhao, Q.; Sun, H.; Li, T.; Li, Z.; Wang, H.; Zhang, L.; Zhang, C.; Xu, S. Development of an organic acid compound disinfectant to control food-borne pathogens and its application in chicken slaughterhouses. Poultry Sci. 2022, 101, 101842. [Google Scholar] [CrossRef]
- Rouger, A.; Tresse, O.; Zagorec, M. Bacterial contaminants of poultry meat: Sources, species, and dynamics. Microorganisms 2017, 5, 50. [Google Scholar] [CrossRef]
- Zeng, H.; De Reu, K.; Gabriël, S.; Mattheus, W.; De Zutter, L.; Rasschaert, G. Salmonella prevalence and persistence in industrialized poultry slaughterhouses. Poultry Sci. 2021, 100, 100991. [Google Scholar] [CrossRef] [PubMed]
- Ripolles-Avila, C.; Hascoët, A.S.; Martínez-Suárez, J.V.; Capita, R.; Rodríguez-Jerez, J.J. Evaluation of the microbiological contamination of food processing environments through implementing surface sensors in an Iberian pork processing plant: An approach towards the control of Listeria monocytogenes. Food Control 2019, 99, 40–47. [Google Scholar] [CrossRef]
- Botta, C.; Ferrocino, I.; Pessione, A.; Cocolin, L.; Rantsiou, K. Spatiotemporal distribution of the environmental microbiota in food processing plants as impacted by cleaning and sanitizing procedures: The case of slaughterhouses and gaseous ozone. Appl. Environ. Microbiol. 2020, 86, e01861-20. [Google Scholar] [CrossRef]
- Song, X.; Wang, H.; Xu, X. Investigation of microbial contamination in a chicken slaughterhouse environment. J. Food Sci. 2021, 86, 3598–3610. [Google Scholar] [CrossRef] [PubMed]
- Abreu, R.; Semedo-Lemsaddek, T.; Cunha, E.; Tavares, L.; Oliveira, M. Antimicrobial drug resistance in poultry production: Current status and innovative strategies for bacterial control. Microorganisms 2023, 11, 953. [Google Scholar] [CrossRef] [PubMed]
- Capita, R.; Alonso-Calleja, C. Antibiotic-resistant bacteria: A challenge for the food industry. Crit. Rev. Food Sci. Nutr. 2013, 53, 11–48. [Google Scholar] [CrossRef] [PubMed]
- Yengui, M.; Trabelsi, R.; Khannous, L.; Mathlouthi, N.E.; Adnan, M.; Siddiqui, A.J.; Noumi, E.; Snoussi, M.; Gdoura, R. Rapid detection of beta-lactamases genes among Enterobacterales in urine samples by using real-time PCR. BioMed. Res. Int. 2022, 2022, 8612933. [Google Scholar] [CrossRef] [PubMed]
- Maciuca, I.E.; Williams, N.J.; Tuchilus, C.; Dorneanu, O.; Guguianu, E.; Carp-Carare, C.; Rimbu, C.; Timofte, D. High prevalence of Escherichia coli-producing CTX-M-15 extended-spectrum beta-lactamases in poultry and human clinical isolates in Romania. Microb. Drug Resist. 2015, 21, 651–662. [Google Scholar] [CrossRef]
- Silva, N.; Carvalho, I.; Currie, C.; Sousa, M.; Igrejas, G.; Poeta, P. Extended-Spectrum-β-Lactamase and Carbapenemase-Producing Enterobacteriaceae in Food-Producing Animals in Europe: An Impact on Public Health? In Antibiotic Drug Resistance; Capelo-Martínez, J.L., Igrejas, G., Eds.; John Wiley & Sons Inc.: Hoboken, NY, USA, 2019; pp. 261–273. [Google Scholar]
- Pérez-Vázquez, M.; Sola Campoy, P.J.; Ortega, A.; Bautista, V.; Monzón, S.; Ruiz-Carrascoso, G.; Mingorance, J.; González-Barberá, E.M.; Gimeno, C.; Aracil, B.; et al. Emergence of NDM-producing Klebsiella pneumoniae and Escherichia coli in Spain: Phylogeny, resistome, virulence and plasmids encoding blaNDM-like genes as determined by WGS. J. Antimicrob. Chemother. 2019, 74, 3489–3496. [Google Scholar] [CrossRef] [PubMed]
- Galán-Sánchez, F.; Aznar-Marín, P.; Marín-Casanova, P.; Rodríguez-Iglesias, M. Diversity of bla genes and low incidence of CTX-M in plasmid-mediated AmpC-producing Escherichia coli clinical isolates. APMIS 2014, 122, 796–799. [Google Scholar] [CrossRef] [PubMed]
- Lopes, E.; Saavedra, M.J.; Costa, E.; de Lencastre, H.; Poirel, L.; Aires-de-Sousa, M. Epidemiology of carbapenemase-producing Klebsiella pneumoniae in northern Portugal: Predominance of KPC-2 and OXA-J. Global Antimicrob. Resist. 2020, 22, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Delshadi, R.; Bahrami, A.; Assadpour, E.; Williams, L.; Jafari, S.M. Nano/microencapsulated natural antimicrobials to control the spoilage microorganisms and pathogens in different food products. Food Control 2021, 128, 108180. [Google Scholar] [CrossRef]
- Møretrø, T.; Langsrud, S. Residential bacteria on surfaces in the food industry and their implications for food safety and quality. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1022–1041. [Google Scholar] [CrossRef]
- Ferone, M.; Gowen, A.; Fanning, S.; Scannell, A.G. Microbial detection and identification methods: Bench top assays to omics approaches. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3106–3129. [Google Scholar] [CrossRef] [PubMed]
- Uzuriaga, M.; Leiva, J.; Guillén-Grima, F.; Rua, M.; Yuste, J.R. Clinical impact of rapid bacterial microbiological identification with the MALDI-TOF MS. Antibiotics 2023, 12, 1660. [Google Scholar] [CrossRef] [PubMed]
- Whyte, P.J.D.C.; Collins, J.D.; McGill, K.; Monahan, C.; O’mahony, H. Distribution and prevalence of airborne microorganisms in three commercial poultry processing plants. J. Food Prot. 2001, 64, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Calleja, C.; Guerrero-Ramos, E.; Capita, R. Hygienic status assessment of two lamb slaughterhouses in Spain. J. Food Prot. 2017, 80, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Takahashi, H.; Kondo, A.; Koike, F.; Kuda, T.; Kimura, B.; Kobayashi, M. Distribution of psychrophilic microorganisms in a beef slaughterhouse in Japan after cleaning. PLoS ONE 2022, 17, e0268411. [Google Scholar] [CrossRef] [PubMed]
- Nortje, G.L.; Nel, L.; Jordaan, E.; Badenhorst, K.; Goedhart, G.; Holzapfel, W.H.; Grimbeek, R.J. A quantitative survey of a meat production chain to determine the microbial profile of the final product. J. Food Prot. 1990, 53, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Gribble, A.; Mills, J.; Brightwell, G. The spoilage characteristics of Brochothrix thermosphacta and two psychrotolerant Enterobacteriacae in vacuum packed lamb and the comparison between high and low pH cuts. Meat Sci. 2014, 97, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Bruzaroski, S.R.; de Souza, R.P.; da Silva Pasquim, P.; Fagnani, R.; de Santana, E.H.W. Influence of storage temperature on the population of microorganisms in raw sheep milk and its physical-chemical profile. Res. Soc. Dev. 2020, 9, e27691210796. [Google Scholar] [CrossRef]
- Haleem, A.M.; Al-bakri, S.A.; Al-Hiyaly, S.A. Determination of microbial content in poultry meat in local Iraqi markets. J. Microbiol. Res. 2013, 3, 205–207. [Google Scholar]
- Hu, Z.; Peng, Z.; Zhang, X.; Li, Z.; Jia, C.; Li, X.; Lv, Y.; Tan, C.; Chen, H.; Wang, X. Prevalence and molecular characterization of antimicrobial-resistant Escherichia coli in pig farms, slaughterhouses, and terminal markets in Henan province of China. Foodborne Path. Dis. 2021, 18, 733–743. [Google Scholar] [CrossRef]
- Vázquez-Villanueva, J.; Vázquez, K.; Martínez-Vázquez, A.V.; Wong-González, A.; Hernández-Escareño, J.; Cabrero-Martínez, O.; Cruz-Pulido, W.L.; Guerrero, A.; Rivera, G.; Bocanegra-García, V. Molecular and antimicrobial susceptibility characterization of Escherichia coli isolates from bovine slaughterhouse process. Antibiotics 2023, 12, 291. [Google Scholar] [CrossRef]
- Lavilla Lerma, L.; Benomar, N.; Gálvez, A.; Abriouel, H. Prevalence of bacteria resistant to antibiotics and/or biocides on meat processing plant surfaces throughout meat chain production. Int. J. Food Microbiol. 2013, 161, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Sebsibe, M.A.; Asfaw, E.T. Occurrence of multi-drug resistant Escherichia coli and Escherichia coli O157: H7 in meat and swab samples of various contact surfaces at abattoir and butcher shops in Jimma town, Southwest district of Ethiopia. Inf. Drug Res. 2020, 13, 3853–3862. [Google Scholar] [CrossRef]
- Agostinho Davanzo, E.F.; Dos Santos, R.L.; Castro, V.H.L.; Palma, J.M.; Pribul, B.R.; Dallago, B.S.L.; Fuga, B.; Medeiros, M.; Titze de Almeida, S.S.; da Costa, H.M.B.; et al. Molecular characterization of Salmonella spp. and Listeria monocytogenes strains from biofilms in cattle and poultry slaughterhouses located in the federal District and State of Goiás, Brazil. PLoS ONE 2021, 16, e0259687. [Google Scholar] [CrossRef]
- Stadtlober, G.A.W.; Fiorentini, Â.M.; Severo, J.; Bernardo, K.B.D.; Carvalho, I.R.; Loro, M.V. Contamination by aerobic mesophilal and Enterobacteriaceae bacteria in a pig refrigerator. Agron. Sci. Biotechnol. 2024, 10, 1–13. [Google Scholar] [CrossRef]
- Moura-Alves, M.; Carvalho, M.; Ribeiro, D.H.B.; Barbosa, J.; Silveira, L.; Pista, Â.; Pinto, H.P.; Saraiva, C.; Teixeira, P.; Esteves, A. Hygiene indicators and salmonellae on surfaces of swine carcasses from two slaughterhouses in northern Portugal. J. Food Prot. 2022, 85, 1566–1575. [Google Scholar] [CrossRef]
- Soares-Santos, V.; Barreto, A.S.; Semedo-Lemsaddek, T. Characterization of enterococci from food and food-related settings. J. Food Prot. 2015, 78, 1320–1326. [Google Scholar] [CrossRef] [PubMed]
- Wambui, J.; Tasara, T.; Njage, P.M.K.; Stephan, R. Species distribution and antimicrobial profiles of Enterococcus spp. isolates from Kenyan small and medium enterprise slaughterhouses. J. Food Prot. 2018, 81, 1445–1449. [Google Scholar] [CrossRef]
- Guerrero-Ramos, E.; Cordero, J.; Molina-González, D.; Poeta, P.; Igrejas, G.; Alonso-Calleja, C.; Capita, R. Antimicrobial resistance and virulence genes in enterococci from wild game meat in Spain. Food Microbiol. 2016, 53, 156–164. [Google Scholar] [CrossRef]
- Guerrero-Ramos, E.; Molina-Gonzalez, D.; Blanco-Moran, S.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C.; Capita, R. Prevalence, antimicrobial resistance, and genotypic characterization of vancomycin-resistant enterococci in meat preparations. J. Food Prot. 2016, 79, 748–756. [Google Scholar] [CrossRef]
- Jang, Y.S.; Moon, J.S.; Kang, H.J.; Bae, D.; Seo, K.H. Prevalence, characterization, and antimicrobial susceptibility of Listeria monocytogenes from raw beef and slaughterhouse environments in Korea. Foodborne Path. Dis. 2021, 18, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Gowda, T.K.; Van Damme, I. Occurrence and antibiotic susceptibility of Listeria species and Staphylococcus aureus in cattle slaughterhouses of Kerala, South India. Foodborne Path. Dis. 2017, 14, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Tasci, F.; Sudagidan, M.; Yavuz, O.; Soyucok, A.; Aydin, A. Virulence properties of Listeria monocytogenes isolated from meat and meat contact surfaces in a slaughterhouse. Pol. J. Vet. Sci. 2024, 27, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Gregova, G.; Kmetova, M.; Kmet, V.; Venglovsky, J.; Feher, A. Antibiotic resistance of Escherichia coli isolated from a poultry slaughterhouse. Ann. Agric. Environ. Med. 2012, 19, 75–79. [Google Scholar] [PubMed]
- Elabbasy, M.T.; Hussein, M.A.; Algahtani, F.D.; Abd El-Rahman, G.I.; Morshdy, A.E.; Elkafrawy, I.A.; Adeboye, A.A. MALDI-TOF MS based typing for rapid screening of multiple antibiotic resistance E. coli and virulent non-O157 shiga toxin-producing E. coli isolated from the slaughterhouse settings and beef carcasses. Foods 2021, 10, 820. [Google Scholar] [CrossRef] [PubMed]
- Savin, M.; Bierbaum, G.; Mutters, N.T.; Schmithausen, R.M.; Kreyenschmidt, J.; García-Meniño, I.; Schmoger, S.; Käsbohrer, A.; Hammerl, J.A. Genetic characterization of carbapenem-resistant Klebsiella spp. from municipal and slaughterhouse wastewater. Antibiotics 2022, 11, 435. [Google Scholar] [CrossRef] [PubMed]
- Homeier-Bachmann, T.; Heiden, S.E.; Lübcke, P.K.; Bachmann, L.; Bohnert, J.A.; Zimmermann, D.; Schaufler, K. Antibiotic-resistant Enterobacteriaceae in wastewater of abattoirs. Antibiotics 2021, 10, 568. [Google Scholar] [CrossRef] [PubMed]
- WHO (World Health Organization). Critically Important Antimicrobials for Human Medicine, 6th ed.; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- OIE (World Organization for Animal Health). OIE List of Antimicrobial Agents of Veterinary Importance; World Organization for Animal Health: Paris, France, 2018. [Google Scholar]
- Nordmann, P.; Poirel, L.; Walsh, T.R.; Livermore, D.M. The emerging NDM carbapenemases. Trends Microbiol. 2011, 19, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.D.; Perez-Bonilla, D.; Hallowell, A.; Redding, L.E. Carbapenem prescribing at a veterinary teaching hospital before an outbreak of carbapenem-resistant Escherichia coli. J. Small Anim. Pract. 2022, 63, 442–446. [Google Scholar] [CrossRef]
- Jiménez-Belenguer, A.I.; Ferrús, M.A.; Hernández, M.; García-Hernández, J.; Moreno, Y.; Castillo, M.Á. Prevalence and characterization of Beta-lactam and Carbapenem-resistant bacteria isolated from organic fresh produce retailed in eastern Spain. Antibiotics 2023, 12, 387. [Google Scholar] [CrossRef]
- Friese, A.; Schulz, J.; Laube, H.; von Salviati, C.; Hartung, J.; Roesler, U. Faecal occurrence and emissions of livestock-associated methicillin-resistant Staphylococcus aureus (laMRSA) and ESbl/AmpC-producing E. coli from animal farms in Germany. Berl. Munch. Tierarztl. Wochenschr. 2013, 126, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Li, Y.; Yang, Y.; Shen, Z.; Cai, C.; Wang, Y.; Wang, S. High prevalence and persistence of carbapenem and colistin resistance in livestock farm environments in China. J. Hazard. Mat. 2021, 406, 124298. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.S.; Choi, D.S.; Kim, Y.J.; Chon, J.W.; Kim, H.S.; Park, H.J.; Moon, J.S.; Wee, S.H.; Seo, K.H. Characterization of Escherichia coli–producing extended-spectrum β-lactamase (ESBL) isolated from chicken slaughterhouses in South Korea. Foodborne Pathog. Dis. 2015, 12, 741–748. [Google Scholar] [CrossRef]
- Wei, B.; Shang, K.; Cha, S.Y.; Zhang, J.F.; Jang, H.K.; Kang, M. Conjugative plasmid-mediated extended spectrum cephalosporin resistance in genetically diverse Escherichia coli from a chicken slaughterhouse. Animals 2021, 11, 2491. [Google Scholar] [CrossRef] [PubMed]
- Adel, W.A.; Ahmed, A.M.; Hegazy, Y.; Torky, H.A.; Shimamoto, T. High prevalence of ESBL and plasmid-mediated quinolone resistance genes in Salmonella enterica isolated from retail meats and slaughterhouses in Egypt. Antibiotics 2021, 10, 881. [Google Scholar] [CrossRef] [PubMed]
- Clemente, L.; Leão, C.; Moura, L.; Albuquerque, T.; Amaro, A. Prevalence and characterization of ESBL/AmpC producing Escherichia coli from fresh meat in Portugal. Antibiotics 2021, 10, 1333. [Google Scholar] [CrossRef] [PubMed]
- Economou, V.; Delis, G.; Stavrou, D.; Gousia, P.; Tsitsos, A.; Mantzios, T.; Choiliara, E.; Kolovos, N.; Soultos, N. Characterization of extended spectrum cephalosporin-resistant Escherichia coli strains isolated from raw poultry carcasses in catering services in Northern Greece. Vet. Sci. 2023, 10, 487. [Google Scholar] [CrossRef] [PubMed]
- Savin, M.; Bierbaum, G.; Schmithausen, R.M.; Heinemann, C.; Kreyenschmidt, J.; Schmoger, S.; Akbaba, I.; Käsbohrer, A.; Hammerl, J.A. Slaughterhouse wastewater as a reservoir for extended-spectrum β-lactamase (ESBL)-producing, and colistin-resistant Klebsiella spp. and their impact in a “One Health” perspective. Sci. Total Environ. 2022, 804, 150000. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Laorden, A.; Arraiz-Fernández, C.; González-Fandos, E. Microbiological quality and safety of fresh turkey meat at retail level, including the presence of ESBL-producing Enterobacteriaceae and methicillin-resistant S. aureus. Foods 2023, 12, 1274. [Google Scholar] [CrossRef]
- Gundran, R.S.; Cardenio, P.A.; Villanueva, M.A.; Sison, F.B.; Benigno, C.C.; Kreausukon, K.; Pichpol, D.; Punyapornwithaya, V. Prevalence and distribution of blaCTX-M, blaSHV, blaTEM genes in extended-spectrum β-lactamase-producing E. coli isolates from broiler farms in the Philippines. BMC Vet. Res. 2019, 15, 227. [Google Scholar] [CrossRef]
- Patil, S.; Chen, X.; Wen, F. Exploring the phenotype and genotype of multi-drug resistant Klebsiella pneumoniae harbouring blaCTX-M group extended-spectrum β-lactamases recovered from paediatric clinical cases in Shenzhen, China. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- Saliu, E.M.; Vahjen, W.; Zentek, J. Types and prevalence of extended–spectrum beta–lactamase producing Enterobacteriaceae in poultry. Animal Health Res. Rev. 2017, 18, 46–57. [Google Scholar] [CrossRef]
- Akpaka, P.E.; Vaillant, A.; Wilson, C.; Jayaratne, P. Extended spectrum beta-lactamase (ESBL) produced by gram-negative bacteria in Trinidad and Tobago. Int. J. Microbiol. 2021, 2021, 5582755. [Google Scholar] [CrossRef] [PubMed]
- Hasman, H.; Mevius, D.; Veldman, K.; Olesen, I.; Aarestrup, F.M. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chem. 2005, 56, 115–121. [Google Scholar] [CrossRef]
- Savin, M.; Alexander, J.; Bierbaum, G.; Hammerl, J.A.; Hembach, N.; Schwartz, T.; Schimthausen, R.M.; Sib, E.; Voigt, A.; Kreyenschmidt, J. Antibiotic-resistant bacteria, antibiotic resistance genes, and antibiotic residues in wastewater from a poultry slaughterhouse after conventional and advanced treatments. Sci. Rep. 2021, 11, 16622. [Google Scholar] [CrossRef] [PubMed]
- Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes and of Listeria spp.—Part 1: Detection Method. UNE-EN ISO 11290-1:2018. Available online: https://www.une.org/encuentra-tu-norma/busca-tu-norma/norma/?c=N0059546 (accessed on 12 May 2024).
- Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp. UNE-EN ISO 6579-1:2017. Available online: https://www.une.org/encuentra-tu-norma/busca-tu-norma/norma/?c=N0058760 (accessed on 12 May 2024).
- Jay, J.M. A review of aerobic and psychrotrophic plate count procedures for fresh meat and poultry products. J. Food Prot. 2002, 65, 1200–1206. [Google Scholar] [CrossRef] [PubMed]
- Cousin, M.A.; Jay, J.M.; Vasavada, P.C. Psychrotrophic microorganisms. In Compendium of Methods for the Microbiological Examination of Foods, 4th ed.; Downes, F.P., Ito, K., Eds.; American Public Health Association: Washington, DC, USA, 2001; pp. 159–166. [Google Scholar]
- Baird, R.M.; Corry, J.E.L.; Curtis, G.D.W. Pharmacopeia of culture media for food microbiology. Int. J. Food Microbiol. 1987, 5, 201–266. [Google Scholar]
- Sørum, M.; Holstad, G.; Lillehaug, A.; Kruse, H. Prevalence of vancomycin resistant enterococci on poultry farms established after the ban of avoparcin. Avian Dis. 2004, 48, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Panera-Martínez, S.; Rodríguez-Melcón, C.; Serrano-Galán, V.; Alonso-Calleja, C.; Capita, R. Prevalence, quantification and antibiotic resistance of Listeria monocytogenes in poultry preparations. Food Control 2021, 135, 108608. [Google Scholar] [CrossRef]
- Fazzeli, H.; Arabestani, M.R.; Esfahani, B.N.; Khorvash, F.; Pourshafie, M.R.; Moghim, S.; Safaei, H.G.; Faghri, J.; Narimani, T. Development of PCR-based method for detection of Enterobacteriaceae in septicemia. J. Res. Med. Sci. 2012, 17, 671–675. [Google Scholar]
- El-Sayed, A.K.A.; Abou Dobara, M.; El-Shihy, E.S. Simultaneous detection of seven foodborne Enterobacteriaceae pathogens using multiplex PCR. J. Egypt. Acad. Soc. Environ. Dev. 2019, 20, 61–78. [Google Scholar] [CrossRef]
- Deasy, B.M.; Rea, M.C.; Fitzgerald, G.F.; Cogan, T.M.; Beresford, T.P. A rapid PCR based method to distinguish between Lactococcus and Enterococcus. Sys. Appl. Microbiol. 2000, 23, 510–522. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Park, S.H.; Yeom, Y.S.; Shrivastav, A.; Lee, S.H.; Kim, Y.R.; Kim, H.Y. Simultaneous detection of Listeria species isolated from meat processed foods using multiplex PCR. Food Control 2013, 32, 659–664. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Clinical and Laboratory Standars Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Weiß, D.; Engelmann, I.; Braun, S.D.; Monecke, S.; Ehricht, R. A multiplex real-time PCR for the direct, fast, economic and simultaneous detection of the carbapenemase genes blaKPC, blaNDM, blaVIM and blaOXA. J. Microbiol. Meth. 2017, 142, 20–26. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development Environment for R. 2019. Available online: http://www.rstudio.com/ (accessed on 15 March 2022).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).