Hospital Wastes as Potential Sources for Multi-Drug-Resistant ESBL-Producing Bacteria at a Tertiary Hospital in Ethiopia
Abstract
1. Introduction
2. Results
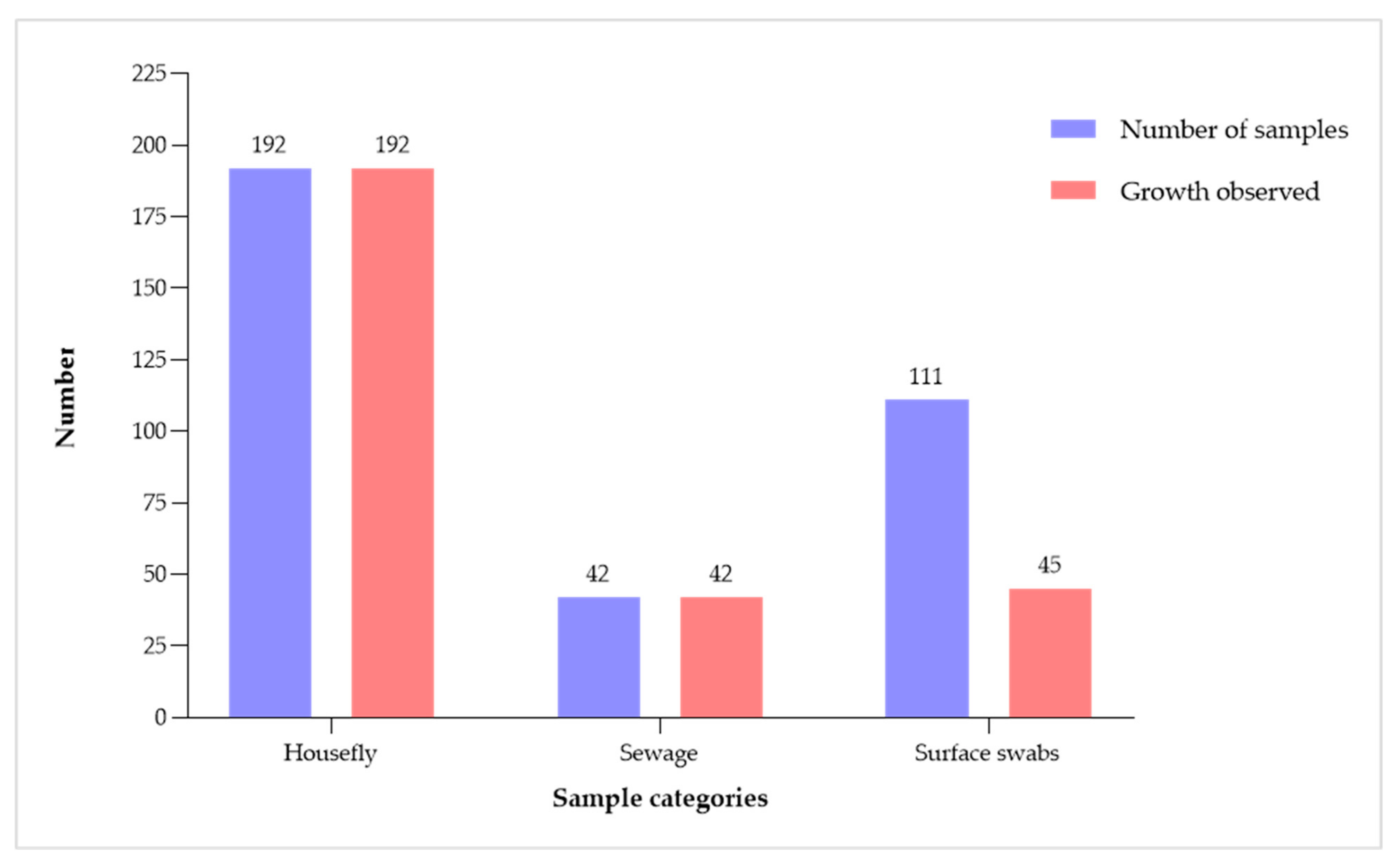
2.1. Proportion of Bacterial Growth
2.2. Profile of Isolated Gram-Negative Bacteria
2.3. Antibiotic Resistance Patterns
2.4. Molecular Epidemiology of ESBLs and Carbapenemase Expression in E. coli Strains
3. Discussion
4. Materials and Methods
4.1. Description of the Hygiene Practice in Study Area
4.2. Study Design, Area, and Period
4.3. Sample Collection
4.4. Bacteria Isolation
4.5. Bacterial Identification
4.6. Antibiotics Susceptibility Test
4.7. Extended Spectrum β-Lactamase Detection
4.8. DNA Extraction
4.9. Molecular Characterization of E. coli Strains
4.10. Data Quality Assurance
4.11. Data Analysis
4.12. Ethical Consideration
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sartorius, B.; Gray, A.P.; Weaver, N.D.; Aguilar, G.R.; Swetschinski, L.R.; Ikuta, K.S.; Mestrovic, T.; Chung, E.; Wool, E.E.; Han, C. The burden of bacterial antimicrobial resistance in the WHO African region in 2019: A cross-country systematic analysis. Lancet Glob. Health 2024, 12, e201–e216. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, K.E.; Joski, P.; Johnston, K.J. Antibiotic-resistant infection treatment costs have doubled since 2002, now exceeding $2 billion annually. Health Aff. 2018, 37, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Irfan, M.; Almotiri, A.; AlZeyadi, Z.A. Antimicrobial Resistance and β-Lactamase Production in Clinically Significant Gram-Negative Bacteria Isolated from Hospital and Municipal Wastewater. Antibiotics 2023, 12, 653. [Google Scholar] [CrossRef] [PubMed]
- Onwugamba, F.C.; Mellmann, A.; Nwaugo, V.O.; Süselbeck, B.; Schaumburg, F. Antimicrobial resistant and enteropathogenic bacteria in ‘filth flies’: A cross-sectional study from Nigeria. Sci. Rep. 2020, 10, 16990. [Google Scholar] [CrossRef] [PubMed]
- Joachim, A.; Manyahi, J.; Issa, H.; Lwoga, J.; Msafiri, F.; Majigo, M. Predominance of Multidrug-Resistant Gram-Negative Bacteria on Contaminated Surfaces at a Tertiary Hospital in Tanzania: A Call to Strengthening Environmental Infection Prevention and Control Measures. Curr. Microbiol. 2023, 80, 148. [Google Scholar] [CrossRef] [PubMed]
- Tufa, T.B.; Fuchs, A.; Wienemann, T.; Eggers, Y.; Abdissa, S.; Schneider, M.; Jensen, B.-E.O.; Bode, J.G.; Pfeffer, K.; Häussinger, D. Carriage of ESBL-producing Gram-negative bacteria by flies captured in a hospital and its suburban surroundings in Ethiopia. Antimicrob. Resist. Infect. Control 2020, 9, 175. [Google Scholar] [CrossRef] [PubMed]
- Davidova-Gerzova, L.; Lausova, J.; Sukkar, I.; Nesporova, K.; Nechutna, L.; Vlkova, K.; Chudejova, K.; Krutova, M.; Palkovicova, J.; Kaspar, J. Hospital and community wastewater as a source of multidrug-resistant ESBL-producing Escherichia coli. Front. Cell. Infect. Microbiol. 2023, 13, 1184081. [Google Scholar] [CrossRef] [PubMed]
- Nakamura-Silva, R.; Dias, L.L.; Sousa, R.C.; Fujimoto, R.Y.; Pitondo-Silva, A. Multidrug-resistant and potentially pathogenic Enterobacteriaceae found in a tertiary hospital sewage in southeastern Brazil. Environ. Monit. Assess. 2022, 194, 782. [Google Scholar] [CrossRef]
- Ma, J.; Sun, H.; Li, B.; Wu, B.; Zhang, X.; Ye, L. Horizontal transfer potential of antibiotic resistance genes in wastewater treatment plants unraveled by microfluidic-based mini-metagenomics. J. Hazard. Mater. 2024, 465, 133493. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Wang, J.; Liu, Y.; Wang, G.; Yang, Y.; Liu, Y.; Kong, Y.; Lin, J.; Li, Q.; Li, G. Dynamics of antibiotic resistance genes and bacterial community during pig manure, kitchen waste, and sewage sludge composting. J. Environ. Manag. 2023, 345, 118651. [Google Scholar] [CrossRef] [PubMed]
- Nayduch, D.; Neupane, S.; Pickens, V.; Purvis, T.; Olds, C. House Flies Are Underappreciated Yet Important Reservoirs and Vectors of Microbial Threats to Animal and Human Health. Microorganisms 2023, 11, 583. [Google Scholar] [CrossRef]
- Offei Addo, S.; Oppong, J.; Monikey Achawe, E.; Baah Nketia, B.; Boateng Agyei, P.; Asiedu Larbi, J. Filth Flies As Carriers of Intestinal Parasites And Fungi in a Tertiary Institution in Ghana. J. Med. Microbiol. Infect. Dis. 2022, 10, 179–185. [Google Scholar] [CrossRef]
- Zeynudin, A.; Pritsch, M.; Schubert, S.; Messerer, M.; Liegl, G.; Hoelscher, M.; Belachew, T.; Wieser, A. Prevalence and antibiotic susceptibility pattern of CTX-M type extended-spectrum β-lactamases among clinical isolates of gram-negative bacilli in Jimma, Ethiopia. BMC Infect. Dis. 2018, 18, 524. [Google Scholar] [CrossRef] [PubMed]
- Sewunet, T.; Asrat, D.; Woldeamanuel, Y.; Ny, S.; Westerlund, F.; Aseffa, A.; Giske, C.G. Polyclonal spread of blaCTX-M-15 through high-risk clones of Escherichia coli at a tertiary hospital in Ethiopia. J. Glob. Antimicrob. Resist. 2022, 29, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Sewunet, T.; Asrat, D.; Woldeamanuel, Y.; Ny, S.; Westerlund, F.; Aseffa, A.; Giske, C.G. High prevalence of bla CTX-M-15 and nosocomial transmission of hypervirulent epidemic clones of Klebsiella pneumoniae at a tertiary hospital in Ethiopia. JAC-Antimicrob. Resist. 2021, 3, dlab001. [Google Scholar] [CrossRef] [PubMed]
- Sewunet, T.; Asrat, D.; Woldeamanuel, Y.; Aseffa, A.; Giske, C.G. Molecular epidemiology and antimicrobial susceptibility of Pseudomonas spp. and Acinetobacter spp. from clinical samples at Jimma medical center, Ethiopia. Front. Microbiol. 2022, 13, 951857. [Google Scholar] [CrossRef] [PubMed]
- Gashaw, M.; Gudina, E.K.; Ali, S.; Gabriele, L.; Seeholzer, T.; Alemu, B.; Kroidl, A. Molecular characterization of carbapenem-resistance in Gram-negative isolates obtained from clinical samples at Jimma Medical Center, Ethiopia. Front. Microbiol. 2024, 15, 1336387. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Birhane, M.; Bekele, S.; Kibru, G.; Teshager, L.; Yilma, Y.; Ahmed, Y.; Fentahun, N.; Assefa, H.; Gashaw, M. Healthcare associated infection and its risk factors among patients admitted to a tertiary hospital in Ethiopia: Longitudinal study. Antimicrob. Resist. Infect. Control 2018, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Alemu, A.Y.; Endalamaw, A.; Bayih, W.A. The burden of healthcare-associated infection in Ethiopia: A systematic review and meta-analysis. Trop. Med. Health 2020, 48, 77. [Google Scholar] [CrossRef] [PubMed]
- Fufa, B.D.; Negao, E.B. Satisfaction of outpatient service consumers and associated factors towards the health service given at Jimma Medical Center, South West Ethiopia. Patient Relat. Outcome Meas. 2019, 10, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Gashaw, M.; Berhane, M.; Bekele, S.; Kibru, G.; Teshager, L.; Yilma, Y.; Ahmed, Y.; Fentahun, N.; Assefa, H.; Wieser, A. Emergence of high drug resistant bacterial isolates from patients with health care associated infections at Jimma University medical center: A cross sectional study. Antimicrob. Resist. Infect. Control 2018, 7, 138. [Google Scholar] [CrossRef] [PubMed]
- Badger-Emeka, L.; Al Rashed, A.S.; Aljindan, R.Y.; Emeka, P.M.; Quadri, S.A.; Almutairi, H.H. Incidence of drug-resistant hospital-associated Gram-negative bacterial infections, the accompanying risk factors, and clinical outcomes with treatment. Antibiotics 2023, 12, 1425. [Google Scholar] [CrossRef] [PubMed]
- Wiktorczyk-Kapischke, N.; Skowron, K.; Kwiecińska-Piróg, J.; Białucha, A.; Wałecka-Zacharska, E.; Grudlewska-Buda, K.; Kraszewska, Z.; Gospodarek-Komkowska, E. Flies as a potential vector of selected alert pathogens in a hospital environment. Int. J. Environ. Health Res. 2022, 32, 1868–1887. [Google Scholar] [CrossRef] [PubMed]
- Karungamye, P.; Rugaika, A.; Mtei, K.; Machunda, R. Antibiotic resistance patterns of Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa isolated from hospital wastewater. Appl. Microbiol. 2023, 3, 867–882. [Google Scholar] [CrossRef]
- Nakamura-Silva, R.; de Sousa, R.C.; Fujimoto, R.Y.; Pitondo-Silva, A. Sewage from a secondary hospital in Ribeirão Preto, southeastern Brazil: A source of multidrug-resistant Enterobacteriaceae. Environ. Monit. Assess. 2023, 195, 204. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Salazar, A.; Martínez-Vázquez, A.V.; Aguilera-Arreola, G.; de Jesus de Luna-Santillana, E.; Cruz-Hernández, M.A.; Escobedo-Bonilla, C.M.; Lara-Ramírez, E.; Sánchez-Sánchez, M.; Guerrero, A.; Rivera, G. Prevalence of ESKAPE Bacteria in Surface Water and Wastewater Sources: Multidrug Resistance and Molecular Characterization, an Updated Review. Water 2023, 15, 3200. [Google Scholar] [CrossRef]
- Addae-Nuku, D.S.; Kotey, F.C.; Dayie, N.T.; Osei, M.-M.; Tette, E.M.; Debrah, P.; Donkor, E.S. Multidrug-resistant bacteria in hospital wastewater of the Korle Bu Teaching Hospital in Accra, Ghana. Environ. Health Insights 2022, 16, 11786302221130613. [Google Scholar] [CrossRef] [PubMed]
- Seguni, N.Z.; Kimera, Z.I.; Msafiri, F.; Mgaya, F.X.; Joachim, A.; Mwingwa, A.; Matee, M.I. Multidrug-resistant Escherichia coli and Klebsiella pneumoniae isolated from hospital sewage flowing through community sewage system and discharging into the Indian Ocean. Bull. Natl. Res. Cent. 2023, 47, 66. [Google Scholar] [CrossRef]
- Hamerlinck, H.; Aerssens, A.; Boelens, J.; Dehaene, A.; McMahon, M.; Messiaen, A.-S.; Vandendriessche, S.; Velghe, A.; Leroux-Roels, I.; Verhasselt, B. Sanitary installations and wastewater plumbing as reservoir for the long-term circulation and transmission of carbapenemase producing Citrobacter freundii clones in a hospital setting. Antimicrob. Resist. Infect. Control 2023, 12, 58. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.-H.; Kelly, P.J.; Wang, C. Flies as vectors and potential sentinels for bacterial pathogens and antimicrobial resistance: A review. Vet. Sci. 2022, 9, 300. [Google Scholar] [CrossRef] [PubMed]
- dos Santos Alves, T.; Lara, G.H.B.; Maluta, R.P.; Ribeiro, M.G.; da Silva Leite, D. Carrier flies of multidrug-resistant Escherichia coli as potential dissemination agent in dairy farm environment. Sci. Total Environ. 2018, 633, 1345–1351. [Google Scholar] [CrossRef] [PubMed]
- Carramaschi, I.N.; Lopes, J.C.O.; Leite, J.A.; Carneiro, M.T.; Barbosa, R.R.; Boas, M.H.V.; Rangel, K.; Chagas, T.P.G.; Queiroz, M.M.; Zahner, V. Surveillance of antimicrobial resistant bacteria in flies (Diptera) in Rio de Janeiro city. Acta Trop. 2021, 220, 105962. [Google Scholar] [CrossRef] [PubMed]
- Poudel, A.; Hathcock, T.; Butaye, P.; Kang, Y.; Price, S.; Macklin, K.; Walz, P.; Cattley, R.; Kalalah, A.; Adekanmbi, F. Multidrug-resistant Escherichia coli, Klebsiella pneumoniae and Staphylococcus spp. in houseflies and blowflies from farms and their environmental settings. Int. J. Environ. Res. Public Health 2019, 16, 3583. [Google Scholar] [CrossRef] [PubMed]
- Freitas, A.R.; Werner, G. Nosocomial pathogens and antimicrobial resistance: Modern challenges and future opportunities. Microorganisms 2023, 11, 1685. [Google Scholar] [CrossRef] [PubMed]
- Saggers, R.T.; Mothibi, L.M.; Irwin, A.D.; Naidoo, K.D. Challenges Facing PICUs in Low-and Middle-Income Countries in the Treatment of Emerging Multidrug-Resistant Organisms: A Review and Perspective from a South African PICU. Curr. Infect. Dis. Rep. 2023, 25, 233–242. [Google Scholar] [CrossRef]
- Mohd, A.B.; Huneiti, N.; Hasan, H.; Mohd, O.B.; Khaity, A.; Albakri, K. Carbapenem-resistance worldwide: A call for action–correspondence. Ann. Med. Surg. 2023, 85, 564. [Google Scholar] [CrossRef] [PubMed]
- Khalid, S.; Ahmad, N.; Ali, S.M.; Khan, A.U. Outbreak of efficiently transferred carbapenem-resistant bla NDM-producing gram-negative bacilli isolated from neonatal intensive care unit of an Indian hospital. Microb. Drug Resist. 2020, 26, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Tanner, W.D.; Atkinson, R.M.; Goel, R.K.; Toleman, M.A.; Benson, L.S.; Porucznik, C.A.; VanDerslice, J.A. Horizontal transfer of the bla NDM-1 gene to Pseudomonas aeruginosa and Acinetobacter baumannii in biofilms. FEMS Microbiol. Lett. 2017, 364, fnx048. [Google Scholar] [CrossRef]
- Manohar, P.; Shanthini, T.; Bozdogan, B.; Lundborg, C.S.; Tamhankar, A.J.; Palaniyar, N.; Ramesh, N. Transfer of antibiotic resistance genes from gram-positive bacterium to gram-negative bacterium. bioRxiv 2020. [CrossRef]
- ISO 14698-1; Cleanrooms and Associated Controlled Environments–Biocontamination Control, Part 1: General Principles and Methods. International Organization for Standardization: Geneva Switzerland, 2003.
- Schaumburg, F.; Onwugamba, F.C.; Akulenko, R.; Peters, G.; Mellmann, A.; Köck, R.; Becker, K. A geospatial analysis of flies and the spread of antimicrobial resistant bacteria. Int. J. Med. Microbiol. 2016, 306, 566–571. [Google Scholar] [CrossRef] [PubMed]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing, Breakpoint Tables for Interpretation of MICs and Zone Diameters; European Society of Clinical Microbiology and Infectious Diseases: Basel, Switzerland, 2021. [Google Scholar]
- Gupta, V.; Singla, N.; Chander, J. Detection of ESBLs using third & fourth generation cephalosporins in double disc synergy test. Indian J. Med. Res. 2007, 126, 486–487. [Google Scholar] [PubMed]
- Check-MDR CT103XL; Guideline, CpU. Molecular Detection and Identification of Carbapenemase, MCR 1-2, AmpC and ESBL Genes; Version 1.1. 2017. Available online: https://check-pointshealth.com/wp-content/uploads/2018/11/Check-MDR_CT103XL_IFU_10-0023_EN-v1.1-20170918.pdf (accessed on 11 February 2024).
| Bacteria | Housefly | Surface Swabs | Sewage | Total | ||||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | |
| E. coli | 32 | 29.6 | 27 | 25.0 | 49 | 45.4 | 108 | 30.4 |
| Klebsiella species | 24 | 54.5 | 2 | 4.5 | 18 | 40.9 | 44 | 12.4 |
| Providencia species | 44 | 100 | - | - | - | - | 44 | 12.4 |
| Proteus species | 40 | 95.2 | 2 | 4.8 | - | - | 42 | 11.8 |
| Enterobacter species | 16 | 69.6 | 1 | 4.3 | 6 | 26.1 | 23 | 6.5 |
| Acinetobacter species | 6 | 31.6 | 4 | 21.1 | 9 | 47.4 | 19 | 5.4 |
| M. morganii | 14 | 100 | - | - | - | - | 14 | 3.9 |
| Aeromonas species | 1 | NA | 7 | NA | 2 | NA | 10 | 2.8 |
| Kluyvera species | 7 | NA | - | - | 2 | NA | 9 | 2.5 |
| R. ornithinolytica | 7 | NA | - | - | 2 | NA | 9 | 2.5 |
| W. chitiniclastica | 9 | NA | - | - | - | - | 9 | 2.5 |
| C. freundii | 5 | NA | - | - | - | - | 5 | 1.4 |
| Pantoea species | 2 | NA | - | - | 1 | NA | 3 | 0.8 |
| P. gergoviae | 1 | NA | - | - | 2 | NA | 3 | 0.8 |
| E. hermannii | 1 | NA | - | - | 1 | NA | 2 | 0.6 |
| L. adecarboxylata | 1 | NA | - | - | 1 | NA | 2 | 0.6 |
| C. sakazakii | - | - | - | - | 1 | NA | 1 | 0.3 |
| E. fergusonii | - | - | 1 | NA | - | - | 1 | 0.3 |
| Hafnia alvei | 1 | NA | - | - | - | - | 1 | 0.3 |
| I. indica | 1 | NA | - | - | - | - | 1 | 0.3 |
| M. wisconsensis | 1 | NA | - | - | - | - | 1 | 0.3 |
| P. carotovorum | 1 | NA | - | - | - | - | 1 | 0.3 |
| P. putida | 1 | NA | - | - | - | - | 1 | 0.3 |
| Salmonella species | - | - | 1 | NA | - | - | 1 | 0.3 |
| S. maltophilia | 1 | NA | - | - | - | - | 1 | 0.3 |
| Total | 216 | 60.8 | 45 | 12.7 | 94 | 26.5 | 355 | 100 |
| Antibiotics | E. coli | Klebsiella spp. | Providencia spp. | Proteus spp. | Enterobacter spp. | Acinetobacter spp. | Ohers | Total |
|---|---|---|---|---|---|---|---|---|
| AMP | 73.1 | 100 | 86.4 | 81.0 | 95.7 | 100 | 91.3 | 61.0 |
| PIP | 65.7 | 100 | 61.4 | 59.5 | 60.9 | IE | 59.6 | 42.2 |
| AMC | 38 | 43.2 | 100 | 21.4 | 91.3 | 100 | 64.3 | 44.0 |
| TZP | 19.4 | 31.8 | 9.1 | 0 | 17.4 | IE | 17.0 | 8.6 |
| CXM | 100 | 100 | - | 100 | - | 100 | 100 | 100 |
| CTX | 34.3 | 50.0 | 65.9 | 64.3 | 69.6 | 100 | 44.6 | 41.1 |
| CAZ | 31.5 | 47.7 | 59.1 | 26.2 | 56.5 | - | 40.4 | 27.9 |
| FEP | 34.3 | 40.9 | 40.9 | 64.3 | 65.2 | - | 42.1 | 30.3 |
| FOX | 13.9 | 13.6 | 15.9 | 0 | 100 | - | 46.4 | 18.5 |
| MEM | 24.1 | 2.3 | 0 | 0 | 4.3 | 31.6 | 4.3 | 3.1 |
| MXF | 35.2 | 38.6 | 61.4 | 76.2 | 56.5 | - | 52.2 | 34.7 |
| CIP | 30.6 | 40.9 | 47.7 | 66.7 | 30.4 | 100 | 37.5 | 33.9 |
| TM | 20.4 | 31.8 | 34.1 | 66.7 | 47.8 | 47.4 | 42.6 | 29.7 |
| GM | 16.7 | 31.8 | 36.4 | 66.7 | 65.2 | 52.6 | 32.6 | 30.1 |
| AN | 1.9 | 2.3 | 0 | 4.8 | 8.7 | 5.3 | 8.5 | 3.1 |
| SXT | 43.5 | 47.7 | 61.4 | 71.4 | 56.5 | 57.9 | 50.0 | 38.7 |
| ESBL | 38.9 | 43.2 | 61.4 | 64.3 | 52.2 | 68.4 | 37.5 | 49.2 |
| Types of Antimicrobial Resistance Gene | Surface Swab (n = 10) | Housefly (n = 20) | Sewage (n = 36) | Total | |
|---|---|---|---|---|---|
| (n = 66) | % | ||||
| Carbapenemase encoding genes | 1 | 5 | 0 | 6 | 5.6 |
| NDM | 1 | 5 | 0 | 6 | 5.6 |
| ESBL encoding genes | 8 | 18 | 15 | 41 | 37.9 |
| CTX-M group 1 type-15 | 6 | 15 | 11 | 33 | 30.6 |
| CTX-M group 1 type-9 | 2 | 0 | 2 | 3 | 2.7 |
| CTX-M group 1, ND * | 0 | 1 | 2 | 3 | 2.8 |
| CTX-M group 1 type-15 + 9 | 0 | 2 | 0 | 2 | 1.8 |
| AMPC encoding genes | 3 | 2 | 1 | 6 | 5.6 |
| CMY II (n = 11) | 0 | 1 | 0 | 1 | 0.9 |
| ACT/MIR (n = 10) | 3 | 0 | 0 | 3 | 2.8 |
| DHA (n = 5) | 0 | 1 | 1 | 2 | 1.9 |
| TEM/SHV encoding genes | 3 | 11 | 23 | 37 | 34.3 |
| blaTEM- (WT) (n = 144) | 3 | 11 | 22 | 36 | 33.4 |
| blaTEM-104K + 164C (n = 1) | 0 | 0 | 1 | 1 | 0.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gashaw, M.; Gudina, E.K.; Tadesse, W.; Froeschl, G.; Ali, S.; Seeholzer, T.; Kroidl, A.; Wieser, A. Hospital Wastes as Potential Sources for Multi-Drug-Resistant ESBL-Producing Bacteria at a Tertiary Hospital in Ethiopia. Antibiotics 2024, 13, 374. https://doi.org/10.3390/antibiotics13040374
Gashaw M, Gudina EK, Tadesse W, Froeschl G, Ali S, Seeholzer T, Kroidl A, Wieser A. Hospital Wastes as Potential Sources for Multi-Drug-Resistant ESBL-Producing Bacteria at a Tertiary Hospital in Ethiopia. Antibiotics. 2024; 13(4):374. https://doi.org/10.3390/antibiotics13040374
Chicago/Turabian StyleGashaw, Mulatu, Esayas Kebede Gudina, Wondwossen Tadesse, Guenter Froeschl, Solomon Ali, Thomas Seeholzer, Arne Kroidl, and Andreas Wieser. 2024. "Hospital Wastes as Potential Sources for Multi-Drug-Resistant ESBL-Producing Bacteria at a Tertiary Hospital in Ethiopia" Antibiotics 13, no. 4: 374. https://doi.org/10.3390/antibiotics13040374
APA StyleGashaw, M., Gudina, E. K., Tadesse, W., Froeschl, G., Ali, S., Seeholzer, T., Kroidl, A., & Wieser, A. (2024). Hospital Wastes as Potential Sources for Multi-Drug-Resistant ESBL-Producing Bacteria at a Tertiary Hospital in Ethiopia. Antibiotics, 13(4), 374. https://doi.org/10.3390/antibiotics13040374






