Abstract
In the present study, a total of 720 samples were collected from retail raw meat from 13 upazilas in Sylhet District, Bangladesh, of which 225 samples were from cattle meat, 210 samples were from goat meat, and 285 samples were from chicken meat. Salmonella enterica serovars Typhimurium and Enteritidis were screened for extended-spectrum β-lactamase (ESBL) genes using multiplex PCR. Among the 720 samples, Salmonella spp. was detected in 28.06% (202 out of 720) of the samples, with S. Enteritidis and S. Typhimurium were identified in 11.53% (83 out of 720) and 12.22% (88 out of 720) of the samples, respectively. It was found that all Salmonella enterica serovars isolated from cattle meat displayed multidrug resistance (MDR) based on antimicrobial susceptibility testing. Notably, a significant proportion of S. Enteritidis isolates and all S. Typhimurium isolates from goat meat demonstrated complete resistance to multiple drugs (ampicillin, cefuroxime, and ceftazidime). Regarding chicken meat, out of 89 isolates encompassing both S. Typhimurium and S. Enteritidis, 57 isolates (64.04%) exhibited MDR. Additionally, blaCTX-M-1 exhibited the highest occurrence at 15.69% for S. Typhimurium and 7.89% for S. Enteritidis in chicken meat. Moreover, blaCTX-M-9 was only detected at 3.92% for S. Enteritidis in chicken meat. Furthermore, blaOXA had the highest prevalence rate of 19.04% for S. Enteritidis and 25.80% for S. Typhimurium in cattle meat, followed by chicken meat. These findings highlight the urgency for monitoring ESBL-producing Salmonella in retail raw meat and the need for strict measure to manage antibiotic use to prevent the spread of multidrug-resistant and ESBL-producing Salmonella strains, thereby protecting humans and reducing public health risks.
1. Introduction
Salmonella remains a leading cause of bacterial foodborne illnesses worldwide [1,2] contributing to approximately 10% of global mortality, predominantly originating from animal sources, resulting in an estimated 33 million deaths annually [3]. Salmonellosis is one of the most common zoonotic diseases, and Salmonella is a common cause of foodborne disease outbreaks. The prevalence of Salmonella spp. infection is a significant public health issue and a continuous threat worldwide [4]. The incidence of Salmonella infections significantly increases the global burden of gastroenteritis, with an estimated 93.8 million cases reported annually, leading to 155,000 deaths [5]. Gram-negative bacteria such as Salmonella employ β-lactamases as a primary defense mechanism against β-lactam antibiotics. The emergence of extended-spectrum β-lactamases (ESBLs) poses a significant global threat, with over 300 distinct types identified [6].
Antimicrobial resistance is a critical global health and development threat, largely driven by the indiscriminate use and overuse of antimicrobial agents in both food animals and humans [7,8]. Countries such as Bangladesh face increased risks of antimicrobial resistance due to significant problems hindering the implementation of antibiotic stewardship, limited regulatory programmed surveillance, and limited monitoring systems regarding antimicrobial use and resistance prevention [9]. Antimicrobial agents are commonly used in cattle, goats, and chickens for disease prevention and treatment, with many of these drugs also being frequently used in human medicine, contributing to potential antimicrobial resistance in both animals and humans [10]. A major risk of zoonotic infections is the spillover of multidrug-resistant Salmonella strains from livestock to humans via food chain including contaminated food, water, direct contact, or the ingestion of contaminated materials derived from infected livestock. Given these concerns, it is imperative to detect Salmonella enterica in livestock meat especially cattle, goat, and chicken and investigate their multidrug resistance (MDR) profiles.
Over the past few decades, S. Typhimurium and S. Enteritidis have become major causes of global salmonellosis outbreaks [11]. The economic impact of Salmonella infections has gained increased attention in developed countries. Animals are exposed to Salmonella through various means, including water, feed, feces, soil, and insects, becoming infected or serving as asymptomatic carriers. Cattle, goats, and chicken meat are significant animal-origin protein sources which are widely consumed in Bangladesh and globally [12,13,14]. However, in many resource-limited regions, these animals are slaughtered in small, unhygienic abattoirs, facilitating microbial contamination, survival, and spread to the surrounding environment and food handlers [15,16,17,18,19]. Several studies have reported the prevalence of Salmonella in meat, with detection rates ranging from 6.79% to 97.6% in chicken meat in India [20] and 21.1% in Bangladesh [21]. Moreover, the prevalence of Salmonella enterica serovar in cattle meat was reported to be 23.3% in Egypt [22], 64.28% in Pakistan [23], 23% in Nigeria [24], 30.55% in Morocco [25], and 29.8% in Tunisia [26]. The reported prevalence rates of Salmonella in goat meat varies globally, with rates as high as 60% reported in Pakistan [23]. The detection of Salmonella was reported from goat meat swabs in different regions of different countries such as a rate of 8.3% in Modjo (Ethiopia), 7.5% in Bishoftu (Ethiopia), 4% in Arusha (Tanzania), and 3.5% in Gujrat (India) [27,28,29].
Antimicrobial resistance in livestock production systems including common meat-producing animals such as cattle, goat, and chicken, particularly multidrug-resistant Salmonella enterica serovars Enteritidis and Typhimurium, remains largely unexplored in Bangladesh. Recent studies have highlighted the significant detection of MDR E. coli in commercial cattle, goats, and poultry [30,31,32]. Thus, the current study aimed to determine and compare the prevalence and antimicrobial resistance of ESBL-producing and MDR Salmonella enterica serovars Enteritidis and Typhimurium in retail meat samples in Bangladesh.
2. Results
2.1. Molecular Detection of Salmonella enterica Serovars
Out of 720 samples, the prevalence of Salmonella spp. was found to be 28.06% (202 out of 720; 95% CI: 24.80–31.49). Specifically, the prevalence rate of S. Enteritidis was 11.53% (83 out of 720; 95% CI: 9.29–14.09), while the prevalence rate of S. Typhimurium was 12.22% (88 out of 720; 95% CI: 9.92–14.84). Out of 225 retail cattle meat samples, the prevalence rates of Salmonella spp., S. Typhimurium, and S. Enteritidis were 26.22%, 13.78%, and 9.33%, respectively (as illustrated in Figure 1 and Figure 2).
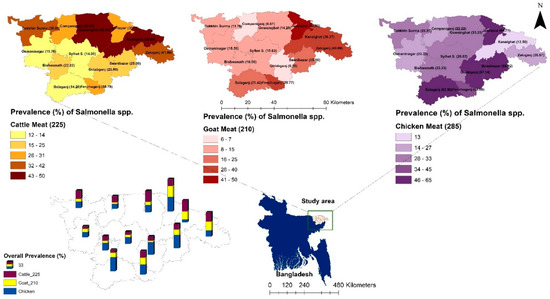
Figure 1.
The location (sub-district) and the prevalence of Salmonella spp. isolated from samples of retail cattle, goat, and chicken meat in Sylhet District, Bangladesh. Bangladesh country map is shown in blue and the box indicates the selected study area in the present study.
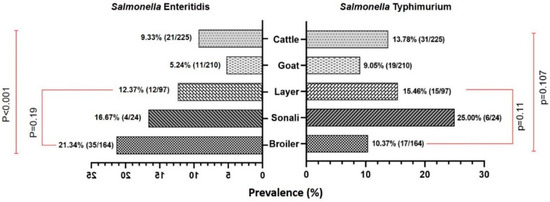
Figure 2.
A bi-directional bar (Mirror bar) diagram showing the prevalence of Salmonella enterica serovar Enteritidis and Typhimurium in cattle and poultry retail meat samples compared to goat meat samples. (χ2-test, Level of Significance p < 0.05).
Analysis of the prevalence of Salmonella spp. across different upazilas within the district revealed that both the Kanaighat and Gowainghat Upazilas had the highest prevalence rate (50.00%) of Salmonella spp., while Osmaninagar displayed the lowest rate of 11.76% (Figure 1). Moreover, Gowainghat Upazila exhibited the highest prevalence of S. Typhimurium in retail cattle meat samples, accounting for 35.71%, whereas Osmaninagar Upazila showed the lowest prevalence rate of 5.88% (Table 1).

Table 1.
The prevalence of S. Typhimurium isolated from retail cattle, goat, and chicken meat samples obtained from 13 upazilas of Sylhet in the present study.
Similarly, Dakkhin Surma Upazila demonstrated the highest prevalence of S. Enteritidis in retail cattle meat samples, accounting for 23.81%, whereas only 2.00% of the samples collected from Sylhet Sadar Upazila were S. Enteritidis positive (Table 2).

Table 2.
The prevalence of Salmonella Enteritidis in different sub-districts (n = 13) of Sylhet, isolated from retail cattle, goat, and chicken meat in the present study.
In the present study, the prevalence of Salmonella spp. in chicken meat samples was 36.84% (105/285) which was found to be higher than that of the prevalence rates in cattle and goat meat samples. Of these isolates, 51 (17.89%) were found to be S. Enteritidis, and 38 (13.33%) were found to be S. Typhimurium. Jaintapur Upazila had the highest prevalence (64.71%) of Salmonella spp., while Kanaighat displayed the lowest (12.50%) (Figure 1). Balaganj Upazila exhibited the highest prevalence of Salmonella Typhimurium, accounting for 25.00% (9.77–46.71%), while Companiganj Upazila showed the lowest prevalence at 5.56% (0.14–27.29%) (Table 1). Similarly, Golapganj Upazila demonstrated the highest (35.71%) prevalence of S. Enteritidis in chicken meat samples, whereas no positive samples were found in Kanaighat Upazila (Table 2). Moreover, the specific prevalence rates of S. Typhimurium in various types of retail chicken meat samples, with broiler, sonali, and layer meats, exhibited prevalence rates of 10.37% (17/164), 25.00% (6/24), and 15.46% (15/97), respectively (Figure 2). Similarly, the prevalence rates for S. Enteritidis in different types of retail chicken meats, with broiler, sonali, and layer meats, demonstrated prevalence rates of 21.34% (35/164), 16.67% (4/24), and 12.37% (12/97), respectively (Figure 2).
The prevalence rate of Salmonella spp. isolated from samples obtained from retail goat meat (Figure 1) was recently reported [33], and it was found to be 18.10% (38/210; 95% CI: 13.13–23.98), which was lower than the prevalence rates of Salmonella spp. in both cattle and meat samples reported in the present study. The highest prevalence was observed in Jaintapur at 50% (6 out of 12), followed by Zakiganj at 40% (4 out of 10). The lowest prevalence was reported in Golapganj at 6.25% (1 out of 16) as previously reported [33]. The distribution of the prevalence of Salmonella enterica serovars in retail goat meat samples across different upazilas of Sylhet District compared to cattle and chicken samples is shown in Table 1 and Table 2. The investigation also revealed that the prevalence rates of S. Typhimurium and S. Enteritidis were 9.05% (95% CI: 5.54–13.77) and 5.24% (95% CI: 2.64–9.18), respectively (Figure 2 and Table 1 and Table 2). The detailed geographical distribution of these prevalence rates is provided in Table 1 and Table 2.
2.2. Antimicrobial Resistance and Phenotypic Correlations
A heatmap was generated using hierarchical cluster analysis (HCA) and illustrated the antibiotic susceptibility pattern. The clustering of antibiotics was represented by a dendrogram (Figure 3A–C).
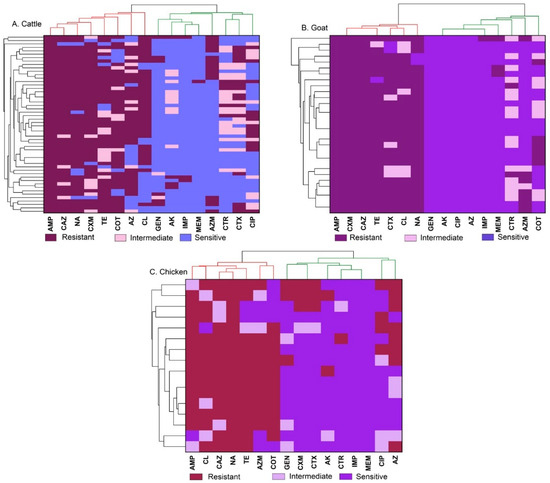
Figure 3.
A heatmap showing the sensitivity pattern of antibiotics with clustering as a dendrogram. The clustering of antibiotics was represented by a dendrogram for cattle (A), goat (B), and chicken (C) meat samples. Ampicillin (AMP), gentamicin (GEN), amikacin (AK), cefuroxime (CXM), ceftriaxone (CTR), cefotaxime (CTX), ceftazidime (CAZ), meropenem (MEM), imipenem (IMP), tetracycline (TE), ciprofloxacin (CIP), azithromycin (AZ), aztreonam (AZM), chloramphenicol (CL), sulfamethoxazole-trimethoprim (COT), and nalidixic acid (NA).
The majority of isolates from cattle meat samples exhibited resistance to ampicillin (100%), ceftazidime, nalidixic acid, among others, while demonstrating high sensitivity to meropenem and imipenem (Figure 3A). Additionally, Salmonella Typhimurium and Salmonella Enteritidis obtained from goat meat samples (Figure 3B) displayed complete resistance to ampicillin, ceftazidime and cefuroxime while 100% sensitive to amikacin, ciprofloxacin and gentamicin. Among Salmonella Enteritidis isolates from chicken meat (Figure 3C), 100% showed resistance to nalidixic acid while demonstrating the highest sensitivity to meropenem, imipenem, and ceftriaxone. Similarly, all Salmonella Typhimurium isolates from chicken meat samples displayed resistance to ampicillin with the isolates demonstrated the highest sensitivity to meropenem, imipenem, and amikacin.
Pearson’s correlation coefficient analysis (Figure 4) depicts the phenotypic correlation between antimicrobial agents, along with the level of significance. In addition, Figure 4 illustrates the phenotypic correlation coefficients (r) among the selected antibiotics, represented by a spectrum ranging from light to deep violet for positive correlations (0 to +1) and light to deep red for negative correlations (0 to −1). A moderately strong positive correlation was observed between CXM and CTX (r = 0.59, p < 0.001). Additionally, moderate positive correlations were noted between CIP and AZ (r = 0.44, p < 0.001), as well as between TE and CAZ (r = 0.36, p < 0.001). Conversely, significantly weak positive correlations were found between CXM and CTR, AZM and GEN, and CTX and CTR, among others. Weak negative correlations were observed between CTX and COT (r = −0.33, p < 0.001), AZM and CXM (r = −0.32, p < 0.01), and CTR and GEN (r = −0.26, p < 0.01), among others. The majority of the relationships were characterized by very weak positive and negative correlations, with further details depicted in Figure 4.
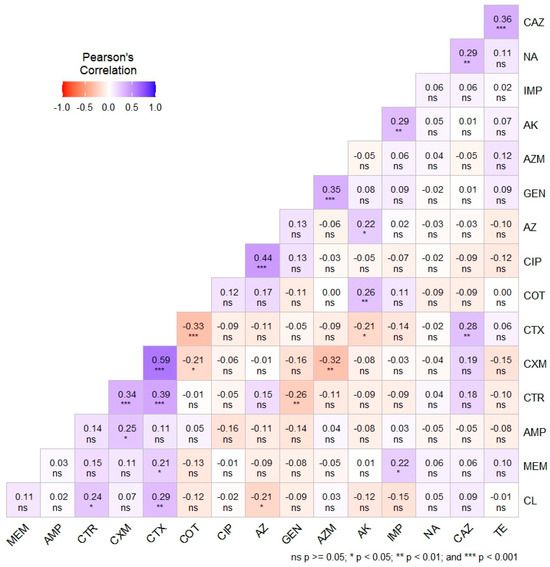
Figure 4.
Pearson’s correlation coefficient showing the phenotypic correlation among the antimicrobial agents with levels of significance. Ampicillin (AMP), gentamicin (GEN), amikacin (AK), cefuroxime (CXM), ceftriaxone (CTR), cefotaxime (CTX), ceftazidime (CAZ), meropenem (MEM), imipenem (IMP), tetracycline (TE), ciprofloxacin (CIP), azithromycin (AZ), aztreonam (AZM), chloramphenicol (CL), sulfamethoxazole-trimethoprim (COT), and nalidixic acid (NA).
2.3. Antibiogram Profile of S. enterica Serovar Typhimurium and Enteritidis
The comprehensive antibiogram profiles of S. enterica Serovar Typhimurium and Enteritidis isolates are shown in Figure 5 and detailed in Supplementary Tables S1 and S2. S. Enteritidis exhibited the highest resistance to ampicillin (92.5%), followed by nalidixic acid (90%), and tetracycline (87.5%). Conversely, all isolates were sensitive to ceftriaxone (100%), followed by meropenem (97.5%), and imipenem (95%). S. Typhimurium showed complete resistance to ampicillin and a high resistance to ceftazidime (98.28%). In contrast, isolates were generally sensitive to gentamicin (98.28%), imipenem (96.55%) and meropenem (94.83%) among the 16 tested antimicrobial agents.
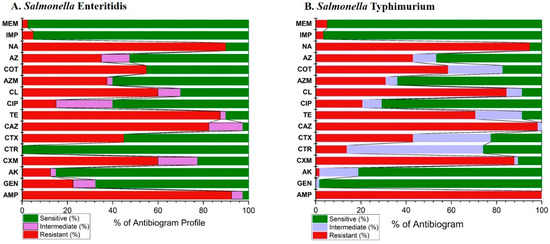
Figure 5.
The overall antibiogram profiles of S. enterica Serovar Typhimurium and Enteritidis isolates in the present study. Ampicillin (AMP), gentamicin (GEN), amikacin (AK), cefuroxime (CXM), ceftriaxone (CTR), cefotaxime (CTX), ceftazidime (CAZ), meropenem (MEM), imipenem (IMP), tetracycline (TE), ciprofloxacin (CIP), azithromycin (AZ), aztreonam (AZM), chloramphenicol (CL), sulfamethoxazole-trimethoprim (COT), and nalidixic acid (NA).
2.4. MAR Index and MDR Profile
The scatter plot correlation matrix, along with histograms, illustrates the status and correlation of the MARI of Salmonella enterica serovars Enteritidis and Typhimurium among cattle, goat, and chicken meat samples (Figure 6A,B).
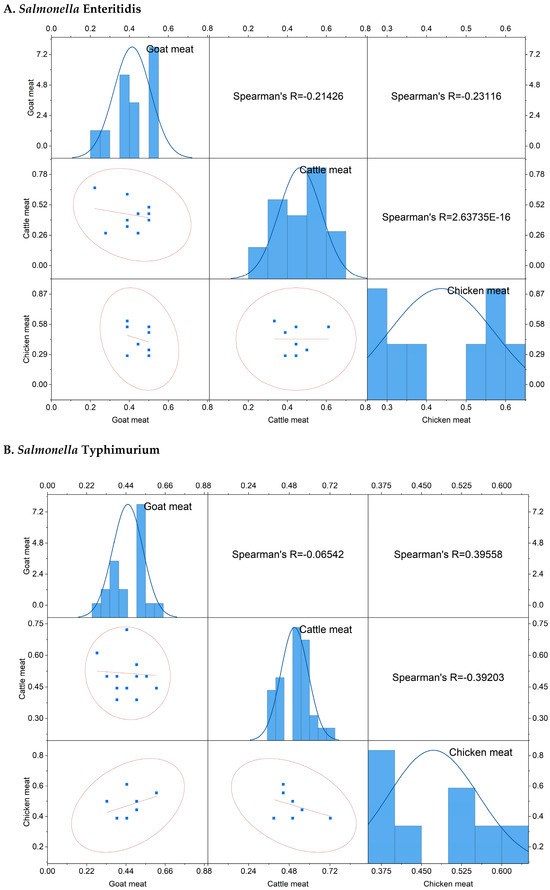
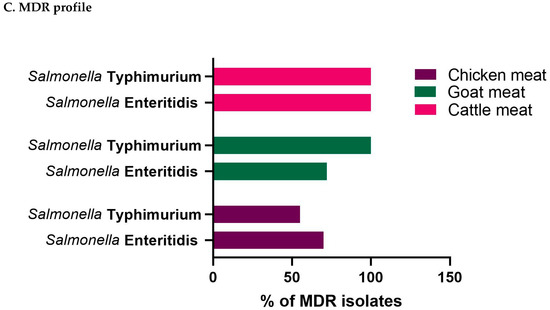
Figure 6.
The scatter plot correlation matrix with a histogram showing the multiple antibiotic resistance index of Salmonella enterica serovars (A) S. Enteritidis, (B) S. Typhimurium, and (C) the multidrug resistance (MDR) profiles of the isolates in different livestock (cattle, goat, and chicken) meat samples investigated in the present study.
The MARI of S. Enteritidis isolated from goat meat samples ranged from 0.22 to 0.50 (mean: 0.41), however MARI ranged from 0.28–0.67 (mean: 0.47) in cattle meat samples, and 0.28–0.61 (mean: 0.44) in chicken meat samples. Regarding S. Typhimurium, the MARI ranged from 0.28 to 0.61 (mean: 0.45) in goat meat samples, 0.39–0.72 (mean: 0.51) in cattle meat samples, and 0.39–0.61 (mean: 0.47) in chicken meat samples. The MARI value of S. Enteritidis isolated from goat meat samples exhibited a weak negative correlation with both cattle and chicken meat samples. Conversely, the MARI value of S. Typhimurium in goat meat showed a negligibly negative correlation with cattle meat but a moderately positive correlation (r = 0.396) with chicken meat samples. Additionally, a moderately negative correlation (r = −0.39) was observed between cattle and chicken meat samples.
The MDR profiles of S. Typhimurium and S. Enteritidis originating from cattle, goat, and chicken meat samples are illustrated in Figure 6C. It was observed that all Salmonella enterica serovars isolated from cattle meat samples displayed MDR. Notably, a significant proportion of S. Enteritidis isolates (72.73%; 8/11; 95% CI: 39.03–93.98) and all S. Typhimurium isolates (100%; 95% CI: 82.35–100.00) from goat meat demonstrated complete resistance to multiple drugs as was previously reported (34). Regarding chicken meat samples, out of 89 isolates including both S. Typhimurium (55.26%; 39.45–71.07) and S. Enteritidis (70.59%; 58.08–83.09), 57 isolates (64.04%) exhibited MDR.
2.5. Molecular Detection of ESBL Genes
In the current study, we screened the presence of ESBL genes in Salmonella enterica serovars isolated from cattle, goat, and chicken meat samples obtained from retail meat (Table 3).

Table 3.
The frequency of ESBL and β-lactam resistance genes in Salmonella enterica Serovars isolated from retail cattle, goat, and chicken meat in the present study.
In the present study, significantly elevated frequencies of ESBLs, particularly associated with the blaTEM genes, were detected using multiplex PCR in both S. Typhimurium (70.96%) and S. Enteritidis (76.19%) isolates (p < 0.001) obtained from cattle meat samples, followed by goat meat and chicken meat samples. The PCR results showed that blaSHV-1 was the predominant ESBL gene identified among Salmonella isolates with 14.28% of S. Enteritidis isolates from cattle meat samples and 21.05% of S. Typhimurium isolates from goat meat samples. Additionally, blaCTX-M-1 exhibited the highest occurrence at 15.69% for S. Enteritidis and 7.89% for S. Typhimurium in chicken meat samples, while not detected in both cattle and goat meat samples in the present study. Moreover, blaCTX-M-9 was only detected at 3.92% for S. Enteritidis in chicken meat samples, with no detection in S. Typhimurium and it was not detected in cattle and goat meat samples. Furthermore, blaOXA had the highest prevalence at 19.04% for S. Enteritidis and 25.80% for S. Typhimurium in cattle meat samples, followed by chicken meat samples, while not detected in goat meat samples.
3. Discussion
The findings presented in the current study shed light on several important aspects of Salmonella contamination, antimicrobial resistance, MDR, and the presence of ESBL genes in the retail meat obtained from livestock in Sylhet District, Bangladesh. The findings have significant implications for public health, food safety, and antimicrobial stewardship efforts.
The prevalence rates of Salmonella spp., particularly S. Enteritidis and S. Typhimurium, in retail meat samples from cattle, goats, and chickens are increasingly alarming. The high prevalence rates indicate a widespread contamination of meat products, posing a significant risk to consumers. These findings highlight the urgent need for improved hygienic practices during meat production, processing, and handling to mitigate the risk of Salmonella contamination.
In the current study, the prevalence of Salmonella spp. in retail cattle meat samples in Sylhet District, Bangladesh was 26.22%, which was closely similar to findings from studies in Egypt (23.3%), Pakistan (21.8%), and Nigeria (23%) [22,23,24]. Notably, the prevalence of Salmonella enterica in the current study was lower than that reported in previous studies in Morocco (30.55%), Tunisia (29.8%), and Vietnam (64.1%) [25,26,34], while being higher than those documented in Turkey (20%), Egypt (8.8%), Iran (4.35%), and Ecuador (11.47%) [35,36,37,38]. These variations in prevalence can be attributed to differences in sampling size, identification methods, and the selection of target genes.
The findings of the present study demonstrated that S. Typhimurium is the dominant serovar among other Salmonella spp. identified in retail cattle meat samples, consistent with findings from previous studies [22,34,36,37]. The prevalence also varied among different upazilas, with the highest prevalence recorded at 50% in Kanaighat and Gowainghat, and the lowest prevalence at 11.76% in Osmaninagar. The prevalence of Salmonella in goat meat was determined to be 18.10%, which was lower than the findings reported in a study from Pakistan [23] and higher than those from studies conducted in India, Modjo, Bishoftu, Arusha (Tanzania), and Gujarat (India) based on goat carcass swabs [27,28,29]. In the present study, S. Typhimurium was identified in cattle meat as the dominant serovar among other Salmonella spp. found in retail goat meat, consistent with findings from previous study [30].
The overall prevalence of Salmonella spp. was found to be 36.84%, a rate comparable to findings from studies conducted in Spain (35.83%) and Russia (38.5%) in chicken meat [39,40]. However, this prevalence rate was higher than the rates reported from Bangladesh, India, Iran, Vietnam, Trinidad, and Malaysia [41,42,43,44,45,46]. Salmonella spp. was detected in various types of retail chicken meat samples, including broiler, spent hen, and sonali, with both S. Typhimurium and S. Enteritidis being identified, consistent with global research findings. Studies conducted in different regions of Bangladesh have also shown the presence of S. Typhimurium and S. Enteritidis in commercial broiler, layer, and breeder farms [41,47]. Similar serotypes of S. Typhimurium and S. Enteritidis have been reported in raw chicken meat sold in Iranian supermarkets [4]. In Turkey, S. Enteritidis (21.9%) and S. Typhimurium (9.4%) were found to be the most prevalent serotypes in poultry [48]. Salmonella enterica serovars have also been identified in backyard chicken flocks in India [49]. Moreover, in Egypt, both S. Enteritidis and S. Typhimurium were detected in chicken meat [50]. In the present study, it was also found that the prevalence rate of S. Typhimurium in sonali chicken meat was notably higher, at 25%, while S. Enteritidis was higher in broiler meat, at 21.34%. These findings differ somewhat from other studies conducted in Bangladesh, where the highest prevalence rates were observed in broiler and layer chickens [51,52]. These variations in the prevalence rates could be attributed to factors such as seasonal changes, environmental conditions, or the lack of adequate farm biosecurity measures. Furthermore, S. Enteritidis emerged as the dominant serovar in retail chicken meat samples, a trend consistent with research findings from Iran, where S. Enteritidis was prevalent in both retail chicken carcasses and meat in Turkey [48,53]. Analysis of the prevalence of salmonellosis in chicken meat across different sampling sites revealed that Jaintapur Upazila had the highest prevalence of 64.71% of Salmonella spp., while Kanaighat displayed the lowest prevalence of 12.50%. These discrepancies regarding salmonellosis in retail cattle, goat, and chicken meat samples in the present study may be explained due to various factors including the method of sample collection, the cleanliness of the farms, or the personal hygienic practices during meat processing and food handlers [36,54]. The varying prevalence rates could also be influenced by the sampling of meat from wet markets, where cattle, goats, and chickens may be eviscerated in contaminated areas with intestinal contents and slaughtered under unhygienic conditions [55]. Additionally, discrepancies may arise from differences in the hygiene practices in the retail shops, the awarenesses of food handlers and workers about contamination risks, the infrastructure and diameters of the shops, and urban versus rural location of the markets [56]. It is important to note that in numerous instances, at the time of the current study, no designated slaughterhouse was pinpointed within the retail meat market. Instead, meat vendors frequently slaughter cattle, goats, and poultry near their shops, increasing the likelihood of meat contamination with environmental pathogenic microorganisms. In Sylhet District, Bangladesh, a notable practice involves selling the guts and intestines of slaughtered animals alongside the meat, posing an additional considerable risk of contamination by enteric microorganisms such as Salmonella.
The antibiotic resistance patterns of Salmonella enterica in the present study are closely aligned with those of previous studies [36,46,57,58]. In contrast to previous findings [59], the findings of the current study showed high sensitivity of Salmonella Typhimurium and Salmonella Enteritidis to gentamicin, ceftriaxone, meropenem, and imipenem, consistent with other reports [46,57,60,61,62].
It was noted that every Salmonella enterica serovar isolated from cattle meat exhibited multidrug resistance (MDR), while a significant majority of S. Enteritidis isolates (72.73%; 8/11) and all S. Typhimurium isolates (100%) from goat meat demonstrated complete resistance to multiple drugs. The lower rates of MDR phenotype between S. Typhimurium (55.26%) and S. Enteritidis (70.59%) isolated from chicken meat, compared to those from cattle and goat meat, could be attributed to several factors. One plausible explanation is the shorter rearing time of chickens, typically five to seven weeks, compared to cattle and goats. This shorter period may result in less exposure to antimicrobial agents, leading to a lower MDR phenotype in chicken-origin isolates. Additionally, differences in the management practices, such as the use of antimicrobial agents in feed or water, may vary between poultry and livestock production systems, contributing to differences in MDR prevalence among bacterial isolates. Moreover, genetic factors within bacterial populations and selective pressure exerted by antimicrobial usage patterns could also influence an MDR status. These findings are closely aligned with those of a study conducted on Egyptian buffalo meat, which reported MDR rate of 79.2% in Salmonella isolates [63]. However, findings of the present study are different from those of another study in Pakistan, where only 11 (10.0%) isolates were identified as MDR [23]. Similarly, a study in South Africa demonstrated that 43% of isolates from livestock and poultry were resistant to multiple drugs [64]. The findings regarding cattle meat isolates are consistent with those of another study where all isolates were MDR [65].
The findings of the present study revealed notably increased frequencies of ESBL genes, particularly associated with the blaTEM genes, which aligns with previous studies where blaTEM were predominantly detected in Salmonella and Escherichia coli isolated from poultry and food products of animal origin [36,66,67]. The current findings revealed a lower prevalence of blaSHV compared to a previous study which reported a rate of 64% blaSHV in Salmonella serovars in poultry meat products [68].
Additionally, blaCTX-M-1 exhibited occurrences, with 15.69% for S. Enteritidis and 7.89% for S. Typhimurium in chicken meat, which are closely similar to those of previous reports [69,70]. Recent studies reported that 2.7% of S. enterica isolates in chicken meat in Bangladesh were positive for blaCTX-M-1 [65], and blaCTX-M-1 detection rate of 3.2% was reported for S. Heidelberg isolated from poultry production chain in Brazil [71], which partially supports the findings in the current study. Moreover, blaCTX-M-9 was detected at 3.92% for S. Enteritidis in chicken meat samples in the present study. The blaOXA exhibited the highest prevalence, at 19.04% for S. Enteritidis and 25.80% for S. Typhimurium in cattle meat samples, followed by chicken meat samples, which align with a previous study [72]. The variation in the prevalence of ESBL genes, such as blaTEM, blaSHV, and blaCTX-M-1, require further investigations in the context of horizontal gene transfer mechanisms, including integrons and gene cassettes which are among the limitations of the present study. However, integrating this aspect into the study findings and future investigations using whole genome sequencing analyses will help provide significant in-depth genomic insights into the potential mechanisms driving the dissemination of antimicrobial resistance determinants among ESBL-producing Salmonella strains isolated from non-human livestock origin meat products in retail meat sources in Bangladesh.
4. Materials and Methods
4.1. Study Design, Site, and Sampling Method
A cross-sectional investigation was undertaken across 13 sub-districts located in the Sylhet (3452 km2), Bangladesh. The selected sub-districts were visualized in Figure 7. Geographically, these upazilas spanned from 24°36′ to 25°11′ North latitude and 91°38′ to 92°30′ East longitude, as shown in Figure 7. Sample procurement followed a convenient sampling approach, based on the accessibility of retail outlets vending cattle, goat, and chicken meat.
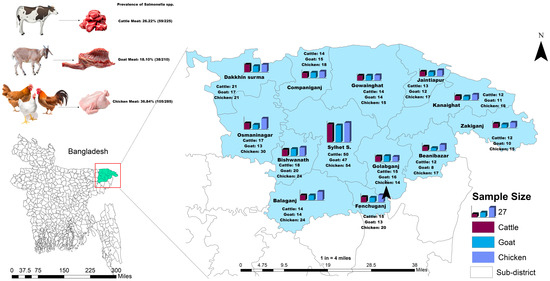
Figure 7.
A geographic map of the study area showing the location of the selected study sites and the sample size of retail cattle, goat, and chicken meat samples in Sylhet District, Bangladesh. The map was created using ArcMap 10.7, ESRI, CA, USA.
4.2. Determination of Sample Size
An appropriate number of samples for prevalence calculation was determined using a mathematical formula as previously reported [73]
where,
Pexp
According to previously published data, the prevalence of Salmonella spp. in meat sold at retail outlet was 51.35% [23]. According to the prior study, Pexp = 0.5135 was utilized to optimize the number of samples. Performing the formula, the minimum number of samples was calculated to be 383.88~384. The current investigation included 720 swabs obtained from retail meat samples from 13 upazilas in Sylhet District.
4.3. Sample Collection and Bacterial Isolation
Seven hundred and twenty samples obtained from meat swabs were collected, comprising 225 samples from cattle meat, 210 samples from goat meat, and 285 samples from chicken meat, respectively. The livestock were sourced from various retail establishments across Sylhet District as shown in Figure 7. Swab samples from the meat were aseptically collected. All retail meat swab samples were pre-enriched by incubating in buffered peptone water medium (Oxoid, UK) at 37 °C for 24 h. Then, the pre-enriched culture medium was inoculated into Modified Semisolid Rappaport Vassiliadis (MSRV) medium enriched with novobiocin selective supplement (SR0181E) and incubated for 24 ± 3 h at 42 °C. Then the culture was streaked onto Salmonella Shigella agar plates (Sigma-Aldrich, Darmstadt, Germany) following appropriate methods. The presumptive positive colonies were subcultured onto xylose-lysine deoxycholate (XLD) agar medium. The suspected colonies of Salmonella were detected by their black-centered red hue characteristics on XLD agar plates (Oxoid, UK) [54,74,75]. The suspected colonies were further inoculated onto nutrient agar, followed by MacConkey agar (Oxoid, UK) plates, in accordance with the procedures outlined in the ISO 6579 manual as previously reported [76]. Biochemical tests, including Sugar Fermentation, Citrate, Methyl-Red-Voges Proskauer, Motility, Indole, and Urease tests, were conducted. Polymerase chain reactions (PCR) were performed to identify Salmonella enterica serovars and to screen for ESBL resistance genes as previously reported [77].
4.4. Identification of S. enterica Serovars and Detection of ESBL-Resistance Genes
DNA extraction was carried out using a DNA extraction kit, following the manufacturer’s instructions (AddBio Incorporated Limited, Daejeon, Republic of Korea). Table 4 lists the primers used for detecting Salmonella enterica serovars and ESBL-resistance genes as previously reported [74,77,78,79].

Table 4.
The primers used in the identification of ESBL and β-lactams resistance in Salmonella enterica serovars isolated from retail cattle, goat, and chicken meat in the present study.
4.5. Antimicrobial Susceptibility Testing (AST)
S. Enteritidis and S. Typhimurium isolates were tested for antimicrobial susceptibility in vitro using the Kirby-Bauer disk diffusion method on Mueller-Hinton Agar plates, and results were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute [80,81]. The AST was performed as previously reported [33] using 16 different antimicrobial disks (Oxoid, UK) belonging to 10 different antimicrobial categories including Penicillins: ampicillin (AMP, 10 μg); Tetracyclines: tetracycline (TE, 30 μg); Non-extended spectrum cephalosporins: cefuroxime (CXM, 30 μg); Extended-spectrum cephalosporins: ceftriaxone (CTR, 30 μg), ceftazidime (CAZ, 30 μg), cefotaxime (CTX, 30 μg); Quinolones: ciprofloxacin (CIP, 5 μg), nalidixic acid (NA, 30 μg); Macrolides: azithromycin (AZM, 15 μg); Folate pathway antagonists: trimethoprim- sulfamethoxazole (COT, 1.25/23.75 μg); Aminoglycosides: gentamicin (GEN, 10 μg), amikacin (AK, 30 μg); Monobactams: aztreonam (AT, 30 μg); Carbapenems: meropenem (MEM, 10 μg), imipenem (IMP, 10 μg); Phenicols: chloramphenicol (CL, 30 μg). The selection of the antibiotic disks in the present study was based on several prescriptions by local veterinarians targeting Salmonella infections in local cattle, goat, and chicken farms.
4.6. Molecular Detection of ESBL Genes
Multiplex polymerase chain reaction was performed as previously reported [82] to detect antimicrobial resistance genes, namely blaTEM, blaSHV, blaOXA, blaCTX-M1, blaCTX-M2, blaCTX-M9, MultiCaseACC, and MultiCaseDHA, in antibiotic-resistant Salmonella enterica serovars Enteritidis and Typhimurium isolates. The PCR utilized specific oligonucleotide primers outlined in Table 4. The PCR amplification conditions included an initial denaturation at 95 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 62 °C for 90 s, and 72 °C for 60 s and also final extension at 72 °C for 10 min. Positive bands were visualized using agarose gel electrophoresis. Negative control (Nuclease free water) was used in all PCR reactions.
4.7. Determination of Multiple Antibiotic Resistance Index (MARI) and MDR
The MAR index was calculated and evaluated using the method described by [76] with the formula: MAR = The count of antimicrobial agents to which the isolate exhibits resistance divided by the number of antimicrobial agents tested. Resistance to one or more antimicrobial agent in three or more antimicrobial categories was defined as MDR (multidrug-resistance) as previously reported [83,84]. The MAR index of 0.20 or higher was deemed indicative of a high-risk source for bacterial contamination or substantial “resistance”.
4.8. Statistical Analysis
A univariate analysis was conducted using a Chi-squared test to assess the associations among various explanatory variables. Fisher’s Exact Test was conducted when more than 20% of the cells had an expected count below 5. p value < 0.05 considered as level of significance. Data analysis and visualization were conducted using SPSS version 26 (Version 26.0., IBM Corp., Armonk, NY, USA), GraphPad Prism 8.4.2 (GraphPad Software, Boston, MA, USA), and R 4.3.2 version.
4.9. Geospatial Mapping and Plotting
The geographical mapping of the study location was conducted using ArcGIS software (ArcMap 10.8, ESRI, Redlands, CA, USA), with shapefile data sourced from (www.diva-gis.org, accessed on 12 March 2024). These maps effectively portrayed choropleth representations, illustrating both the study area and the prevalence of key explanatory variables, alongside corresponding sample sizes. Additionally, a bi-directional bar (Mirror bar) diagram was employed to visually depict the prevalence of Salmonella enterica serovar Enteritidis and Typhimurium among livestock and poultry retail meat samples. To emphasize the antibiogram profile of the isolates, Origin-Pro 2024 (www.originlab.com, accessed on 14 March 2024) was used to generate heat maps with a dendrogram, providing a comprehensive overview of the data. This included heatmaps with hierarchical cluster analysis (HCA), illustrating the sensitivity pattern of antibiotics, with clustering depicted as a dendrogram. Additionally, a scatter plot correlation matrix with histograms was generated to display MARI value of Salmonella enterica serovars, utilizing Origin-Pro 2024 (www.originlab.com, accessed on 15 March 2024). For the portrayal of MDR and the antibiogram profile of the isolates, GraphPad Prism 8.4.2 (GraphPad Software, Boston, MA, USA) was utilized, presenting the data through a Stack bar diagram. The heatmap was created using OriginPro 2024 with “Heatmap with Dendrogram” packages, and the correlation plot was created using the “metan” package on R and RStudio 4.3.2 version.
5. Conclusions
The prevalence of Salmonella spp., particularly S. Enteritidis and S. Typhimurium, underscores the potential health risks associated with contaminated livestock-origin meat consumption. Notably, all Salmonella isolates in the present study from cattle meat samples demonstrated multidrug resistance, while a significant proportion of isolates from goat and chicken meat samples demonstrated complete resistance to multiple drugs. The high prevalence of ESBL genes, including blaTEM, blaSHV, blaCTX-M-1, and blaOXA, highlights the urgent need for enhanced surveillance and antimicrobial stewardship measures to mitigate the dissemination of antibiotic-resistant Salmonella serovars in raw meat, thus safeguarding public health. Moreover, the differential prevalence of ESBL resistance determinants detected in Salmonella serovars isolated from meat samples in the current study emphasizes the importance of implementing specific measures tailored to livestock production systems. Future investigations including genomic surveillance will help delineate the underlying antimicrobial resistance mechanisms in Salmonella serovars and will help provide insights to develop effective strategies to combat the extensive and multi-drug resistance and ESBL production in livestock-origin meat for human consumption. Effective intersectoral and interdisciplinary collaboration utilizing One health approach among healthcare authorities, veterinary professionals, environmental agencies, food producers, and consumers is critically required to better address the pressing problems of public safety and to reduce the continuous threats posed by the emergence and spread of antibiotic-resistant bacteria in the food distribution chain.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics13070586/s1, Table S1: Antibiogram profile of S. enterica Serovar Enteritidis isolated from cattle, goat and chicken meat samples in the present study; Table S2: Antibiogram profile of S. enterica Serovar Typhimurium isolated from cattle, goat and chicken meat samples in the present study.
Author Contributions
Conceptualization, M.M.R., Y.A.H. and M.E.E.Z.; methodology, H.H., M.S.R.C. and M.M.H.; software, H.H., M.M.H., M.M.R. and M.E.E.Z.; validation, M.M.H., M.M.R., Y.A.H., R.B. and M.E.E.Z.; formal analysis, M.M.R., H.H., M.M.R., Y.A.H. and M.E.E.Z.; investigation, M.M.R., H.H. and M.S.R.C.; resources, M.M.R. and M.E.E.Z.; data curation, H.H., M.S.R.C., M.M.R., Y.A.H. and M.E.E.Z.; writing—original draft preparation, M.M.R., H.H., M.S.R.C. and M.E.E.Z.; writing—review and editing, H.H., M.S.R.C., M.M.R., A.N., R.B., A.S., Y.A.H. and M.E.E.Z.; visualization, M.M.R. and M.E.E.Z.; supervision, M.M.R., Y.A.H. and M.E.E.Z.; project administration, M.M.R. and M.E.E.Z.; funding acquisition, M.M.R., R.B., Y.A.H. and M.E.E.Z.; Critical proofreading and review: M.M.R., H.H. and M.E.E.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported in part by funding from the Sylhet Agricultural University Research System (SAURES) under the auspices of the University Grant Commission (UGC) of Bangladesh. The research was supported in part by discretionary funding from Prof. ME El Zowalaty and Dr. Yosra A. Helmy. This research was funded by the Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2024R304), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
The animal experimentation and ethics committee (AEEC) of Sylhet Agricultural University, Bangladesh, approved the study with the following protocols: Protocol #AUP2022031 reference number SAU/Ethical committee/AUP/22/31, Protocol #AUP2022032 reference number SAU/Ethical committee/AUP/22/32, and Protocol #AUP2022034 reference number SAU/Ethical committee/AUP/22/34.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data supporting the findings of this study are included in the manuscript. Additional data are available from the corresponding authors upon reasonable request.
Acknowledgments
The authors would like to thank the farm owners for their cooperation and support during the execution of the study. Authors would like to thank students and staff members from the Faculty of Veterinary, Animal and Biomedical Sciences, Sylhet Agricultural University for their technical support and cooperation during the execution of the project. Authors thank the anonymous reviewers for their insightful comments which significantly improved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Helmy, Y.A.; Taha-Abdelaziz, K.; Hawwas, H.A.E.H.; Ghosh, S.; AlKafaas, S.S.; Moawad, M.M.M.; Saied, E.M.; Kassem, I.I.; Mawad, A.M.M. Antimicrobial Resistance and Recent Alternatives to Antibiotics for the Control of Bacterial Pathogens with an Emphasis on Foodborne Pathogens. Antibiotics 2023, 12, 274. [Google Scholar] [CrossRef]
- Lamichhane, B.; Mawad, A.M.M.; Saleh, M.; Kelley, W.G.; Harrington, P.J.; Lovestad, C.W.; Amezcua, J.; Sarhan, M.M.; El Zowalaty, M.E.; Ramadan, H.; et al. Salmonellosis: An Overview of Epidemiology, Pathogenesis, and Innovative Approaches to Mitigate the Antimicrobial Resistant Infections. Antibiotics 2024, 13, 76. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Vargas, F.M.; Abu-El-Haija, M.A.; Gómez-Duarte, O.G. Salmonella Infections: An Update on Epidemiology, Management, and Prevention. Travel Med. Infect. Dis. 2011, 9, 263–277. [Google Scholar] [CrossRef]
- Carrasco, E.; Morales-Rueda, A.; García-Gimeno, R.M. Cross-Contamination and Recontamination by Salmonella in Foods: A Review. Food Res. Int. 2012, 45, 545–556. [Google Scholar] [CrossRef]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef]
- Sharma, M.; Pathak, S.; Srivastava, P. Prevalence and Antibiogram of Extended Spectrum β-Lactamase (ESBL) Producing Gram Negative Bacilli and Further Molecular Characterization of ESBL Producing Escherichia coli and Klebsiella spp. J. Clin. Diagn. Res. 2013, 7, 2173–2177. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.A.; Domínguez, L.; Teshager, T.; Herrero, I.A.; Porrero, M.C. Antibiotic Resistance Monitoring: The Spanish Programme. Int. J. Antimicrob. Agents 2000, 14, 285–290. [Google Scholar] [CrossRef]
- Okeke, I.N.; Lamikanra, A.; Edelman, R. Socioeconomic and Behavioral Factors Leading to Acquired Bacterial Resistance to Antibiotics in Developing Countries. Emerg. Infect. Dis. 1999, 5, 18–27. [Google Scholar] [CrossRef]
- Hoque, R.; Ahmed, S.M.; Naher, N.; Islam, M.A.; Rousham, E.K.; Islam, B.Z.; Hassan, S. Tackling Antimicrobial Resistance in Bangladesh: A Scoping Review of Policy and Practice in Human, Animal and Environment Sectors. PLoS ONE 2020, 15, e0227947. [Google Scholar] [CrossRef]
- Chowdhury, S.; Ghosh, S.; Aleem, M.A.; Parveen, S.; Islam, M.A.; Rashid, M.M.; Akhtar, Z.; Chowdhury, F. Antibiotic Usage and Resistance in Food Animal Production: What Have We Learned from Bangladesh? Antibiotics 2021, 10, 1032. [Google Scholar] [CrossRef]
- Thung, T.Y.; Lee, E.; Mahyudin, N.A.; Anuradha, K.; Mazlan, N.; Kuan, C.H.; Pui, C.F.; Ghazali, F.M.; Mahmud Ab Rashid, N.K.; Rollon, W.D.; et al. Evaluation of a Lytic Bacteriophage for Bio-Control of Salmonella Typhimurium in Different Food Matrices. LWT 2019, 105, 211–214. [Google Scholar] [CrossRef]
- Lalhriatpuii, M.; Kumar Singh, A. Goat Meat: No Less Source of Protein in Comparison to Other Meat for Human Consumption. In Goat Science—Environment, Health and Economy; IntechOpen: London, UK, 2023. [Google Scholar]
- Sarma, P.K.; Raha, S.K.; Mia, M.I.A.; Jorgensen, H. Value Chain Analysis of Beef Cattle in Selected Areas of Northern Bangladesh. Bangladesh J. Political Econ. 2017, 31, 345–358. [Google Scholar]
- Rahman, M.; Chowdhury, E.H.; Parvin, R. Small-Scale Poultry Production in Bangladesh: Challenges and Impact of COVID-19 on Sustainability. Ger. J. Vet. Res. 2021, 1, 19–27. [Google Scholar] [CrossRef]
- Chandra, M.; Singh, B.R.; Shankar, H.; Agarwal, M.; Agrawal, R.K.; Sharma, G.; Babu, N. Study on Prevalence of Salmonella Infection in Goats. Small Rumin. Res. 2006, 65, 24–30. [Google Scholar] [CrossRef]
- Guarnido-López, P.; Resconi, V.C.; del Campo, M.M.; Guerrero, A.; María, G.A.; Olleta, J.L. Slaughtering of Heifers in a Local or an Industrial Abattoir: Animal Welfare and Meat Quality Consequences. Livest. Sci. 2022, 259, 104904. [Google Scholar] [CrossRef]
- Fegan, N.; Vanderlinde, P.; Higgs, G.; Desmarchelier, P. Quantification and Prevalence of Salmonella in Beef Cattle Presenting at Slaughter. J. Appl. Microbiol. 2004, 97, 892–898. [Google Scholar] [CrossRef]
- Ferdous, J.; Uddin, M.H.; Mahmud, R.; Hennessey, M.; Al Sattar, A.; Das Gupta, S.; Gibson, J.S.; Alders, R.; Henning, J.; Fournié, G.; et al. Mapping of Dressed and Processed Poultry Products in Bangladesh: Identifying the Food Safety Risks for Policy Intervention. Vet. Res. Commun. 2023, 47, 1991–2002. [Google Scholar] [CrossRef]
- Bhandare, S.G.; Sherikar, A.T.; Paturkar, A.M.; Waskar, V.S.; Zende, R.J. A Comparison of Microbial Contamination on Sheep/Goat Carcasses in a Modern Indian Abattoir and Traditional Meat Shops. Food Control 2007, 18, 854–858. [Google Scholar] [CrossRef]
- Soumet, C.; Ermel, G.; Rose, N.; Rose, V.; Drouin, P.; Salvat, G.; Colin, P. Evaluation of a Multiplex PCR Assay for Simultaneous Identification of Salmonella Sp., Salmonella Enteritidis and Salmonella Typhimurium from Environmental Swabs of Poultry Houses. Lett. Appl. Microbiol. 1999, 28, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, M.S.; Bari, M.L.; Hossain, M.A. Prevalence of Salmonella Serovars and Antimicrobial Resistance Profiles in Poultry of Savar Area, Bangladesh. Foodborne Pathog. Dis. 2011, 8, 1111–1118. [Google Scholar] [CrossRef]
- Sallam, K.I.; Mohammed, M.A.; Hassan, M.A.; Tamura, T. Prevalence, Molecular Identification and Antimicrobial Resistance Profile of Salmonella Serovars Isolated from Retail Beef Products in Mansoura, Egypt. Food Control 2014, 38, 209–214. [Google Scholar] [CrossRef]
- Fatima, A.; Saleem, M.; Nawaz, S.; Khalid, L.; Riaz, S.; Sajid, I. Prevalence and Antibiotics Resistance Status of Salmonella in Raw Meat Consumed in Various Areas of Lahore, Pakistan. Sci. Rep. 2023, 13, 22205. [Google Scholar] [CrossRef] [PubMed]
- Uzeh, R.E.; Ihekire, V.C.; Smith, S.I.; Fowora, M.A. Phenotypic and Molecular Detection of Multi-Drug Resistant Salmonella Enteritidis, Salmonella Typhimurium and Salmonella Species in Retail Raw Beef and Chicken. Asian Pac. J. Trop. Dis. 2017, 7, 482–485. [Google Scholar] [CrossRef]
- Ed-dra, A.; Filali, F.R.; Karraouan, B.; El Allaoui, A.; Aboulkacem, A.; Bouchrif, B. Prevalence, Molecular and Antimicrobial Resistance of Salmonella Isolated from Sausages in Meknes, Morocco. Microb. Pathog. 2017, 105, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Abbassi-Ghozzi, I.; Jaouani, A.; Hammami, S.; Martinez-Urtaza, J.; Boudabous, A.; Gtari, M. Molecular Analysis and Antimicrobial Resistance of Salmonella Isolates Recovered from Raw Meat Marketed in the Area of “Grand Tunis”, Tunisia. Pathol. Biol. 2012, 60, e49–e54. [Google Scholar] [CrossRef] [PubMed]
- Woldemariam, E.; Molla, B.; Alemayehu, D.; Muckle, A. Prevalence and Distribution of Salmonella in Apparently Healthy Slaughtered Sheep and Goats in Debre Zeit, Ethiopia. Small Rumin. Res. 2005, 58, 19–24. [Google Scholar] [CrossRef]
- Gaspary, O.M.; Joram, B.; Benardether, T.R.; Catherine, L.; Rehema, M.; Beatus, l.; Murugan, S.; Douglas, R.C. Recovery and Prevalence of Antibiotic-Resistant Salmonella from Fresh Goat Meat in Arusha, Tanzania. Afr. J. Microbiol. Res. 2016, 10, 1315–1321. [Google Scholar] [CrossRef]
- Makwana, P.P.; Nayak, J.B.; Brahmbhatt, M.N.; Chaudhary, J.H. Detection of Salmonella Spp. from Chevon, Mutton and Its Environment in Retail Meat Shops in Anand City (Gujarat), India. Vet. World 2015, 8, 388–392. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Zhang, Z.; Liu, Y.; Zhou, Y.; Wu, C.; Huang, W.; Chen, N.; Zhu, Z. Phenotypic and Molecular Characterizations of Multidrug-Resistant Diarrheagenic E. Coli of Calf Origin. Anim. Dis. 2021, 1, 14. [Google Scholar] [CrossRef]
- Rahman, M.M.; Husna, A.; Elshabrawy, H.A.; Alam, J.; Runa, N.Y.; Badruzzaman, A.T.M.; Banu, N.A.; Al Mamun, M.; Paul, B.; Das, S.; et al. Isolation and Molecular Characterization of Multidrug-Resistant Escherichia Coli from Chicken Meat. Sci. Rep. 2020, 10, 21999. [Google Scholar] [CrossRef]
- Pomwised, R.; Naknaen, A.; Surachat, K.; Issuriya, A.; Prochantasene, S.; Wiriyaprom, R.; Ngasaman, R. Antibiotic-Resistant Escherichia Coli from Goat Farms and the Potential Treatment by Acalypha Indica L. Extract. Small Rumin. Res. 2023, 219, 106889. [Google Scholar] [CrossRef]
- Naser, J.A.; Hossain, H.; Chowdhury, M.S.A.; Liza, N.S.; Lasker, R.M.; Rahman, A.; Haque, M.A.; Hossain, M.A.; Rahman, M.A. Exploring of spectrum beta lactamase producing multidrug-resistant Salmonella enterica serovars in goat meat markets of Bangladesh. Vet. Animal Sci. 2024, 25, 100367. [Google Scholar] [CrossRef]
- Nhung, N.T.; Van, N.T.B.; Van Cuong, N.; Duong, T.T.Q.; Nhat, T.T.; Hang, T.T.T.; Nhi, N.T.H.; Kiet, B.T.; Hien, V.B.; Ngoc, P.T.; et al. Antimicrobial Residues and Resistance against Critically Important Antimicrobials in Non-Typhoidal Salmonella from Meat Sold at Wet Markets and Supermarkets in Vietnam. Int. J. Food Microbiol. 2018, 266, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Siriken, B.; Al, G.; Erol, I. Prevalence and Antibiotic Resistance of Salmonella Enteritidis and Salmonella Typhimurium in Ground Beef and Meatball Samples in Samsun, Turkey. Microb. Drug Resist. 2020, 26, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Moawad, A.A.; Hotzel, H.; Awad, O.; Tomaso, H.; Neubauer, H.; Hafez, H.M.; El-Adawy, H. Occurrence of Salmonella Enterica and Escherichia Coli in Raw Chicken and Beef Meat in Northern Egypt and Dissemination of Their Antibiotic Resistance Markers. Gut Pathog. 2017, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Amini, K.; Salehi, T.Z.; Nikbakht, G.; Ranjbar, R.; Amini, J. Molecular Detection of InvA and Spv Virulence Genes in Salmonella Enteritidis Isolated from Human and Animals in Iran. Afr. J. Microb. Res. 2010, 4, 2202–2210. [Google Scholar]
- Herrera, B. Isolation And Molecular Identification Of Escherichia Coli And Salmonella Spp., From Pork, Beef And Chicken Meat Collected From Different Markets In Guaranda, Ecuador. Int. J. Pharm. Res. 2021, 13, 14–19. [Google Scholar] [CrossRef]
- Domínguez, C.; Gómez, I.; Zumalacárregui, J. Prevalence of Salmonella and Campylobacter in Retail Chicken Meat in Spain. Int. J. Food Microbiol. 2002, 72, 165–168. [Google Scholar] [CrossRef]
- Alali, W.Q.; Gaydashov, R.; Petrova, E.; Panin, A.; Tugarinov, O.; Kulikovskii, A.; Mamleeva, D.; Walls, I.; Doyle, M.P. Prevalence of Salmonella on Retail Chicken Meat in Russian Federation. J. Food Prot. 2012, 75, 1469–1473. [Google Scholar] [CrossRef]
- Siddiky, N.A.; Sarker, M.S.; Khan, M.S.R.; Begum, R.; Kabir, M.E.; Karim, M.R.; Rahman, M.T.; Mahmud, A.; Samad, M.A. Virulence and Antimicrobial Resistance Profiles of Salmonella Enterica Serovars Isolated from Chicken at Wet Markets in Dhaka, Bangladesh. Microorganisms 2021, 9, 952. [Google Scholar] [CrossRef]
- Kaushik, P.; Anjay; Kumari, S.; Bharti, S.K.; Dayal, S. Isolation and Prevalence of Salmonella from Chicken Meat and Cattle Milk Collected from Local Markets of Patna, India. Vet. World 2014, 7, 62–65. [Google Scholar] [CrossRef][Green Version]
- Sodagari, H.R.; Mashak, Z.; Ghadimianazar, A. Prevalence and Antimicrobial Resistance of Salmonella Serotypes Isolated from Retail Chicken Meat and Giblets in Iran. J. Infect. Dev. Ctries 2015, 9, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Van, T.T.H.; Moutafis, G.; Istivan, T.; Tran, L.T.; Coloe, P.J. Detection of Salmonella Spp. in Retail Raw Food Samples from Vietnam and Characterization of Their Antibiotic Resistance. Appl. Environ. Microbiol. 2007, 73, 6885–6890. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.S.; Georges, K.; Rahaman, S.; Abdela, W.; Adesiyun, A.A. Prevalence and Serotypes of Salmonella Spp. on Chickens Sold at Retail Outlets in Trinidad. PLoS ONE 2018, 13, e0202108. [Google Scholar] [CrossRef] [PubMed]
- Thung, T.Y.; Radu, S.; Mahyudin, N.A.; Rukayadi, Y.; Zakaria, Z.; Mazlan, N.; Tan, B.H.; Lee, E.; Yeoh, S.L.; Chin, Y.Z.; et al. Prevalence, Virulence Genes and Antimicrobial Resistance Profiles of Salmonella Serovars from Retail Beef in Selangor, Malaysia. Front. Microbiol. 2018, 8, 2697. [Google Scholar] [CrossRef] [PubMed]
- Barua, H.; Biswas, P.K.; Olsen, K.E.P.; Shil, S.K.; Christensen, J.P. Molecular Characterization of Motile Serovars of Salmonella Enterica from Breeder and Commercial Broiler Poultry Farms in Bangladesh. PLoS ONE 2013, 8, e57811. [Google Scholar] [CrossRef] [PubMed]
- Arkali, A.; Çetinkaya, B. Molecular Identification and Antibiotic Resistance Profiling of Salmonella Species Isolated from Chickens in Eastern Turkey. BMC Vet. Res. 2020, 16, 205. [Google Scholar] [CrossRef] [PubMed]
- Samanta, I.; Joardar, S.N.; Das, P.K.; Das, P.; Sar, T.K.; Dutta, T.K.; Bandyopadhyay, S.; Batabyal, S.; Isore, D.P. Virulence Repertoire, Characterization, and Antibiotic Resistance Pattern Analysis of Escherichia Coli Isolated from Backyard Layers and Their Environment in India. Avian Dis. 2014, 58, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Tarabees, R.; Elsayed, M.S.A.; Shawish, R.; Basiouni, S.; Shehata, A.A. Isolation and Characterization of Salmonella Enteritidis and Salmonella Typhimurium from Chicken Meat in Egypt. J. Infect. Dev. Ctries 2017, 11, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Mamun, M.A.; Islam, K.M.; Rahman, M. Occurrence of Poultry Diseases at Kishoregonj District of Bangladesh. MOJ Proteom. Bioinform. 2019, 8, 7–12. [Google Scholar]
- Naurin, S.; Islam, M.A.; Khatun, M.M. Prevalence of Salmonella in Apparently Healthy Chickens in Mymensingh, Bangladesh. Microbes Health 2013, 1, 30–33. [Google Scholar] [CrossRef]
- Afshari, A.; Baratpour, A.; Khanzade, S.; Jamshidi, A. Salmonella Enteritidis and Salmonella Typhimorium Identification in Poultry Carcasses. Iran. J. Microbiol. 2018, 10, 45–50. [Google Scholar] [PubMed]
- Pullinger, G.D.; Van Diemen, P.M.; Dziva, F.; Stevens, M.P. Role of Two-Component Sensory Systems of Salmonella Enterica Serovar Dublin in the Pathogenesis of Systemic Salmonellosis in Cattle. Microbiology 2010, 156, 3108–3122. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fossler, C.P.; Wells, S.J.; Kaneene, J.B.; Ruegg, P.L.; Warnick, L.D.; Bender, J.B.; Eberly, L.E.; Godden, S.M.; Halbert, L.W. Herd-Level Factors Associated with Isolation of Salmonella in a Multi-State Study of Conventional and Organic Dairy Farms: I. Salmonella Shedding in Cows. Prev. Vet. Med. 2005, 70, 257–277. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kim, K.S.; Kwon, Y.K.; Tak, R. bin Biochemical Characteristics and Antimicrobials Susceptibility of Salmonella Gallinarum Isolated in Korea. J. Vet. Sci. 2003, 4, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elghany, S.M.; Sallam, K.I.; Abd-Elkhalek, A.; Tamura, T. Occurrence, Genetic Characterization and Antimicrobial Resistance of Salmonella Isolated from Chicken Meat and Giblets. Epidemiol. Infect. 2015, 143, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.B.; Mahmud, M.; Akter, R.; Hasan, M.; Sobur, A.; Nazir, N.H.; Noreddin, A.; Rahman, M.; El Zowalaty, M.E.; Rahman, T. Molecular Detection of Multidrug Resistant Salmonella Species Isolated from Broiler Farm in Bangladesh. Pathogens 2020, 9, 201. [Google Scholar] [CrossRef] [PubMed]
- Saha, G.K.; Paul, A.K.; Samad, M.A.; Islam, M.A.; Khan, M.S.R. Prevalence of Salmonella Associated With Goats in Bangladesh. Suranaree J. Sci. Technol. 2014, 21, 193–199. [Google Scholar]
- Bhatta, D.R.; Bangtrakulnonth, A.; Tishyadhigama, P.; Saroj, S.D.; Bandekar, J.R.; Hendriksen, R.S.; Kapadnis, B.P. Serotyping, PCR, Phage-Typing and Antibiotic Sensitivity Testing of Salmonella Serovars Isolated from Urban Drinking Water Supply Systems of Nepal. Lett. Appl. Microbiol. 2007, 44, 588–594. [Google Scholar] [CrossRef]
- Rayamajhi, N.; Jung, B.Y.; Cha, S.B.; Shin, M.K.; Kim, A.; Kang, M.S.; Lee, K.M.; Yoo, H.S. Antibiotic Resistance Patterns and Detection of BlaDHA-1 Salmonella Species Isolates from Chicken Farms in South Korea. Appl. Environ. Microbiol. 2010, 76, 4760–4764. [Google Scholar] [CrossRef][Green Version]
- Perin, A.P.; Martins, B.T.F.; Barreiros, M.A.B.; Yamatogi, R.S.; Nero, L.A.; dos Santos Bersot, L. Occurrence, Quantification, Pulse Types, and Antimicrobial Susceptibility of Salmonella Sp. Isolated from Chicken Meat in the State of Paraná, Brazil. Braz. J. Microbiol. 2020, 51, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elghany, S.M.; Fathy, T.M.; Zakaria, A.I.; Imre, K.; Morar, A.; Herman, V.; Pașcalău, R.; Șmuleac, L.; Morar, D.; Imre, M.; et al. Prevalence of Multidrug-Resistant Salmonella Enterica Serovars in Buffalo Meat in Egypt. Foods 2022, 11, 2924. [Google Scholar] [CrossRef] [PubMed]
- Mthembu, T.P.; Zishiri, O.T.; El Zowalaty, M.E. Molecular Detection of Multidrug-Resistant Salmonella Isolated from Livestock Production Systems in South Africa. Infect. Drug Resist. 2019, 12, 3537–3548. [Google Scholar] [CrossRef] [PubMed]
- Parvin, S.; Hasan, M.; Ali, Y.; Chowdhury, E.H.; Rahman, T.; Islam, T. Prevalence and Multidrug Resistance Pattern of Salmonella Carrying Extended-Spectrum b-Lactamase in Frozen Chicken Meat in Bangladesh. J. Food Prot. 2020, 83, 2107–2121. [Google Scholar] [CrossRef] [PubMed]
- Gottapu, G.C.; Suresh, B. Multidrug Resistance and ESBL Profile of Salmonella Serovars Isolated from Poultry Birds and Foods of Animal Origin. Pharma Innov. J. 2019, 8, 277–282. [Google Scholar]
- Crecencio, R.B.; Brisola, M.C.; Bitner, D.; Frigo, A.; Rampazzo, L.; Borges, K.A.; Furian, T.Q.; Salle, C.T.P.; Moraes, H.L.S.; Faria, G.A.; et al. Antimicrobial Susceptibility, Biofilm Formation and Genetic Profiles of Escherichia Coli Isolated from Retail Chicken Meat. Infect. Genet. Evol. 2020, 84, 104355. [Google Scholar] [CrossRef] [PubMed]
- Forgaciu, A.; Tabaran, A.; Colobatiu, L.; Mihaiu, R.; Dan, S.D.; Mihaiu, M. Concerning Increase in Antimicrobial Resistance Patterns of Pathogenic Strains of Salmonella Isolated in Poultry Meat Products. Antibiotics 2022, 11, 1469. [Google Scholar] [CrossRef] [PubMed]
- Adel, W.A.; Ahmed, A.M.; Hegazy, Y.; Torky, H.A.; Shimamoto, T. High Prevalence of Esbl and Plasmid-Mediated Quinolone Resistance Genes in Salmonella Enterica Isolated from Retail Meats and Slaughterhouses in Egypt. Antibiotics 2021, 10, 881. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Shimamoto, T.; Shimamoto, T. Characterization of Integrons and Resistance Genes in Multidrug-Resistant Salmonella Enterica Isolated from Meat and Dairy Products in Egypt. Int. J. Food Microbiol. 2014, 189, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Souza, A.I.S.; Saraiva, M.M.S.; Casas, M.R.T.; Oliveira, G.M.; Cardozo, M.V.; Benevides, V.P.; Barbosa, F.O.; Freitas Neto, O.C.; Almeida, A.M.; Junior, A.B. High Occurrence of β-Lactamase-Producing Salmonella Heidelberg from Poultry Origin. PLoS ONE 2020, 15, e0230676. [Google Scholar] [CrossRef]
- Orole, O.O.; Lamini, J.N.; Chuku, A. Phylogenetic Characterization of Resistant Salmonella Strains in Typhoid Fever Patients in Nigeria. Bioinform. Biol. Insights 2024, 18, 11779322231220194. [Google Scholar] [CrossRef] [PubMed]
- Mahen, M.S.K.; Chowdhury, M.S.R.; Hossain, H.; Hossain, M.M.; Islam, M.R.; Rahman, M.M. Investigating the infection dynamics and molecular detection of Cryptosporidium in Buffaloes in Sylhet, Bangladesh. Vet. Parasitol. Reg. Stud. Rep. 2024, 52, 101043. [Google Scholar] [CrossRef] [PubMed]
- ISO 6579-1:2017; (Annex D) Microbiology of the Food Chain, Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella Spp. Part 1 Detect. Salmonella Spp. Elsevier: Amsterdam, The Netherlands, 2017; Volume 2017, pp. 2022–2023. Available online: https://www.iso.org/standard/56712.html (accessed on 6 April 2024).
- Hailu, W.; Helmy, Y.A.; Carney-Knisely, G.; Kauffman, M.; Fraga, D.; Rajashekara, G. Prevalence and Antimicrobial Resistance Profiles of Foodborne Pathogens Isolated from Dairy Cattle and Poultry Manure Amended Farms in Northeastern Ohio, the United States. Antibiotics 2021, 10, 1450. [Google Scholar] [CrossRef] [PubMed]
- Mir, R.; Salari, S.; Najimi, M.; Rashki, A. Determination of Frequency, Multiple Antibiotic Resistance Index and Resistotype of Salmonella Spp. in Chicken Meat Collected from Southeast of Iran. Vet. Med. Sci. 2022, 8, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Samad, A.; Abbas, F.; Tanveer, Z.; Ahmad, Z.; Ahmad, I.; Patching, S.G.; Nawaz, N.; Asmat, M.T.; Raziq, A.; Lah, A.; et al. Prevalence of Salmonella Spp. in Chicken Meat from Quetta Retail Outlets and Typing through Multiplex PCR. Rom. Biotechnol. Lett. 2019, 24, 271–279. [Google Scholar] [CrossRef]
- Dallenne, C.; da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important Beta-Lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Villegas, K.J.; Herrera-Sánchez, M.P.; Beltrán-Martínez, M.A.; Cárdenas-Moscoso, S.; Rondón-Barragán, I.S. Molecular Detection of Virulence Factors in Salmonella Serovars Isolated from Poultry and Human Samples. Vet. Med. Int. 2023, 2023, 1875253. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, M.; Poelstra, J.W.; Kauffman, M.; Varghese, B.; Helmy, Y.A.; Scaria, J.; Rajashekara, G. Genomic Diversity, Antimicrobial Resistance, Plasmidome, and Virulence Profiles of Salmonella Isolated from Small Specialty Crop Farms Revealed by Whole-Genome Sequencing. Antibiotics 2023, 12, 1637. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Teklu, D.S.; Negeri, A.A.; Legese, M.H.; Bedada, T.L.; Woldemariam, H.K.; Tullu, K.D. Extended-Spectrum Beta-Lactamase Production and Multi-Drug Resistance among Enterobacteriaceae Isolated in Addis Ababa, Ethiopia. Antimicrob. Resist. Infect. Control 2019, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria:An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Sweeney, M.T.; Lubbers, B.V.; Schwarz, S.; Watts, J.L. Applying definitions for multidrug resistance, extensive drug resistance and pandrug resistance to clinically significant livestock and companion animal bacterial pathogens. J. Antimicrob. Chemother. 2018, 73, 1460–1463. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).