Correction: Furugaito et al. Antimicrobial Susceptibility to 27 Drugs and the Molecular Mechanisms of Macrolide, Tetracycline, and Quinolone Resistance in Gemella sp. Antibiotics 2023, 12, 1538
Text Correction
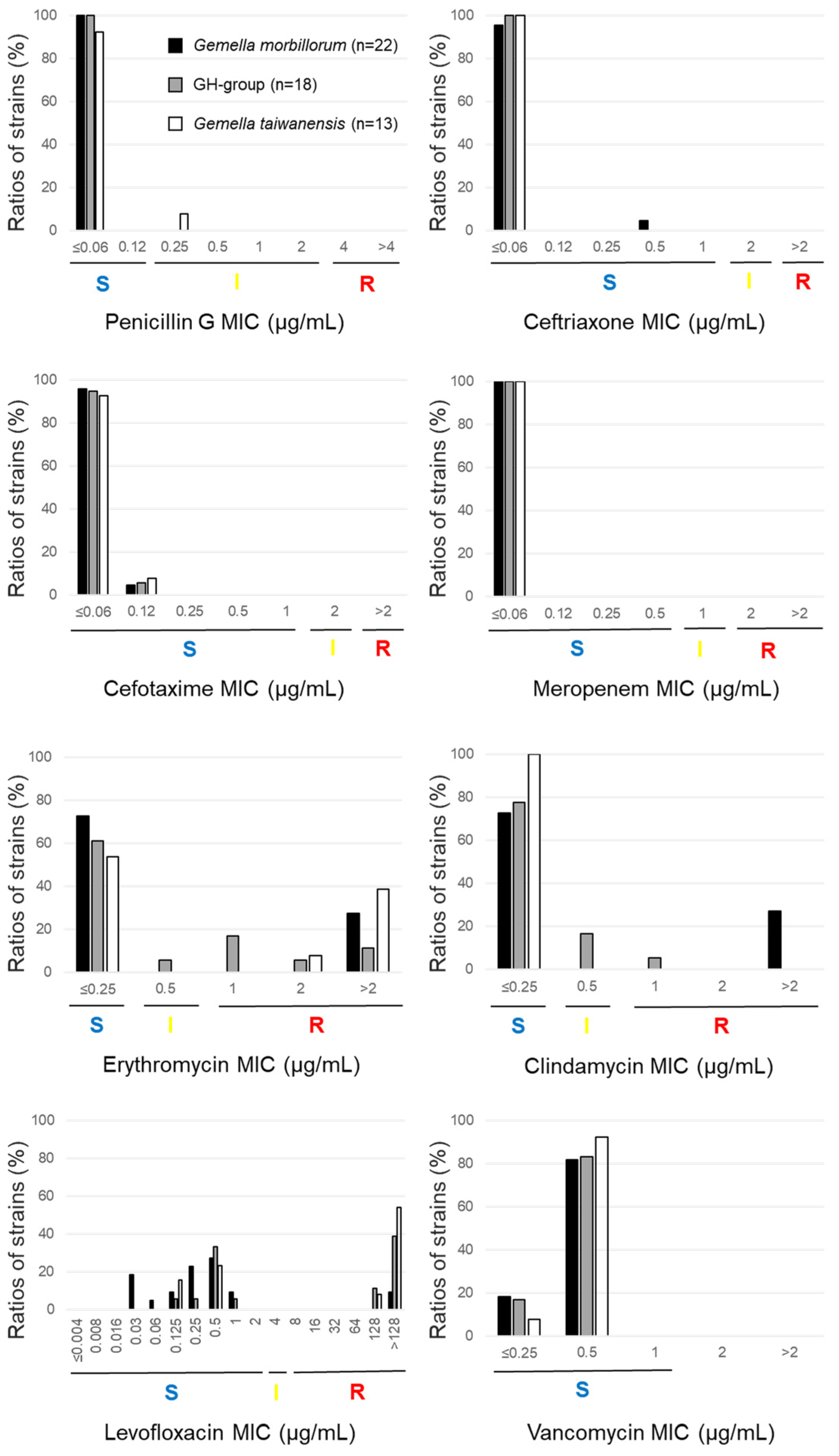
- Abstract: Gemella is a catalase-negative, facultative anaerobic, Gram-positive coccus that is commensal in humans but can become opportunistic and cause severe infectious diseases, such as infective endocarditis. Few studies have tested the antimicrobial susceptibility of Gemella. We tested its antimicrobial susceptibility to 27 drugs and defined the resistant genes using PCR in 58 Gemella strains, including 52 clinical isolates and 6 type strains. The type strains and clinical isolates comprised 22 G. morbillorum, 18 G. haemolysans (GH) group (genetically indistinguishable from G. haemolysans and G. parahaemolysans), 13 G. taiwanensis, three G. sanguinis, and two G. bergeri. No strain was resistant to beta-lactams and vancomycin. In total, 6/22 (27.3%) G. morbillorum strains were erythromycin- and clindamycin-resistant ermB-positive, whereas 5/18 (27.8%) in the GH group, 6/13 (46.2%) G. taiwanensis, and 1/3 (33.3%) of the G. sanguinis strains were erythromycin-non-susceptible mefE- or mefA-positive and clindamycin-susceptible. The MIC90 of minocycline and the ratios of tetM-positive strains varied across the different species—G. morbillorum: 2 µg/mL and 27.3% (6/22); GH group: 8 µg/mL and 22.2% (4/18); G. taiwanensis: 8 µg/mL and 53.8% (7/13), respectively. Levofloxacin resistance was significantly higher in G. taiwanensis (8/13, 61.5%) than in G. morbillorum (2/22, 9.1%). Levofloxacin resistance was associated with a substitution at serine 83 for leucine, phenylalanine, or tyrosine in GyrA. The mechanisms of resistance to erythromycin and clindamycin differed across Gemella species. In addition, the rate of susceptibility to levofloxacin differed across Gemella spp., and the quinolone resistance mechanism was caused by mutations in GyrA alone.
2.2.2. Susceptibility to Erythromycin
2.2.3. Susceptibility to Clindamycin
2.2.4. Susceptibility to Levofloxacin
2.2.6. Susceptibility to Other Antimicrobial Agents
2.3. Phenotypes and Genotypes of Macrolide-Resistant Strains
2.4. Tetracycline Resistance
2.5. Mutations in gyrA and gyrB

| Antimicrobial Agents/ Gemella spp. | MIC (μg/mL) | Interpretive Breakpoint (μg/mL) a or % of Isolates | ||||
|---|---|---|---|---|---|---|
| Range | MIC50 | MIC90 | Susceptible | Intermediate | Resistant | |
| Penicillin G c | ≤0.06–>4 | ≤0.12 | 0.25–2 | ≥4 | ||
| Gemella morbillorum | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| GH group | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella taiwanensis | ≤0.06–0.25 | ≤0.06 | ≤0.06 | 92.3 | 7.7 | 0.0 |
| Gemella sanguinis | ≤0.06 | – | – | 100.0 | 0.0 | 0.0 |
| Gemella bergeri | ≤0.06 | – | – | 100.0 | 0.0 | 0.0 |
| Total | ≤0.06–0.25 | ≤0.06 | ≤0.06 | 98.3 | 1.7 | 0.0 |
| Ampicillin | ≤0.12–>4 | |||||
| Gemella morbillorum | ≤0.12–0.25 | ≤0.12 | ≤0.12 | NA b | NA | NA |
| GH group | ≤0.12 | ≤0.12 | ≤0.12 | NA | NA | NA |
| Gemella taiwanensis | ≤0.12–0.5 | ≤0.12 | ≤0.12 | NA | NA | NA |
| Gemella sanguinis | ≤0.12 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.12 | – | – | NA | NA | NA |
| Total | ≤0.12–0.5 | ≤0.12 | ≤0.12 | NA | NA | NA |
| Amoxicillin–clavulanic acid | ≤0.25/0.12–>4/2 | |||||
| Gemella morbillorum | ≤0.25/0.12 | ≤0.25/0.12 | ≤0.25/0.12 | NA | NA | NA |
| GH group | ≤0.25/0.12 | ≤0.25/0.12 | ≤0.25/0.12 | NA | NA | NA |
| Gemella taiwanensis | ≤0.25/0.12 | ≤0.25/0.12 | ≤0.25/0.12 | NA | NA | NA |
| Gemella sanguinis | ≤0.25/0.12 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.25/0.12 | – | – | NA | NA | NA |
| Total | ≤0.25/0.12 | ≤0.25/0.12 | ≤0.25/0.12 | NA | NA | NA |
| Sulbactam–ampicillin | ≤0.06/0.12–>2/4 | |||||
| Gemella morbillorum | ≤0.06/0.12 | ≤0.06/0.12 | ≤0.06/0.12 | NA | NA | NA |
| GH group | ≤0.06/0.12 | ≤0.06/0.12 | ≤0.06/0.12 | NA | NA | NA |
| Gemella taiwanensis | ≤0.06/0.12–0.25/0.5 | ≤0.06/0.12 | ≤0.06/0.12 | NA | NA | NA |
| Gemella sanguinis | ≤0.06/0.12 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.06/0.12 | – | – | NA | NA | NA |
| Total | ≤0.06/0.12–0.25/0.5 | ≤0.06/0.12 | ≤0.06/0.12 | NA | NA | NA |
| Cefazolin | ≤0.25–>2 | |||||
| Gemella morbillorum | ≤0.25 | ≤0.25 | ≤0.25 | NA | NA | NA |
| GH group | ≤0.25–0.5 | ≤0.25 | 0.5 | NA | NA | NA |
| Gemellataiwanensis | ≤0.25–0.5 | ≤0.25 | 0.5 | NA | NA | NA |
| Gemella sanguinis | ≤0.25 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.25 | – | – | NA | NA | NA |
| Total | ≤0.25–0.5 | ≤0.25 | ≤0.25 | NA | NA | NA |
| Cefdinir | ≤0.25–>1 | |||||
| Gemella morbillorum | ≤0.25 | ≤0.25 | ≤0.25 | NA | NA | NA |
| GH group | ≤0.25 | ≤0.25 | ≤0.25 | NA | NA | NA |
| Gemella taiwanensis | ≤0.25 | ≤0.25 | ≤0.25 | NA | NA | NA |
| Gememlla sanguinis | ≤0.25–0.5 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.25 | – | – | NA | NA | NA |
| Total | ≤0.25–0.5 | ≤0.25 | ≤0.25 | NA | NA | NA |
| Ceftriaxone c | ≤0.06–>2 | ≤1 | 2 | ≥4 | ||
| Gemella morbillorum | ≤0.06–0.5 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| GH group | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella taiwanensis | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella sanguinis | 0.25–1 | – | – | 100.0 | 0.0 | 0.0 |
| Gemella bergeri | ≤0.06 | 100.0 | 0.0 | 0.0 | ||
| Total | ≤0.06–1 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Cefotaxime c | ≤0.06–>2 | ≤1 | 2 | ≥4 | ||
| Gemella morbillorum | ≤0.06–0.12 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| GH group | ≤0.06–0.12 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella taiwanensis | ≤0.06–0.12 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella sanguinis | 0.25–1 | – | – | 100.0 | 0.0 | 0.0 |
| Gemella bergeri | ≤0.06 | – | – | 100.0 | 0.0 | 0.0 |
| Total | ≤0.06–1 | ≤0.06 | 0.12 | 100.0 | 0.0 | 0.0 |
| Cefepime | ≤0.06–>2 | |||||
| Gemella morbillorum | ≤0.06–0.5 | ≤0.06 | ≤0.06 | NA | NA | NA |
| GH group | ≤0.06–0.12 | ≤0.06 | 0.12 | NA | NA | NA |
| Gemella taiwanensis | ≤0.06–0.12 | ≤0.06 | ≤0.06 | NA | NA | NA |
| Gemella sanguinis | 0.25–1 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.06 | – | – | NA | NA | NA |
| Total | ≤0.06–1 | ≤0.06 | 0.12 | NA | NA | NA |
| Imipenem | ≤0.06–>4 | |||||
| Gemella morbillorum | ≤0.06 | ≤0.06 | ≤0.06 | NA | NA | NA |
| GH group | ≤0.06 | ≤0.06 | ≤0.06 | NA | NA | NA |
| Gemella taiwanensis | ≤0.06 | ≤0.06 | ≤0.06 | NA | NA | NA |
| Gemella sanguinis | ≤0.06 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.06 | – | – | NA | NA | NA |
| Total | ≤0.06 | ≤0.06 | ≤0.06 | NA | NA | NA |
| Meropenem c | ≤0.06–>2 | ≤0.5 | 1 | ≥2 | ||
| Gemella morbillorum | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| GH group | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella taiwanensis | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Gemella sanguinis | ≤0.06 | – | – | 100.0 | 0.0 | 0.0 |
| Gemella bergeri | ≤0.06 | – | – | 100.0 | 0.0 | 0.0 |
| Total | ≤0.06 | ≤0.06 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Erythromycin c | ≤0.25–>2 | ≤0.25 | 0.5 | ≥1 | ||
| Gemella morbillorum | ≤0.25–>2 | ≤0.25 | >2 | 72.7 | 0.0 | 27.3 |
| GH group | ≤0.25–>2 | ≤0.25 | >2 | 61.1 | 5.6 | 33.3 |
| Gemella taiwanensis | ≤0.25–>2 | ≤0.25 | >2 | 53.8 | 0.0 | 46.2 |
| Gemella sanguinis | ≤0.25–1 | – | – | 66.7 | 0.0 | 33.3 |
| Gemella bergeri | ≤0.25 | – | – | 100.0 | 0.0 | 0.0 |
| Total | ≤0.25–>2 | ≤0.25 | >2 | 65.5 | 1.7 | 32.8 |
| Clarithromycin | ≤0.12–>16 | |||||
| Gemella morbillorum | ≤0.12–>16 | ≤0.12 | >16 | NA | NA | NA |
| GH group | ≤0.12–16 | ≤0.12 | 8 | NA | NA | NA |
| Gemella taiwanensis | ≤0.12–8 | ≤0.12 | 2 | NA | NA | NA |
| Gemella sanguinis | ≤0.12–0.25 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.12 | – | – | NA | NA | NA |
| Total | ≤0.12–>16 | ≤0.12 | 8 | NA | NA | NA |
| Azithromycin | ≤0.12–>4 | |||||
| Gemella morbillorum | ≤0.12–>4 | ≤0.12 | >4 | NA | NA | NA |
| GH group | ≤0.12–>4 | ≤0.12 | >4 | NA | NA | NA |
| Gemella taiwanensis | ≤0.12–>4 | 0.25 | >4 | NA | NA | NA |
| Gemella sanguinis | 0.25–4 | – | – | NA | NA | NA |
| Gemella bergeri | 0.25 | – | – | NA | NA | NA |
| Total | ≤0.12–>4 | ≤0.12 | >4 | NA | NA | NA |
| Clindamycin c | ≤0.25–>2 | ≤0.25 | 0.5 | ≥1 | ||
| Gemella morbillorum | ≤0.25–>2 | ≤0.25 | >2 | 72.7 | 0.0 | 27.3 |
| GH group | ≤0.25–1 | ≤0.25 | 0.5 | 77.8 | 16.7 | 5.6 |
| Gemella taiwanensis | ≤0.25 | ≤0.25 | ≤0.25 | 100.0 | 0.0 | 0.0 |
| Gemella sanguinis | ≤0.25 | – | – | 100.0 | 0.0 | 0.0 |
| Gemella bergeri | ≤0.25 | – | – | 100.0 | 0.0 | 0.0 |
| Total | ≤0.25–>2 | ≤0.25 | >2 | 82.8 | 5.2 | 12.1 |
| Erythromycin/clindamycin | ≤1/0.5–>1/0.5 | |||||
| G. morbillorum | ≤1/0.5–>1/0.5 | ≤1/0.5 | >1/0.5 | NA | NA | NA |
| GH group | ≤1/0.5–>1/0.5 | ≤1/0.5 | ≤1/0.5 | NA | NA | NA |
| G. taiwanensis | ≤1/0.5 | ≤1/0.5 | ≤1/0.5 | NA | NA | NA |
| G. sanguinis | ≤1/0.5 | – | – | NA | NA | NA |
| Gemella bergeri | ≤1/0.5 | – | – | NA | NA | NA |
| Total | ≤1/0.5–>1/0.5 | ≤1/0.5 | >1/0.5 | NA | NA | NA |
| Levofloxacin c | ≤0.004–>128 | ≤2 | 4 | ≥8 | ||
| Gemella morbillorum | 0.03–>128 | 0.25 | 1 | 90.9 | 0.0 | 9.1 |
| GH group | 0.125–>128 | 1 | >128 | 50.0 | 0.0 | 50.0 |
| Gemella taiwanensis | 0.125–>128 | >128 | >128 | 38.5 | 0.0 | 61.5 |
| Gemella sanguinis | 0.5–>128 | – | – | 33.3 | 0.0 | 66.7 |
| Gemella bergeri | 0.5 | – | – | 100.0 | 0.0 | 0.0 |
| Total | 0.03–>128 | 0.5 | >128 | 63.8 | 0.0 | 36.2 |
| Moxifloxacin | ≤0.5–>2 | |||||
| Gemella morbillorum | ≤0.5–>2 | ≤0.5 | >2 | NA | NA | NA |
| GH group | ≤0.5–>2 | ≤0.5 | >2 | NA | NA | NA |
| Gemella taiwanensis | ≤0.5–>2 | >2 | >2 | NA | NA | NA |
| Gemella sanguinis | ≤0.5–>2 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.5 | – | – | NA | NA | NA |
| Total | ≤0.5–>2 | ≤0.5 | >2 | NA | NA | NA |
| Minocycline | ≤1–>8 | |||||
| Gemella morbillorum | ≤1–>8 | ≤1 | 2 | NA | NA | NA |
| GH group | ≤1–8 | ≤1 | 8 | NA | NA | NA |
| Gemella taiwanensis | ≤1–8 | ≤1 | 8 | NA | NA | NA |
| Gemella sanguinis | ≤1 | – | – | NA | NA | NA |
| Gemella bergeri | ≤1 | – | – | NA | NA | NA |
| Total | ≤1 | ≤1 | 8 | NA | NA | NA |
| Sulfamethoxazole– trimethoprim | ≤9.5/0.5–>38/2 | |||||
| Gemella morbillorum | ≤9.5/0.5–>38/2 | 19/1 | >38/2 | NA | NA | NA |
| GH group | ≤9.5/0.5–>38/2 | 38/2 | >38/2 | NA | NA | NA |
| Gemella taiwanensis | ≤9.5/0.5–>38/2 | 19/1 | 19/1 | NA | NA | NA |
| Gemella sanguinis | 19/1–>38/2 | – | – | NA | NA | NA |
| Gemella bergeri | ≤9.5/0.5 | – | – | NA | NA | NA |
| Total | ≤9.5/0.5–>38/2 | 19/1 | >38/2 | NA | NA | NA |
| Gentamicin | ≤1–>8 | |||||
| Gemella morbillorum | ≤1–8 | 2 | 8 | NA | NA | NA |
| GH group | ≤1–2 | ≤1 | 2 | NA | NA | NA |
| Gemella taiwanensis | ≤1–4 | 2 | 4 | NA | NA | NA |
| Gemella sanguinis | ≤1–8 | – | – | NA | NA | NA |
| Gemella bergeri | 2, 4 | – | – | NA | NA | NA |
| Total | ≤1–8 | 2 | 8 | NA | NA | NA |
| Gentamicin 500 | ≤500–>500 | |||||
| Gemella morbillorum | ≤500 | ≤500 | ≤500 | NA | NA | NA |
| GH group | ≤500 | ≤500 | ≤500 | NA | NA | NA |
| Gemella taiwanensis | ≤500 | ≤500 | ≤500 | NA | NA | NA |
| Gemella sanguinis | ≤500 | – | – | NA | NA | NA |
| Gemella bergeri | ≤500 | – | – | NA | NA | NA |
| Total | ≤500 | ≤500 | ≤500 | NA | NA | NA |
| Arbekacin | ≤1–>8 | |||||
| Gemella morbillorum | ≤1–8 | 8 | >8 | NA | NA | NA |
| GH group | ≤1–8 | 4 | 8 | NA | NA | NA |
| Gemella taiwanensis | 2–>8 | 4 | 8 | NA | NA | NA |
| Gemella sanguinis | 4–>8 | – | – | NA | NA | NA |
| Gemella bergeri | 4, >8 | – | – | NA | NA | NA |
| Total | ≤1–8 | 4 | >8 | NA | NA | NA |
| Fosfomycin | ≤16–>128 | |||||
| Gemella morbillorum | ≤16–32 | ≤16 | ≤16 | NA | NA | NA |
| GH group | ≤16 | ≤16 | ≤16 | NA | NA | NA |
| Gemella taiwanensis | ≤16 | ≤16 | ≤16 | NA | NA | NA |
| Gemalla sanguinis | ≤16 | – | – | NA | NA | NA |
| Gemella bergeri | ≤16 | – | – | NA | NA | NA |
| Total | ≤16–32 | ≤16 | ≤16 | NA | NA | NA |
| Rifampicin | ≤0.5–>2 | |||||
| Gemella morbillorum | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| GH group | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Gemella taiwanensis | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Gemella sanguinis | ≤0.5 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.5 | NA | NA | NA | ||
| Total | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Vancomycin c | ≤0.25–>2 | ≤1 | ||||
| Gemella morbillorum | ≤0.25–0.5 | 0.5 | 0.5 | 100.0 | 0.0 | 0.0 |
| GH group | ≤0.25–0.5 | 0.5 | 0.5 | 100.0 | 0.0 | 0.0 |
| Gemella taiwanensis | ≤0.25–0.5 | 0.5 | 0.5 | 100.0 | 0.0 | 0.0 |
| Gemella sanguinis | ≤0.25–0.5 | – | – | 100.0 | 0.0 | 0.0 |
| Gemella bergeri | 0.5 | – | – | 100.0 | 0.0 | 0.0 |
| Total | ≤0.25–0.5 | 0.5 | 0.5 | 100.0 | 0.0 | 0.0 |
| Teicoplanin | ≤0.5–>16 | |||||
| Gemella morbillorum | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| GH group | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Gemella taiwanensis | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Gemella sanguinis | ≤0.5 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.5 | – | – | NA | NA | NA |
| Total | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Linezolid | ≤0.5–>4 | |||||
| Gemella morbillorum | ≤0.5–1 | ≤0.5 | 1 | NA | NA | NA |
| GH group | ≤0.5–1 | ≤0.5 | 1 | NA | NA | NA |
| Gemella taiwanensis | ≤0.5 | ≤0.5 | ≤0.5 | NA | NA | NA |
| Gemella sanguinis | ≤0.5–1 | – | – | NA | NA | NA |
| Gemella bergeri | ≤0.5, 2 | – | – | NA | NA | NA |
| Total | ≤0.5 | ≤0.5 | 1 | NA | NA | NA |
| Daptomycin | ≤0.25–>4 | |||||
| Gemella morbillorum | ≤0.25–4 | 2 | 2 | NA | NA | NA |
| GH group | 0.5–2 | 1 | 2 | NA | NA | NA |
| Gemella taiwanensis | ≤0.25–2 | 1 | 2 | NA | NA | NA |
| Gemella sanguinis | 1–4 | – | – | NA | NA | NA |
| Gemella bergeri | 2, 4 | – | – | NA | NA | NA |
| Total | ≤0.25–4 | 1 | 2 | NA | NA | NA |
| Strain No. | Identification | MIC (μg/mL) | Macrolide Phenotype a,b | mefA/E | erm | msrA | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Erythromycin | Clindamycin | Erythromycin/Clindamycin | Clarithromycin | Azithromycin | ||||||
| TWCC 57201 | Gemella morbillorum | >2 | >2 | >1/0.5 | 8 | >4 | cMLSB | - | ermB | - |
| TWCC 57818 | Gemella morbillorum | >2 | >2 | >1/0.5 | >16 | >4 | cMLSB | - | ermB | - |
| TWCC 57944 | Gemella morbillorum | >2 | >2 | >1/0.5 | >16 | >4 | cMLSB | - | ermB | - |
| TWCC 59111 | Gemella morbillorum | >2 | >2 | >1/0.5 | 8 | >4 | cMLSB | - | ermB | - |
| TWCC 71703 | Gemella morbillorum | >2 | >2 | >1/0.5 | >16 | >4 | cMLSB | - | ermB | - |
| TWCC 72266 | Gemella morbillorum | >2 | >2 | >1/0.5 | >16 | >4 | cMLSB | - | ermB | - |
| TWCC 52027 | GH group | 0.5 | ≤0.25 | ≤1/0.5 | 8 | 2 | M | mefE | - | - |
| TWCC 59566 | GH group | 2 | ≤0.25 | ≤1/0.5 | 2 | >4 | M | mefE | - | - |
| TWCC 59567 | GH group | >2 | 1 | >1/0.5 | 16 | >4 | M | mefE | - | - |
| TWCC 59795 | GH group | 1 | 0.5 | ≤1/0.5 | 0.5 | 2 | M | mefE | - | - |
| TWCC 70939 | GH group | >2 | ≤0.25 | ≤1/0.5 | 2 | >4 | M | mefE | - | - |
| TWCC 71200 | GH group | 1 | ≤0.25 | ≤1/0.5 | 2 | 2 | M | mefA | - | - |
| TWCC 71814 | GH group | 1 | ≤0.25 | ≤1/0.5 | 0.5 | 1 | M | mefE | - | - |
| TWCC 55344 | Gemella taiwanensis | >2 | ≤0.25 | ≤1/0.5 | 8 | >4 | M | mefE | - | - |
| TWCC 58522 | Gemella taiwanensis | >2 | ≤0.25 | ≤1/0.5 | 2 | 4 | M | mefE | - | - |
| TWCC 70386 | Gemella taiwanensis | >2 | ≤0.25 | ≤1/0.5 | 2 | 4 | M | mefE | - | - |
| TWCC 72085 | Gemella taiwanensis | >2 | ≤0.25 | ≤1/0.5 | 2 | >4 | M | mefE | - | - |
| TWCC 70387L | Gemella taiwanensis | 2 | ≤0.25 | ≤1/0.5 | 0.5 | >4 | M | mefE | - | - |
| TWCC 70387S | Gemella taiwanensis | >2 | ≤0.25 | ≤1/0.5 | 2 | >4 | M | mefE | - | - |
| TWCC 54965 | Gemella sanguinis | 1 | ≤0.25 | ≤1/0.5 | 0.25 | 4 | M | mefE | - | - |
| TWCC 70419 | Gemella sanguinis | ≤0.25 c | ≤0.25 | ≤1/0.5 | ≤0.12 | 0.25 | not M | mefE | - | - |
| Strain No. | Identification | tetM | Minocycline MIC (μg/mL) | ermB |
|---|---|---|---|---|
| TWCC 57944 | Gemella morbillorum | + | 2 | + |
| TWCC 57987 | Gemella morbillorum | + | ≤1 | − |
| TWCC 59111 | Gemella morbillorum | + | 2 | + |
| TWCC 70937 | Gemella morbillorum | + | >8 | − |
| TWCC 71703 | Gemella morbillorum | + | 2 | + |
| TWCC 72266 | Gemella morbillorum | + | ≤1 | + |
| TWCC 51800 | GH group | + | 8 | − |
| TWCC 59795 | GH group | + | ≤1 | − |
| TWCC 70939 | GH group | + | 8 | − |
| TWCC 71814 | GH group | + | 2 | − |
| TWCC 53044 | Gemella taiwanensis | + | 8 | − |
| TWCC 56546 | Gemella taiwanensis | + | 2 | − |
| TWCC 58522 | Gemella taiwanensis | + | 8 | − |
| TWCC 70386 | Gemella taiwanensis | + | 8 | − |
| TWCC 72085 | Gemella taiwanensis | + | 8 | − |
| TWCC 70387L | Gemella taiwanensis | + | ≤1 | − |
| TWCC 70387S | Gemella taiwanensis | + | ≤1 | − |
| Strain | n | MIC (μg/mL) | GyrA Amino Acid Substitutions a | |
|---|---|---|---|---|
| Levofloxacin | Moxifloxacin | |||
| Gemella morbillorum | 2 | >128 | >2 | Ser83 > Leu83 (n = 2) |
| GH group | 9 | 128–>128 | >2 | Ser83 > Phe83 (n = 7), Ser83 > Tyr83 (n =2) |
| Gemella taiwanensis | 8 | 128–>128 | >2 | Ser83 > Phe83 (n = 7), Ser83 > Tyr83 (n = 1) |
| Gemella sanguinis | 2 | 128–>128 | >2 | Ser83 > Phe83 (n = 2) |
Reference
- Furugaito, M.; Arai, Y.; Uzawa, Y.; Kamisako, T.; Ogura, K.; Okamoto, S.; Kikuchi, K. Antimicrobial Susceptibility to 27 Drugs and the Molecular Mechanisms of Macrolide, Tetracycline, and Quinolone Resistance in Gemella sp. Antibiotics 2023, 12, 1538. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furugaito, M.; Arai, Y.; Uzawa, Y.; Kamisako, T.; Ogura, K.; Okamoto, S.; Kikuchi, K. Correction: Furugaito et al. Antimicrobial Susceptibility to 27 Drugs and the Molecular Mechanisms of Macrolide, Tetracycline, and Quinolone Resistance in Gemella sp. Antibiotics 2023, 12, 1538. Antibiotics 2024, 13, 515. https://doi.org/10.3390/antibiotics13060515
Furugaito M, Arai Y, Uzawa Y, Kamisako T, Ogura K, Okamoto S, Kikuchi K. Correction: Furugaito et al. Antimicrobial Susceptibility to 27 Drugs and the Molecular Mechanisms of Macrolide, Tetracycline, and Quinolone Resistance in Gemella sp. Antibiotics 2023, 12, 1538. Antibiotics. 2024; 13(6):515. https://doi.org/10.3390/antibiotics13060515
Chicago/Turabian StyleFurugaito, Michiko, Yuko Arai, Yutaka Uzawa, Toshinori Kamisako, Kohei Ogura, Shigefumi Okamoto, and Ken Kikuchi. 2024. "Correction: Furugaito et al. Antimicrobial Susceptibility to 27 Drugs and the Molecular Mechanisms of Macrolide, Tetracycline, and Quinolone Resistance in Gemella sp. Antibiotics 2023, 12, 1538" Antibiotics 13, no. 6: 515. https://doi.org/10.3390/antibiotics13060515
APA StyleFurugaito, M., Arai, Y., Uzawa, Y., Kamisako, T., Ogura, K., Okamoto, S., & Kikuchi, K. (2024). Correction: Furugaito et al. Antimicrobial Susceptibility to 27 Drugs and the Molecular Mechanisms of Macrolide, Tetracycline, and Quinolone Resistance in Gemella sp. Antibiotics 2023, 12, 1538. Antibiotics, 13(6), 515. https://doi.org/10.3390/antibiotics13060515





