In Vitro/Vivo Mechanisms of Antibacterial Peptide NZ2114 against Staphylococcus pseudintermedius and Its Biofilms
Abstract
1. Introduction
2. Results
2.1. In Vitro Antibacterial Assay
2.1.1. Minimal Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) Determination
2.1.2. Dose-Killing Curve Assays
2.1.3. Bactericidal Effect Observation by Fluorescence Microscope
2.2. Antibacterial Mechanism of NZ2114
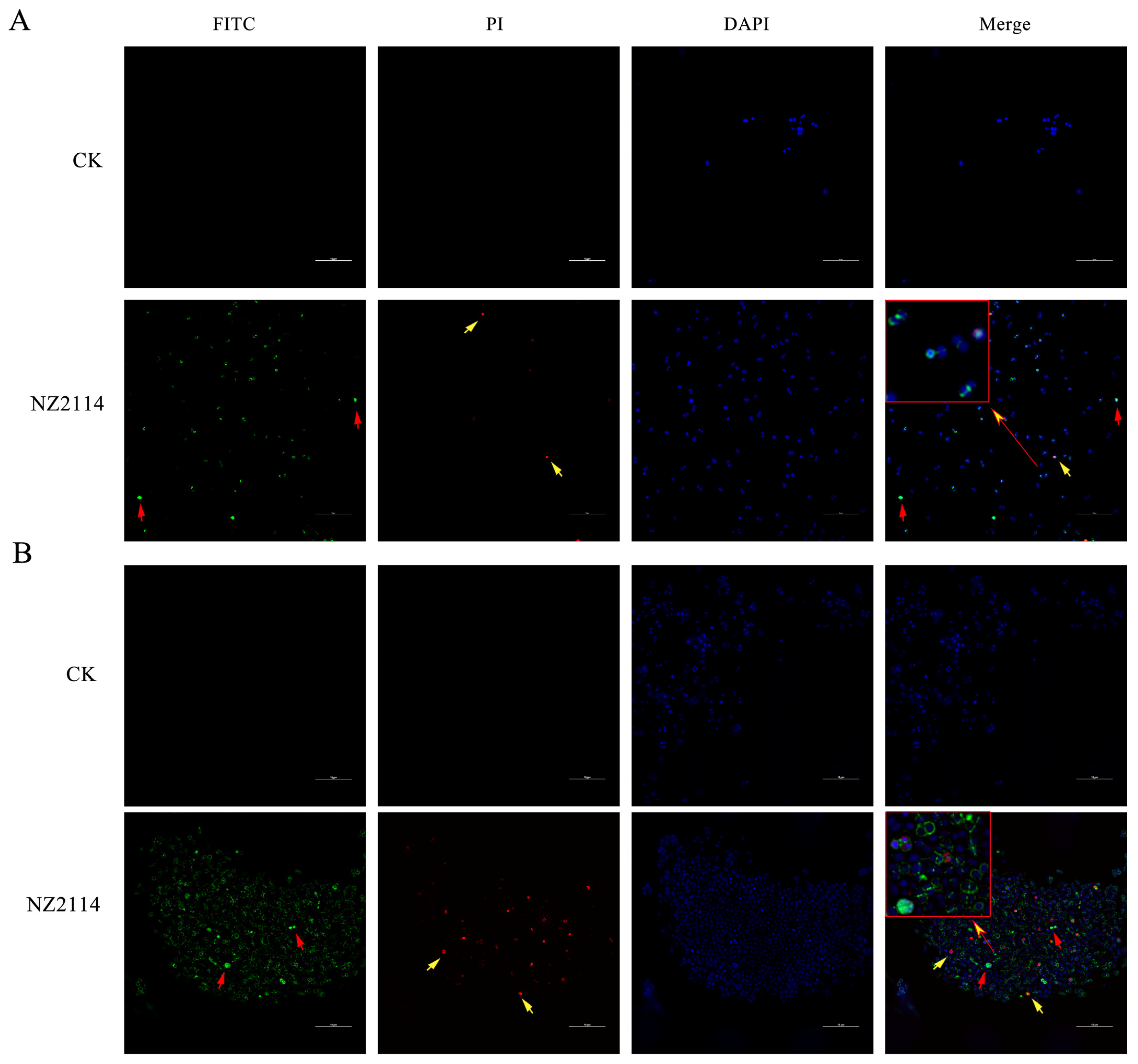
2.2.1. Effect of NZ2114 on Membrane Morphology and Cell Ultrastructure
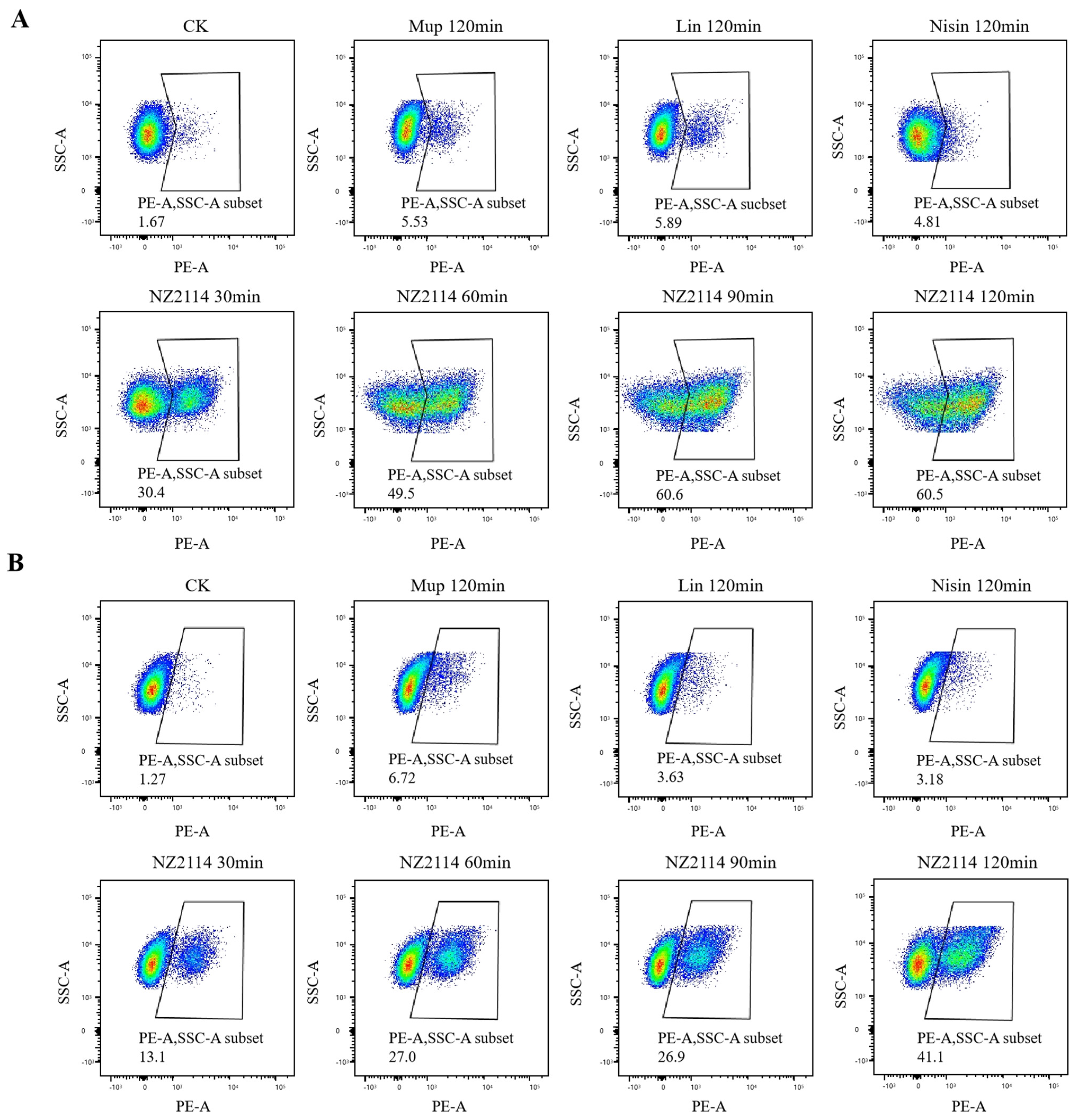
2.2.2. Membrane Integrity Analysis
2.2.3. Super-Resolution Microscopy (SRM) Observation
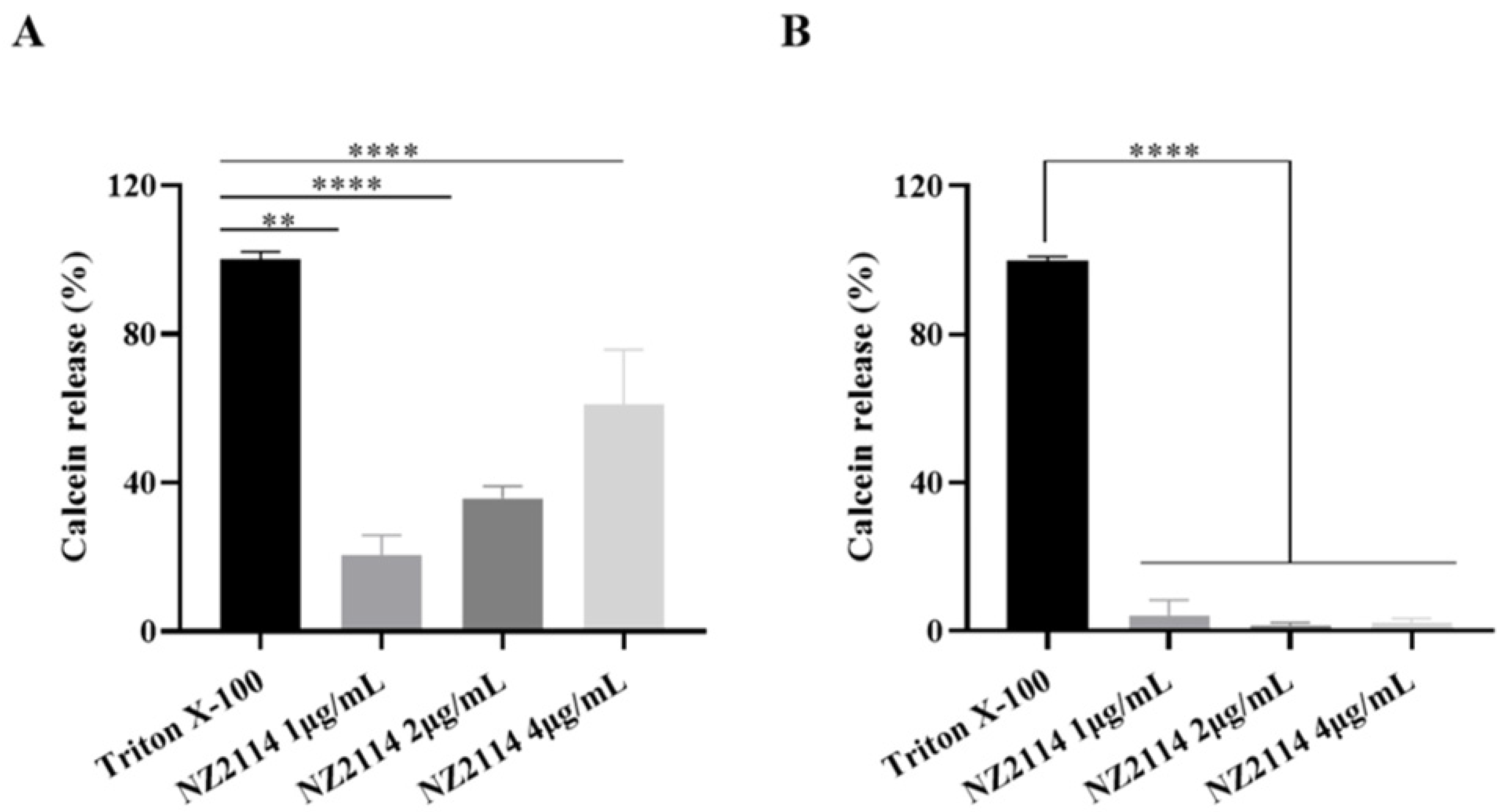
2.2.4. Calcein Leakage Assay
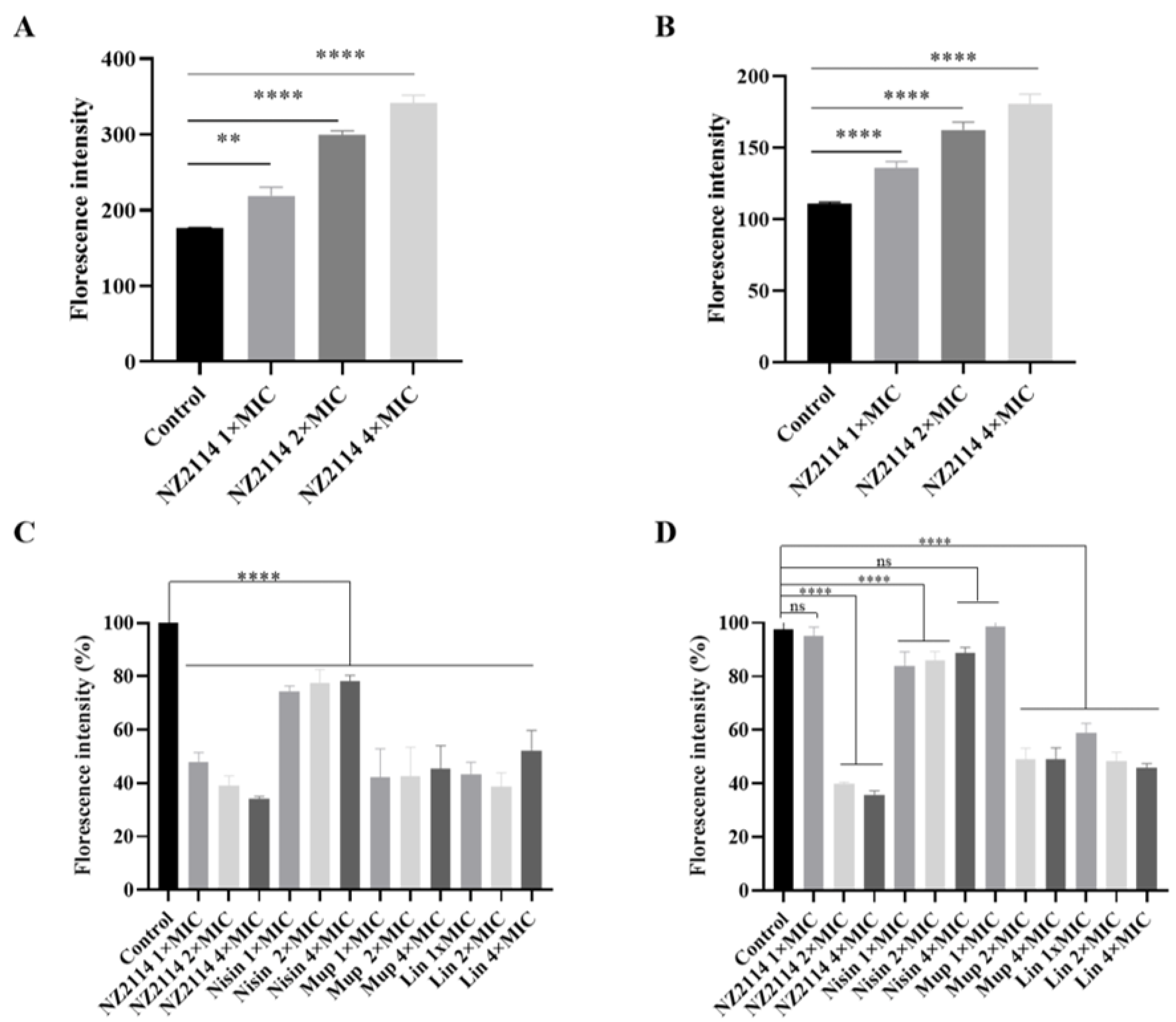
2.2.5. Fluorescence Detection of Intracellular ROS Activity
2.2.6. Alamar Blue Detection of Cell Metabolic Activity
2.3. Effects of NZ2114 on Biofilm
2.3.1. Inhibitory Effects of NZ2114 on Biofilm Formation
2.3.2. Biofilm Observation by Confocal Laser Scanning Microscopy (CLSM)
2.4. Efficacy of NZ2114 in Mice
3. Discussion
4. Materials and Methods
4.1. Strains, Mice, and Reagents
4.2. In Vitro Antibacterial Assay
4.2.1. Determination of Antimicrobial Activity
4.2.2. Dose-Killing Curve Assays
4.2.3. Bactericidal Effect Observation by Fluorescence Microscope
4.3. Antibacterial Mechanism of NZ2114
4.3.1. Electron Microscopy Observation
4.3.2. Membrane Integrity Analysis by Flow Cytometry
4.3.3. Super-Resolution Microscopy (SRM) Observation
4.3.4. Preparation of Lipids
4.3.5. Calcein Leakage Assay
4.3.6. Fluorescence Detection of Intracellular ROS Activity
4.3.7. Alamar Blue Detection Cell Metabolic Activity
4.4. Effect of NZ2114 on Biofilm
4.4.1. Effect of NZ2114 on Inhibit Biofilm Formation
4.4.2. Biofilm Observation by CLSM
4.5. Mouse In Vivo Test
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhooshan, S.; Negi, V.; Khatri, P.K. Staphylococcus pseudintermedius: An undocumented, emerging pathogen in humans. GMS Hyg. Infect. Control 2020, 15, Doc32. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.E.; Chirino-Trejo, M. Prevalence, sites of colonization, and antimicrobial resistance among Staphylococcus pseudintermedius isolated from healthy dogs in Saskatoon. Can. J. Vet. Diagn. Investig. 2011, 23, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Gharajalar, S.N.; Tanhaee, S.; Omidzadeh, M.; Onsori, M. Detection of antimicrobial resistance and biofilm production among Staphylococcus pseudintermedius from canine skin lesions. Microb. Drug Resist. 2023. [Google Scholar] [CrossRef] [PubMed]
- Bourguignon, E.; Viçosa, G.N.; Corsini, C.M.M.; Moreira, M.A.S.; Nero, L.A.; Conceição, L.G. Description of Methicillin-resistant Staphylococcus pseudintermedius from canine pyoderma in Minas Gerais state, Brazil. Arq. Bras. Med. Vet. Zootec. 2016, 68, 299–306. [Google Scholar] [CrossRef]
- Srednik, M.E.; Perea, C.A.; Giacoboni, G.I.; Hicks, J.A.; Foxx, C.L.; Harris, B.; Schlater, L.K. Genomic features of antimicrobial resistance in Staphylococcus pseudintermedius isolated from dogs with pyoderma in Argentina and the United States: A comparative study. Int. J. Mol. Sci. 2023, 24, 11361. [Google Scholar] [CrossRef]
- Marchegiani, A.; Fruganti, A.; Bazzano, M.; Cerquetella, M.; Dini, F.; Spaterna, A. Fluorescent light energy in the management of multi drug resistant canine pyoderma: A prospective exploratory study. Pathogens 2022, 11, 1197. [Google Scholar] [CrossRef]
- Pesset, C.M.; Fonseca, C.O.D.; Antunes, M.; Santos, A.L.L.D.; Teixeira, I.M.; Ribeiro, T.A.N.; Sachs, D.; Penna, B. Characterizing biofilm formation of Staphylococcus pseudintermedius in different suture materials. Microb. Pathog. 2022, 172, 105796. [Google Scholar] [CrossRef]
- Summers, J.F.; Hendricks, A.; Brodbelt, D.C. Prescribing practices of primary-care veterinary practitioners in dogs diagnosed with bacterial pyoderma. BMC Vet. Res. 2014, 10, 240. [Google Scholar] [CrossRef]
- Loeffler, A.; Lloyd, D.H. What has changed in canine pyoderma? A narrative review. Vet. J. 2018, 235, 73–82. [Google Scholar] [CrossRef]
- Yang, L.; Tian, Z.; Zhou, L.; Zhu, L.; Sun, C.; Huang, M.; Peng, J.; Guo, G. In vitro antifungal activity of a novel antimicrobial peptide AMP-17 against planktonic cells and biofilms of Cryptococcus neoformans. Infect. Drug Resist. 2022, 15, 233–248. [Google Scholar] [CrossRef]
- Sabokkhiz, M.A.; Tanhaeian, A.; Mamarabadi, M. Study on antiviral activity of two recombinant antimicrobial peptides against Tobacco Mosaic Virus. Probiotics Antimicrob. Proteins 2019, 11, 1370–1378. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.K.H.M.; Jayathilaka, E.H.T.T.; Edirisinghe, S.L.; Lim, J.W.; Nikapitiya, C.; Kang, S.Y.; Whang, I.; De Zoysa, M. In-vitro immunomodulatory responses and antiviral activities of antimicrobial peptide octominin against fish pathogenic viruses. Fish Shellfish Immunol. 2023, 142, 109129. [Google Scholar] [CrossRef] [PubMed]
- de Moura, G.A.; de Oliveira, J.R.; Rocha, Y.M.; de Oliveira Freitas, J.; Rodrigues, J.P.V.; Ferreira, V.P.G.; Nicolete, R. Antitumor and antiparasitic activity of antimicrobial peptides derived from snake venom: A systematic review approach. Curr. Med. Chem. 2022, 29, 5358–5368. [Google Scholar] [CrossRef] [PubMed]
- Duarte-Mata, D.I.; Salinas-Carmona, M.C. Antimicrobial peptides’ immune modulation role in intracellular bacterial infection. Front. Immunol. 2023, 14, 1119574. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Zhang, Q.; Mao, R.; Hao, Y.; Ma, X.; Teng, D.; Fan, H.; Wang, J. Effect of NZ2114 against Streptococcus dysgalactiae biofilms and its application in murine mastitis model. Front. Microbiol. 2022, 13, 1010148. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Wang, J.; de la Fuente-Nunez, C.; Franco, O.L. Editorial: Antimicrobial peptides: Molecular design, structure-function relationship, and biosynthesis optimization. Front. Microbiol. 2022, 13, 888540. [Google Scholar] [CrossRef]
- Zhang, Y.; Teng, D.; Mao, R.; Wang, X.; Xi, D.; Hu, X.; Wang, J. High expression of a plectasin-derived peptide NZ2114 in Pichia pastoris and its pharmacodynamics, postantibiotic and synergy against Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2014, 98, 681–694. [Google Scholar] [CrossRef]
- Klein, K.; Grønnemose, R.B.; Alm, M.; Brinch, K.S.; Kolmos, H.J.; Andersen, T.E. Controlled release of plectasin NZ2114 from a hybrid silicone-hydrogel material for inhibition of Staphylococcus aureus biofilm. Antimicrob. Agents Chemother. 2017, 61, e00604-17. [Google Scholar] [CrossRef]
- Yang, N.; Huang, Y.; Li, Y.; Teng, D.; Mao, R.; Hao, Y.; Wei, L.; Wang, J. Efficiency of NZ2114 on superficial pyoderma infected with Staphylococcus pseudintermedius. Pharmaceuticals 2024, 17, 277. [Google Scholar] [CrossRef]
- Jarosiewicz, M.; Garbacz, K.; Neubauer, D.; Kamysz, W. In vitro efficiency of antimicrobial peptides against Staphylococcal pathogens associated with canine pyoderma. Animals 2020, 10, 470. [Google Scholar] [CrossRef]
- Zhao, F.; Yang, N.; Wang, X.; Mao, R.; Hao, Y.; Li, Z.; Wang, X.; Teng, D.; Fan, H.; Wang, J. In vitro/vivo mechanism of action of MP1102 with low/nonresistance against Streptococcus suis type 2 strain CVCC 3928. Front. Cell Infect. Microbiol. 2019, 9, 48. [Google Scholar] [CrossRef]
- Zheng, X.; Teng, D.; Mao, R.; Hao, Y.; Yang, N.; Hu, F.; Wang, J. A study on fungal defensin against multi-drug-resistant Clostridium perfringens and its treatment on infected poultry. Appl. Microbiol. Biotechnol. 2021, 105, 7265–7282. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.H.; Wang, C.H.; Liao, Y.T.; Chan, F.Y.; Kanaoka, Y.; Uchihashi, T.; Kato, K.; Lai, L.; Chang, Y.W.; Ho, M.C.; et al. Visualizing the membrane disruption action of antimicrobial peptides by cryo-electron tomography. Nat. Commun. 2023, 14, 5464. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.J.; Nam, S.H.; Lee, B.J. Engineering approaches for the development of antimicrobial peptide-based antibiotics. Antibiotics 2022, 11, 1338. [Google Scholar] [CrossRef] [PubMed]
- Cassone, M.; Otvos, L., Jr. Synergy among antibacterial peptides and between peptides and small-molecule antibiotics. Expert. Rev. Anti-Infect. Ther. 2010, 8, 703–716. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, P.; Ismail, B.B.; Yang, Z.; Zou, Z.; Suo, Y.; Ye, X.; Liu, D.; Guo, M. Chickpea-derived modified antimicrobial peptides KTA and KTR inactivate Staphylococcus aureus via disrupting cell membrane and interfering with peptidoglycan synthesis. J. Agric. Food Chem. 2024, 72, 2727–2740. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.H.; Zeng, P.; Liu, J.; Wong, K.Y.; Chan, E.W.C.; Lin, Y.B.; Chan, K.F.; Chen, S. Antimicrobial peptide zp37 inhibits Escherichia coli O157:H7 in alfalfa sprouts by inflicting damage in cell membrane and binding to DNA. LWT-Food Sci. 2021, 146, 111392. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apid-ianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Schäfer, A.B.; Wenzel, M. A how-to guide for mode of action analysis of antimicrobial peptides. Front. Cell Infect. Microbiol. 2020, 10, 540898. [Google Scholar] [CrossRef]
- Fang, Y.; Yang, G.; Wu, X.; Qin, B.; Xie, Y.; Zhuang, L. Sub-MIC antibiotics affect microbial ferrihydrite reduction by extracellular membrane vesicles. J. Hazard Mater. 2023, 458, 131876. [Google Scholar] [CrossRef]
- Maredia, R.; Devineni, N.; Lentz, P.; Dallo, S.F.; Yu, J.; Guentzel, N.; Chambers, J.; Arulanandam, B.; Haskins, W.E.; Weitao, T. Vesiculation from Pseudomonas aeruginosa under SOS. Sci. World J. 2012, 2012, 402919. [Google Scholar] [CrossRef]
- Andreoni, F.; Toyofuku, M.; Menzi, C.; Kalawong, R.; Shambat, S.M.; François, P.; Zinkernagel, A.S.; Eberl, L. Antibiotics stimulate formation of vesicles in Staphylococcus aureus in both phage-dependent and -independent fashions and via different routes. Antimicrob. Agents Chemother. 2019, 63, e01439-18. [Google Scholar] [CrossRef]
- Omardien, S.; Drijfhout, J.W.; van Veen, H.; Schachtschabel, S.; Riool, M.; Hamoen, L.W.; Brul, S.; Zaat, S.A.J. Synthetic antimicrobial peptides delocalize membrane bound proteins thereby inducing a cell envelope stress response. Biochim. Biophys. Acta Biomembr. 2018, 1860, 2416–2427. [Google Scholar] [CrossRef]
- Medeiros-Silva, J.; Jekhmane, S.; Breukink, E.; Weingarth, M. Towards the native binding modes of antibiotics that target lipid II. ChemBioChem 2019, 20, 1731–1738. [Google Scholar] [CrossRef]
- Su, M.; Liu, F.; Luo, Z.; Wu, H.; Zhang, X.; Wang, D.; Zhu, Y.; Sun, Z.; Xu, W.; Miao, Y. The antibacterial activity and mechanism of chlorogenic acid against foodborne pathogen Pseudomonas aeruginosa. Foodborne Pathog. Dis. 2019, 16, 823–830. [Google Scholar] [CrossRef]
- Seyedjavadi, S.S.; Khani, S.; Eslamifar, A.; Ajdary, S.; Goudarzi, M.; Halabian, R.; Akbari, R.; Zare-Zardini, H.; Fooladi, A.A.I.; Amani, J.; et al. The antifungal peptide MCh-AMP1 derived from Matricaria chamomilla inhibits Candida albicans growth via inducing ROS generation and altering fungal cell membrane permeability. Front. Microbial. 2020, 10, 3150. [Google Scholar] [CrossRef]
- Oyinloye, B.E.; Adenowo, A.F.; Kappo, A.P. Reactive oxygen species, apoptosis, antimicrobial peptides and human inflammatory diseases. Pharmaceuticals 2015, 8, 151–175. [Google Scholar] [CrossRef]
- Singh, B.P.; Ghosh, S.; Chauhan, A. Development, dynamics and control of antimicrobial-resistant bacterial biofilms: A review. Chem. Lett. 2021, 19, 1983–1993. [Google Scholar] [CrossRef]
- Rajput, A.; Bhamare, K.T.; Thakur, A.; Kumar, M. Anti-biofilm: Machine learning assisted prediction of IC50 activity of chemicals against biofilms of microbes causing antimicrobial resistance and implications in drug repurposing. J. Mol. Biol. 2023, 435, 168115. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Tochio, T.; Nishifuji, K. Erythritol alters gene transcriptome signatures, cell growth, and biofilm formation in Staphylococcus pseudintermedius. BMC Vet. Res. 2023, 19, 146. [Google Scholar] [CrossRef] [PubMed]
- Jantorn, P.; Tipmanee, V.; Wanna, W.; Prapasarakul, N.; Visutthi, M.; Sotthibandhu, D.S. Potential natural antimicrobial and antibiofilm properties of Piper betle L. against Staphylococcus pseudintermedius and methicillin-resistant strains. J. Ethnopharmacol. 2023, 317, 116820. [Google Scholar] [CrossRef] [PubMed]
- Parducho, K.R.; Beadell, B.; Ybarra, T.K.; Bush, M.; Escalera, E.; Trejos, A.T.; Chieng, A.; Mendez, M.; Anderson, C.; Park, H.; et al. The antimicrobial peptide human beta-defensin 2 inhibits biofilm production of Pseudomonas aeruginosa without compromising metabolic activity. Front. Immunol. 2022, 11, 805. [Google Scholar] [CrossRef]
- Rajapaksha, D.C.; Edirisinghe, S.L.; Nikapitiya, C.; Whang, I.; De Zoysa, M. The antimicrobial peptide octopromycin suppresses biofilm formation and quorum sensing in Acinetobacter baumannii. Antibiotics 2023, 12, 623. [Google Scholar] [CrossRef]
- Arima, S.; Ochi, H.; Mitsuhashi, M.; Kibe, R.; Takahashi, K.; Kataoka, Y. Staphylococcus pseudintermedius biofilms secrete factors that induce inflammatory reactions in vitro. Lett. Appl. Microbiol. 2018, 67, 214–219. [Google Scholar] [CrossRef]
- Karched, M.; Bhardwaj, R.G.; Qudeimat, M.; Al-Khabbaz, A.; Ellepola, A. Proteomic analysis of the periodontal pathogen Prevotella intermedia secretomes in biofilm and planktonic lifestyles. Sci. Rep. 2022, 12, 5636. [Google Scholar] [CrossRef] [PubMed]
- Charafeddine, R.A.; Nosanchuk, J.D.; Sharp, D.J. Targeting microtubules for wound repair. Adv. Wound Care 2016, 5, 444–454. [Google Scholar] [CrossRef]
- Kamr, A.; Arbaga1, A.; El-Bahrawy, A.; Elsify, A.; Khaled, H.; Hassan, H. The therapeutic efficacy of Aloe vera gel ointment on staphylococcal pyoderma in dogs. Vet. World 2020, 13, 2371–2380. [Google Scholar] [CrossRef]
- Xie, T.; Lin, J.; Lin, D.; Zhang, D.; Xu, X.; Zhu, N.; Lin, J. In vitro and in vivo antibacterial studies of volatile oil from Atractylodis Rhizoma against Staphylococcus pseudintermedius and multidrug resistant Staphylococcus pseudintermedius strains from canine pyoderma. J. Ethnopharmacol. 2024, 319, 117326. [Google Scholar] [CrossRef] [PubMed]
- Phensri, P.; Thummasema, K.; Sukatta, U.; Morand, S.; Pruksakorn, C. In vitro antimicrobial activity of Piper betle leaf extract and some topical agents against methicillin-resistant and methicillin-susceptible Staphylococcus strains from canine pyoderma. Animals 2022, 12, 3203. [Google Scholar] [CrossRef]
- Zhang, Q.; Yang, N.; Mao, R.; Hao, Y.; Ma, X.; Teng, D.; Fan, H.; Wang, J. A recombinant fungal defensin-like peptide-P2 combats Streptococcus dysgalactiae and biofilms. Appl. Microbiol. Biotechnol. 2021, 105, 1489–1504. [Google Scholar] [CrossRef]
- Brinch, K.S.; Tulkens, P.M.; Van Bambeke, F.; Frimodt-Møller, N.; Høiby, N.; Kristensen, H.H. Intracellular activity of the peptide antibiotic NZ2114: Studies with Staphylococcus aureus and human THP-1 monocytes, and comparison with daptomycin and vancomycin. J. Antimicrob. Chemother. 2010, 65, 1720–1724. [Google Scholar] [CrossRef] [PubMed]
- Rangel, K.; Cabral, F.O.; Lechuga, G.C.; Carvalho, J.P.R.S.; Villas-Bôas, M.H.S.; Midlej, V.; De-Simone, S.G. Detrimental effect of ozone on pathogenic bacteria. Microorganisms 2021, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yang, N.; Mao, R.; Hao, Y.; Teng, D.; Wang, J. In vitro pharmacodynamics and bactericidal mechanism of fungal defensin-derived peptides NZX and P2 against Streptococcus agalactiae. Microorganisms 2022, 10, 881. [Google Scholar] [CrossRef] [PubMed]
- Shang, D.; Zhang, Q.; Dong, W.; Liang, H.; Bi, X. The effects of LPS on the activity of Trp-containing antimicrobial peptides against Gram-negative bacteria and endotoxin neutralization. Acta Biomater. 2016, 33, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Hong, W.; Zhao, Y.; Guo, Y.; Huang, C.; Qiu, P.; Zhu, J.; Chu, C.; Shi, H.; Liu, M. PEGylated self-assembled nano-bacitracin A: Probing the antibacterial mechanism and real-time tracing of target delivery in vivo. ACS Appl. Mater. Interfaces 2018, 10, 10688–10705. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Chen, C.; Wang, D.; Wang, Z.; Liu, Y. The antimicrobial peptide LI14 combats multidrug-resistant bacterial infections. Commun. Biol. 2022, 5, 926. [Google Scholar] [CrossRef]
- Lall, N.; Henley-Smith, C.J.; De Canha, M.N.; Oosthuizen, C.B.; Berrington, D. Viability reagent, PrestoBlue, in comparison with other available reagents, utilized in cytotoxicity and antimicrobial assays. Int. J. Microbiol. 2013, 2013, 420601. [Google Scholar] [CrossRef]






| Strains | NZ2114 | Mupirocin | Lincomycin | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | |||||||
| μg/mL | μM | μg/mL | μM | μg/mL | μM | μg/mL | μM | μg/mL | μM | μg/mL | μM | |
| S. pseudintermedius CGMCC 1.90024 | 1 | 0.23 | 2 | 0.46 | 0.25 | 0.5 | 16 | 32 | 32 | 69.41 | 64 | 138.83 |
| S. pseudintermedius CGMCC 1.90005 | 1 | 0.23 | 1 | 0.23 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4.34 | >256 | >555.30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Yang, N.; Mao, R.; Hao, Y.; Teng, D.; Wang, J. In Vitro/Vivo Mechanisms of Antibacterial Peptide NZ2114 against Staphylococcus pseudintermedius and Its Biofilms. Antibiotics 2024, 13, 341. https://doi.org/10.3390/antibiotics13040341
Zhang S, Yang N, Mao R, Hao Y, Teng D, Wang J. In Vitro/Vivo Mechanisms of Antibacterial Peptide NZ2114 against Staphylococcus pseudintermedius and Its Biofilms. Antibiotics. 2024; 13(4):341. https://doi.org/10.3390/antibiotics13040341
Chicago/Turabian StyleZhang, Shuang, Na Yang, Ruoyu Mao, Ya Hao, Da Teng, and Jianhua Wang. 2024. "In Vitro/Vivo Mechanisms of Antibacterial Peptide NZ2114 against Staphylococcus pseudintermedius and Its Biofilms" Antibiotics 13, no. 4: 341. https://doi.org/10.3390/antibiotics13040341
APA StyleZhang, S., Yang, N., Mao, R., Hao, Y., Teng, D., & Wang, J. (2024). In Vitro/Vivo Mechanisms of Antibacterial Peptide NZ2114 against Staphylococcus pseudintermedius and Its Biofilms. Antibiotics, 13(4), 341. https://doi.org/10.3390/antibiotics13040341









